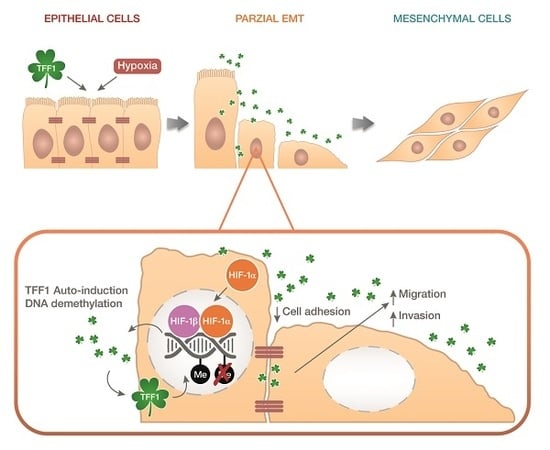
TFF1 Promotes EMT-Like Changes through an Auto-Induction Mechanism
Abstract
:1. Introduction
2. Results
2.1. TFF1 Overexpression Promotes Invasion and “EMT-Like” Molecular Changes
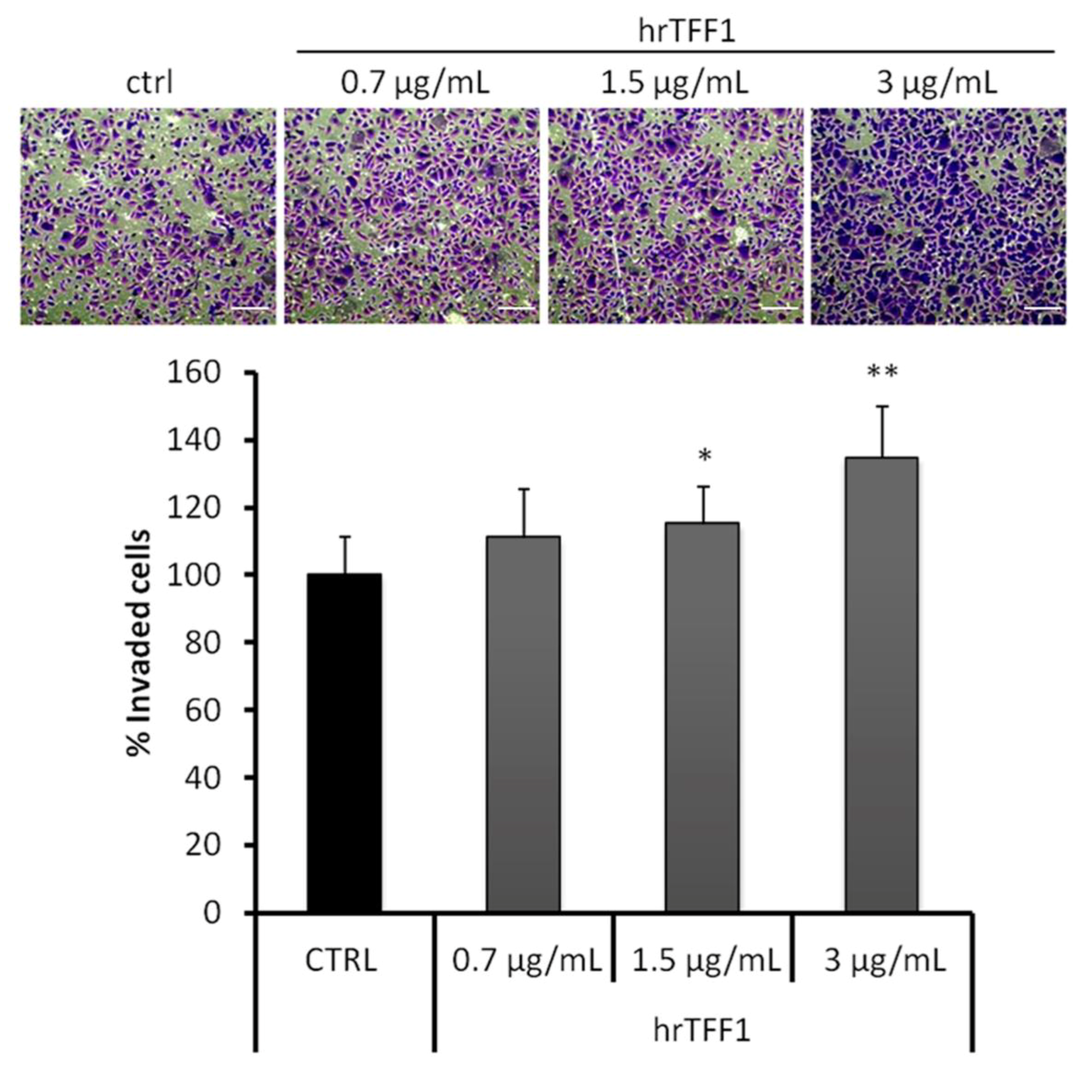
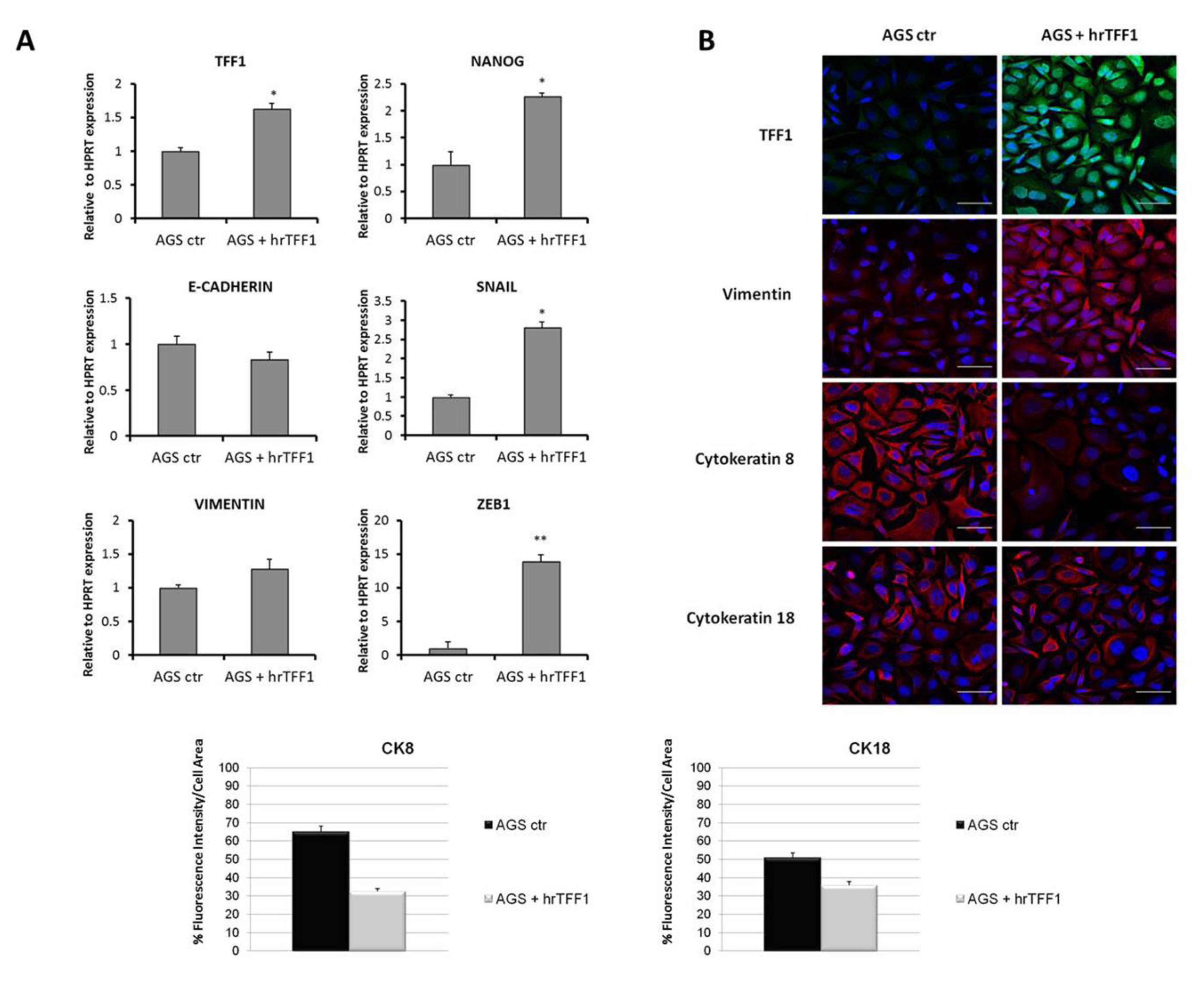
2.2. Exogenous TFF1 Protein Induces Invasion and EMT in AGS Cells
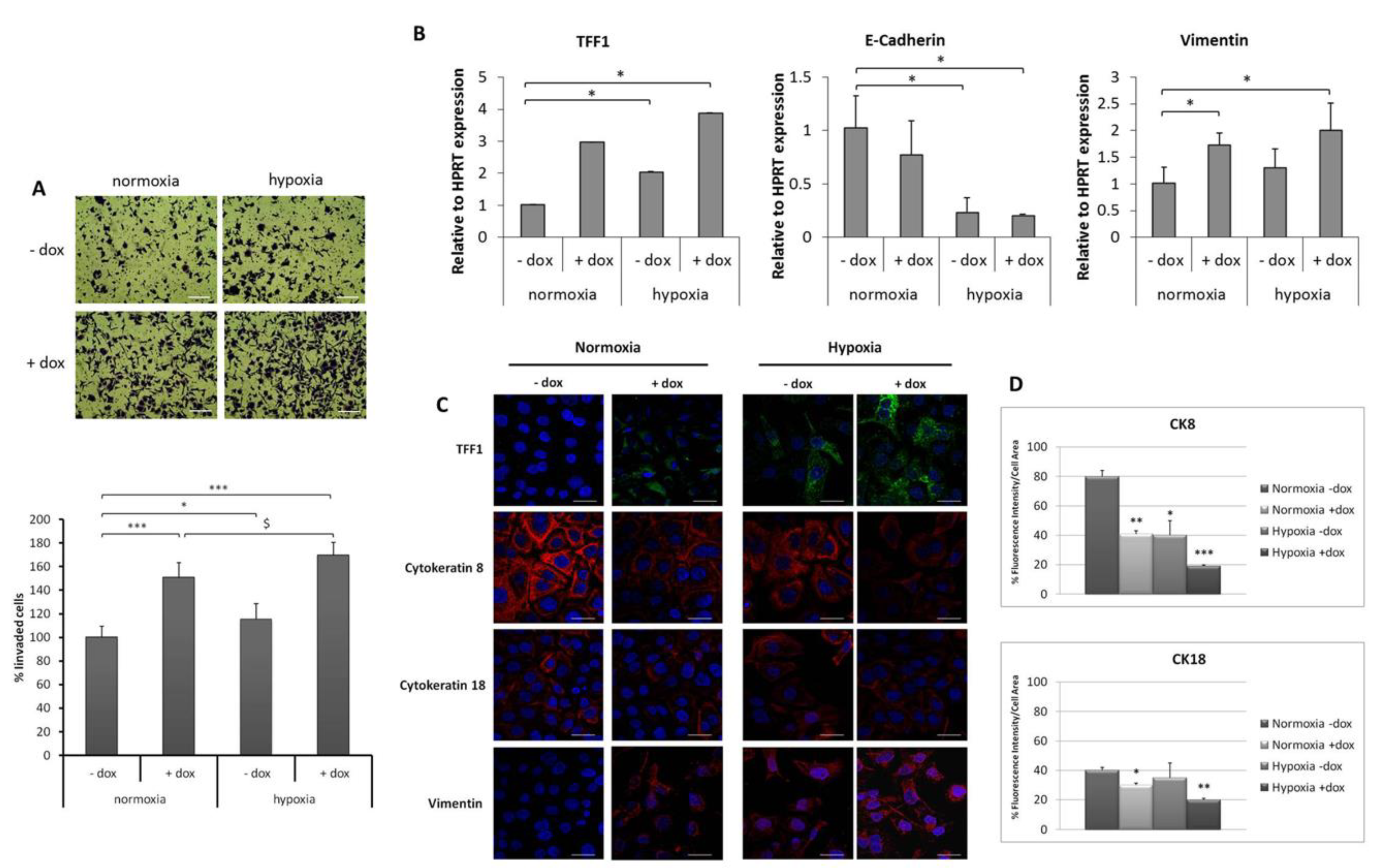
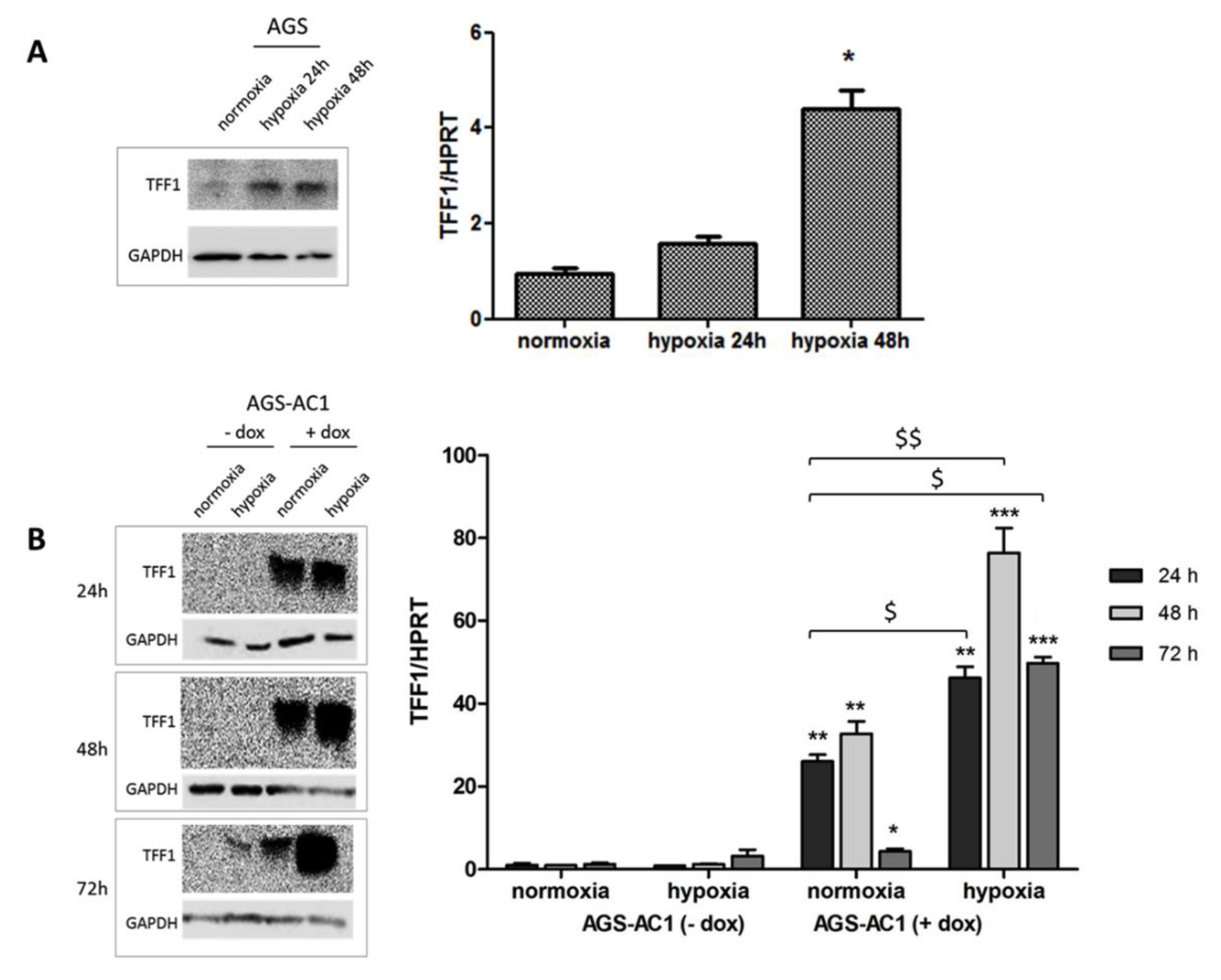
2.3. TFF1 Potentiates Cell Invasiveness and EMT Changes during Hypoxia
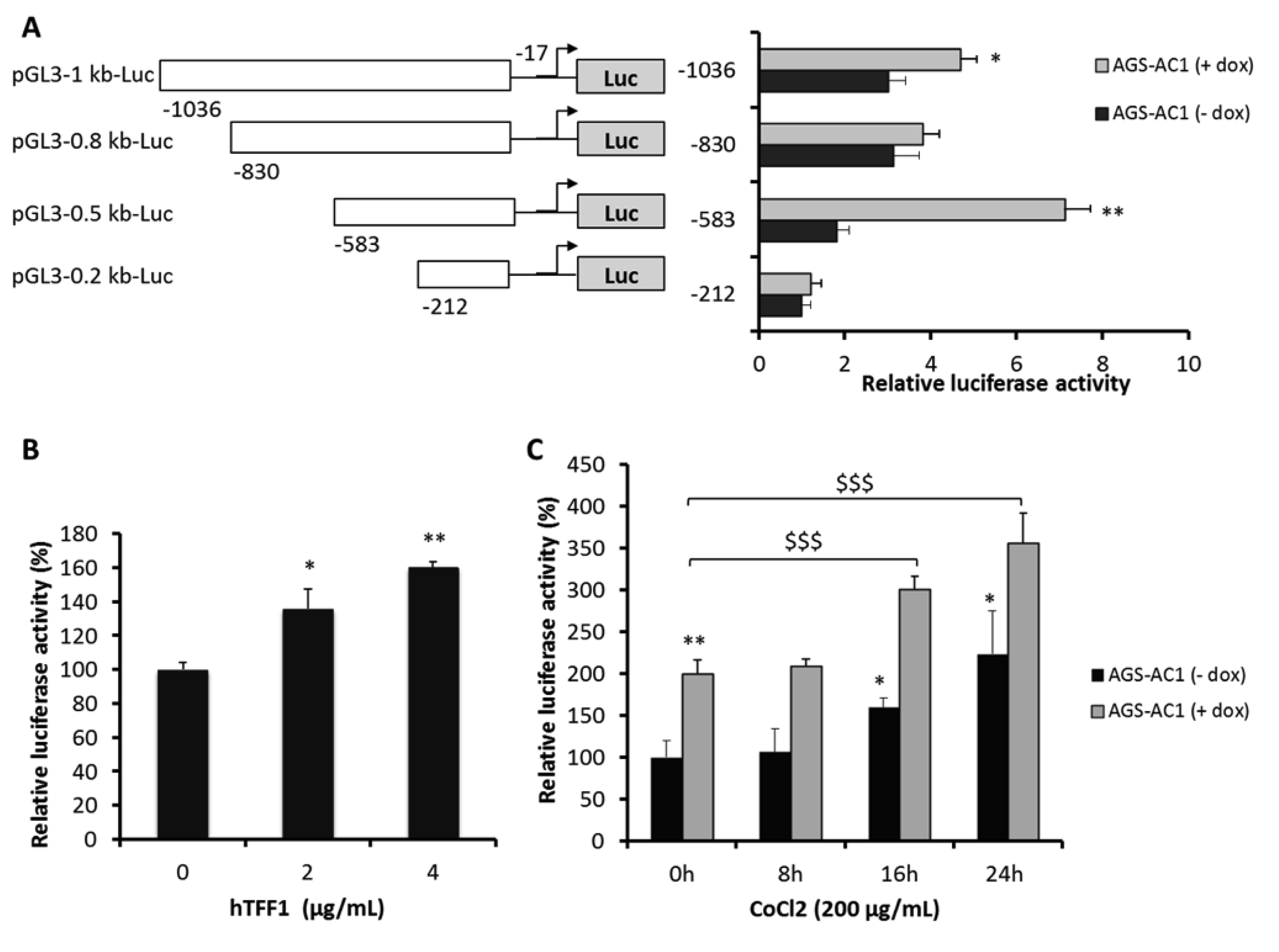
2.4. TFF1 Strengthens Its Own Induction upon Hypoxia Treatment
2.5. TFF1 Shows an Auto-Induction Mechanism
2.6. TFF1 Regulates the Methylation Status of Its DNA
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Conditioned Media
4.3. Hypoxic Culture Conditions
4.4. Western Blot Analysis
4.5. Confocal Microscopy
4.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.7. Trans-Well Invasion Assay
4.8. Plasmid Constructs and Luciferase Assays
4.9. High-Resolution Melting (HRM) Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Taupin, D.; Podolsky, D.K. Trefoil factors: Initiators of mucosal healing. Nat. Rev. Mol. Cell Biol. 2003, 4, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Gespach, C. Trefoil Factors. In Encyclopedia of Cancer; Schwab, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 4652–4658. [Google Scholar]
- Masiakowski, P.; Breathnach, R.; Bloch, J.; Gannon, F.; Krust, A.; Chambon, P. Cloning of cDNA sequences of hormone-regulated genes from the MCF-7 human breast cancer cell line. Nucleic Acids Res. 1982, 10, 7895–7903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prest, S.J.; May, F.E.; Westley, B.R. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. FASEB J. 2002, 16, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Amiry, N.; Kong, X.; Muniraj, N.; Kannan, N.; Grandison, P.M.; Lin, J.; Yang, Y.; Vouyovitch, C.M.; Borges, S.; Perry, J.K.; et al. Trefoil factor-1 (TFF1) enhances oncogenicity of mammary carcinoma cells. Endocrinology 2009, 150, 4473–4483. [Google Scholar] [CrossRef] [PubMed]
- Buache, E.; Etique, N.; Alpy, F.; Stoll, I.; Muckensturm, M.; Reina-San-Martin, B.; Chenard, M.P.; Tomasetto, C.; Rio, M.C. Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity of human breast cancer cells and mammary tumor development in TFF1-knockout mice. Oncogene 2011, 30, 3261–3273. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.A.; Nicholson, S.; Hennessy, C.; Lennard, T.W.; May, F.E.; Westley, B.R. Expression of the oestrogen regulated pNR-2 mRNA in human breast cancer: Relation to oestrogen receptor mRNA levels and response to tamoxifen therapy. Br. J. Cancer 1990, 61, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.; Rodrigue, C.M.; Attoub, S.; Fléjou, J.F.; Bruyneel, E.; Bracke, M.; Emami, S.; Gespach, C. Induction of the adenoma-carcinoma progression and Cdc25A-B phosphatases by the trefoil factor TFF1 in human colon epithelial cells. Oncogene 2006, 25, 6628–6636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arumugam, T.; Brandt, W.; Ramachandran, V.; Moore, T.T.; Wang, H.; May, F.E.; Westley, B.R.; Hwang, R.F.; Logsdon, C.D. Trefoil factor 1 stimulates both pancreatic cancer and stellate cells and increases metastasis. Pancreas 2011, 40, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ma, Y.; Huang, X. Trefoil factor 1 elevates the malignant phenotype of mucinous ovarian cancer cell through Wnt/β-catenin signaling. Int. J. Clin. Exp. Pathol. 2015, 8, 10412–10419. [Google Scholar] [PubMed]
- Lefebvre, O.; Chenard, M.P.; Masson, R.; Linares, J.; Dierich, A.; LeMeur, M.; Wendling, C.; Tomasetto, C.; Chambon, P.; Rio, M.C. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science 1996, 274, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.; Kayademir, T.; Soares, P.; Canedo, P.; Sousa, S.; Oliveira, C.; Leistenschneider, P.; Seruca, R.; Gött, P.; Blin, N.; et al. Loss of heterozygosity and promoter methylation, but not mutation, may underlie loss of TFF1 in gastric carcinoma. Lab. Investig. 2002, 82, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, J.; Yasui, W.; Tahara, H.; Tahara, E.; Kudo, Y.; Yokozaki, H.; Tahara, E. DNA hypermethylation at the pS2 promoter region is associated with early stage of stomach carcinogenesis. Cancer Lett. 2000, 149, 125–134. [Google Scholar] [CrossRef]
- Park, W.S.; Oh, R.R.; Park, J.Y.; Yoo, N.J.; Lee, S.H.; Shin, M.S.; Kim, S.Y.; Kim, Y.S.; Lee, J.H.; Kim, H.S.; et al. Mapping of a new target region of allelic loss at 21q22 in primary gastric cancers. Cancer Lett. 2000, 159, 15–21. [Google Scholar] [CrossRef]
- Machado, J.C.; Nogueira, A.M.M.F.; Carneiro, F.; Reis, C.A.; Sobrinho-Simões, M. Gastric carcinoma exhibits distinct types of cell differentiation: An immunohistochemical study of trefoil peptides (TFF1 and TFF2) and mucins (MUC1, MUC2, MUC5AC, and MUC6). J. Pathol. 2000, 190, 437–443. [Google Scholar] [CrossRef]
- Shi, S.Q.; Cai, J.T.; Yang, J.M. Expression of trefoil factors 1 and 2 in precancerous condition and gastric cancer. World J. Gastroenterol. 2006, 12, 3119–3122. [Google Scholar] [CrossRef] [PubMed]
- Kirikoshi, H.; Katoh, M. Expression of TFF1, TFF2 and TFF3 in gastric cancer. Int. J. Oncol. 2002, 21, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.L.; Luo, J.Y.; Lu, Y.P.; Wang, L.; Shi, H.X. Molecular forms of trefoil factor 1 in normal gastric mucosa and its expression in normal and abnormal gastric tissues. World J. Gastroenterol. 2006, 12, 7361–7364. [Google Scholar] [CrossRef] [PubMed]
- Im, S.; Yoo, C.; Jung, J.H.; Choi, H.J.; Yoo, J.; Kang, C.S. Reduced expression of TFF1 and increased expression of TFF3 in gastric cancer: Correlation with clinicopathological parameters and prognosis. Int. J. Med. Sci. 2013, 10, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Karam, S.M.; Tomasetto, C.; Rio, M.C. Trefoil factor 1 is required for the commitment programme of mouse oxyntic epithelial progenitors. Gut 2004, 53, 1408–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, W. TFF (trefoil factor family) peptides and their potential roles for differentiation processes during airway remodeling. Curr. Med. Chem. 2007, 14, 2716–2719. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Tomikawa, M.; Mohajer, B.; Tarnawski, A.S. Gastrointestinal mucosal regeneration: Role of growth factors. Front. Biosci. 1999, 4, D303–D309. [Google Scholar] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rio, M.C.; Chenard, M.P.; Wolf, C.; Marcellin, L.; Tomasetto, C.; Lathe, R.; Bellocq, J.P.; Chambon, P. Induction of pS2 and hSP genes as markers of mucosal ulceration of the digestive tract. Gastroenterology 1991, 100, 375–379. [Google Scholar] [CrossRef]
- Wright, N.A.; Poulsom, R.; Stamp, G.; Van Noorden, S.; Sarraf, C.; Elia, G.; Ahnen, D.; Jeffery, R.; Longcroft, J.; Pike, C.; et al. Trefoil peptide gene expression in gastrointestinal epithelial cells in inflammatory bowel disease. Gastroenterology 1993, 104, 12–20. [Google Scholar] [CrossRef]
- Esposito, R.; Morello, S.; Vllahu, M.; Eletto, D.; Porta, A.; Tosco, A. Gastric TFF1 Expression from Acute to Chronic Helicobacter Infection. Front. Cell. Infect. Microbiol. 2017, 7, 434. [Google Scholar] [CrossRef] [PubMed]
- Métivier, R.; Gallais, R.; Tiffoche, C.; Le Péron, C.; Jurkowska, R.Z.; Carmouche, R.P.; Ibberson, D.; Barath, P.; Demay, F.; Reid, G.; et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008, 452, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Wright, N.A.; Poulsom, R.; Stamp, G.W.; Hall, P.A.; Jeffery, R.E.; Longcroft, J.M.; Rio, M.C.; Tomasetto, C.; Chambon, P. Epidermal growth factor (EGF/URO) induces expression of regulatory peptides in damaged human gastrointestinal tissues. J. Pathol. 1990, 162, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Al-azzeh, E.D.; Fegert, P.; Blin, N.; Gött, P. Transcription factor GATA-6 activates expression of gastroprotective trefoil genes TFF1 and TFF2. Biochim. Biophys. Acta 2000, 1490, 324–332. [Google Scholar] [CrossRef]
- Lacroix, M.; Leclercq, G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol. Cell. Endocrinol. 2004, 219, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.; Montefusco, S.; Ferro, P.; Monti, M.C.; Baldantoni, D.; Tosco, A.; Marzullo, L. Trefoil Factor 1 is involved in gastric cell copper homeostasis. Int. J. Biochem. Cell Biol. 2015, 59, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Hernández, C.; Santamatilde, E.; McCreath, K.J.; Cervera, A.M.; Díez, I.; Ortiz-Masiá, D.; Martínez, N.; Calatayud, S.; Esplugues, J.V.; Barrachina, M.D. Induction of trefoil factor (TFF)1, TFF2 and TFF3 by hypoxia is mediated by hypoxia inducible factor-1: Implications for gastric mucosal healing. Br. J. Pharmacol. 2009, 156, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Bulitta, C.J.; Fleming, J.V.; Raychowdhury, R.; Taupin, D.; Rosenberg, I.; Wang, T.C. Autoinduction of the trefoil factor 2 (TFF2) promoter requires an upstream cis-acting element. Biochem. Biophys. Res. Commun. 2002, 293, 366–374. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, L.; Zhou, Y.; Mao, X.; Deng, X. Human Trefoil Factor 3 induces the transcription of its own promoter through STAT3. Sci. Rep. 2016, 6, 30421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.W.; Zhang, Z.G.; Hao, Y.X.; Zhao, Y.L.; Qian, F.; Shi, Y.; Li, P.A.; Liu, C.Y.; Yu, P.W. HIF-1α induces the epithelial-mesenchymal transition in gastric cancer stem cells through the Snail pathway. Oncotarget 2017, 8, 9535–9545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosco, A.; Monti, M.C.; Fontanella, B.; Montefusco, S.; D’Andrea, L.; Ziaco, B.; Baldantoni, D.; Rio, M.C.; Marzullo, L. Copper binds the carboxy-terminus of trefoil protein 1 (TFF1), favoring its homodimerization and motogenic activity. Cell. Mol. Life Sci. 2010, 67, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Boyer, B.; Vallés, A.M.; Edme, N. Induction and regulation of epithelial-mesenchymal transitions. Biochem. Pharmacol. 2000, 60, 1091–1099. [Google Scholar] [CrossRef]
- Aigner, K.; Dampier, B.; Descovich, L.; Mikula, M.; Sultan, A.; Schreiber, M.; Mikulits, W.; Brabletz, T.; Strand, D.; Obrist, P.; et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 2007, 26, 6979–6988. [Google Scholar] [CrossRef] [PubMed]
- Marchbank, T.; Westley, B.R.; May, F.E.B.; Calnan, D.P.; Playford, R.J. Dimerization of human ps2 (tff1) plays a key role in its protective/healing effects. J. Pathol. 1998, 185, 153–158. [Google Scholar] [CrossRef]
- Ito, M.; Tanaka, S.; Kim, S.; Kuwai, T.; Matsutani, N.; Kamada, T.; Kitadai, Y.; Sumii, M.; Yoshihara, M.; Haruma, K.; et al. The specific expression of hypoxia inducible factor-1alpha in human gastric mucosa induced by nonsteroidal anti-inflammatory drugs. Aliment. Pharmacol. Ther. 2003, 18 (Suppl. 1), 90–98. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Masiá, D.; Hernández, C.; Quintana, E.; Kuwai, T.; Matsutani, N.; Kamada, T.; Kitadai, Y.; Sumii, M.; Yoshihara, M.; Haruma, K.; et al. iNOS-derived nitric oxide mediates the increase in TFF2 expression associated with gastric damage: Role of HIF-1. FASEB J. 2010, 24, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Tanka, T.; Nakamura, J.; Kitajima, Y.; Kai, K.; Miyake, S.; Hiraki, M.; Ide, T.; Koga, Y.; Noshiro, H. Loss of trefoil factor 1 is regulated by DNA methylation and is an independent predictive factor for poor survival in advanced gastric cancer. Int. J. Oncol. 2013, 42, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Zhang, Y.; Yuan, H.; Bai, R.; Zheng, J.; Zhang, J.; Song, M. DNA methylation of trefoil factor 1 (TFF1) is associated with the tumorigenesis of gastric carcinoma. Mol. Med. Rep. 2014, 9, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Cerda, S.; Weitzman, S.A. Influence of oxygen radical injury on DNA methylation. Mutat. Res. 1997, 386, 141–152. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Kalbacher, H.; Hoffmann, W. TFF1 is differentially expressed in stationary and migratory rat gastric epithelial cells (RGM-1) after in vitro wounding: Influence of TFF1 RNA interference on cell migration. Cell. Physiol. Biochem. 2013, 32, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Szabo, S. “Gastric cytoprotection” is still relevant. J. Gastroenterol. Hepatol. 2014, 29 (Suppl. 4), 124–132. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Takaishi, S.; Menheniott, T.R.; Yang, X.; Shibata, W.; Jin, G.; Betz, K.S.; Kawakami, K.; Minamoto, T.; Tomasetto, C.; et al. Inhibition of gastric carcinogenesis by the hormone gastrin is mediated by suppression of TFF1 epigenetic silencing. Gastroenterology 2011, 140, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Koslowski, M.; Luxemburger, U.; Türeci, O.; Sahin, U. Tumor-associated CpG demethylation augments hypoxia-induced effects by positive autoregulation of HIF-1α. Oncogene 2011, 30, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Chatagnon, A.; Ballestar, E.; Esteller, M.; Dante, R. A role for methyl-CpG binding domain protein 2 in the modulation of the estrogen response of pS2/TFF1 gene. PLoS ONE 2010, 5, e9665. [Google Scholar] [CrossRef] [PubMed]
- Tse, M.Y.; Ashbury, J.E.; Zwingerman, N.; King, W.D.; Taylor, S.A.; Pang, S.C. A refined, rapid and reproducible high resolution melt (HRM)-based method suitable for quantification of global LINE-1 repetitive element methylation. BMC Res. Notes 2011, 4, 565. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romano, E.; Vllahu, M.; Bizzarro, V.; Belvedere, R.; Esposito, R.; Petrella, A.; Tosco, A. TFF1 Promotes EMT-Like Changes through an Auto-Induction Mechanism. Int. J. Mol. Sci. 2018, 19, 2018. https://doi.org/10.3390/ijms19072018
Romano E, Vllahu M, Bizzarro V, Belvedere R, Esposito R, Petrella A, Tosco A. TFF1 Promotes EMT-Like Changes through an Auto-Induction Mechanism. International Journal of Molecular Sciences. 2018; 19(7):2018. https://doi.org/10.3390/ijms19072018
Chicago/Turabian StyleRomano, Elena, Megi Vllahu, Valentina Bizzarro, Raffaella Belvedere, Roberta Esposito, Antonello Petrella, and Alessandra Tosco. 2018. "TFF1 Promotes EMT-Like Changes through an Auto-Induction Mechanism" International Journal of Molecular Sciences 19, no. 7: 2018. https://doi.org/10.3390/ijms19072018
APA StyleRomano, E., Vllahu, M., Bizzarro, V., Belvedere, R., Esposito, R., Petrella, A., & Tosco, A. (2018). TFF1 Promotes EMT-Like Changes through an Auto-Induction Mechanism. International Journal of Molecular Sciences, 19(7), 2018. https://doi.org/10.3390/ijms19072018