Label-Free Quantitative Proteomics Combined with Biological Validation Reveals Activation of Wnt/β-Catenin Pathway Contributing to Trastuzumab Resistance in Gastric Cancer
Abstract
1. Introduction
2. Results
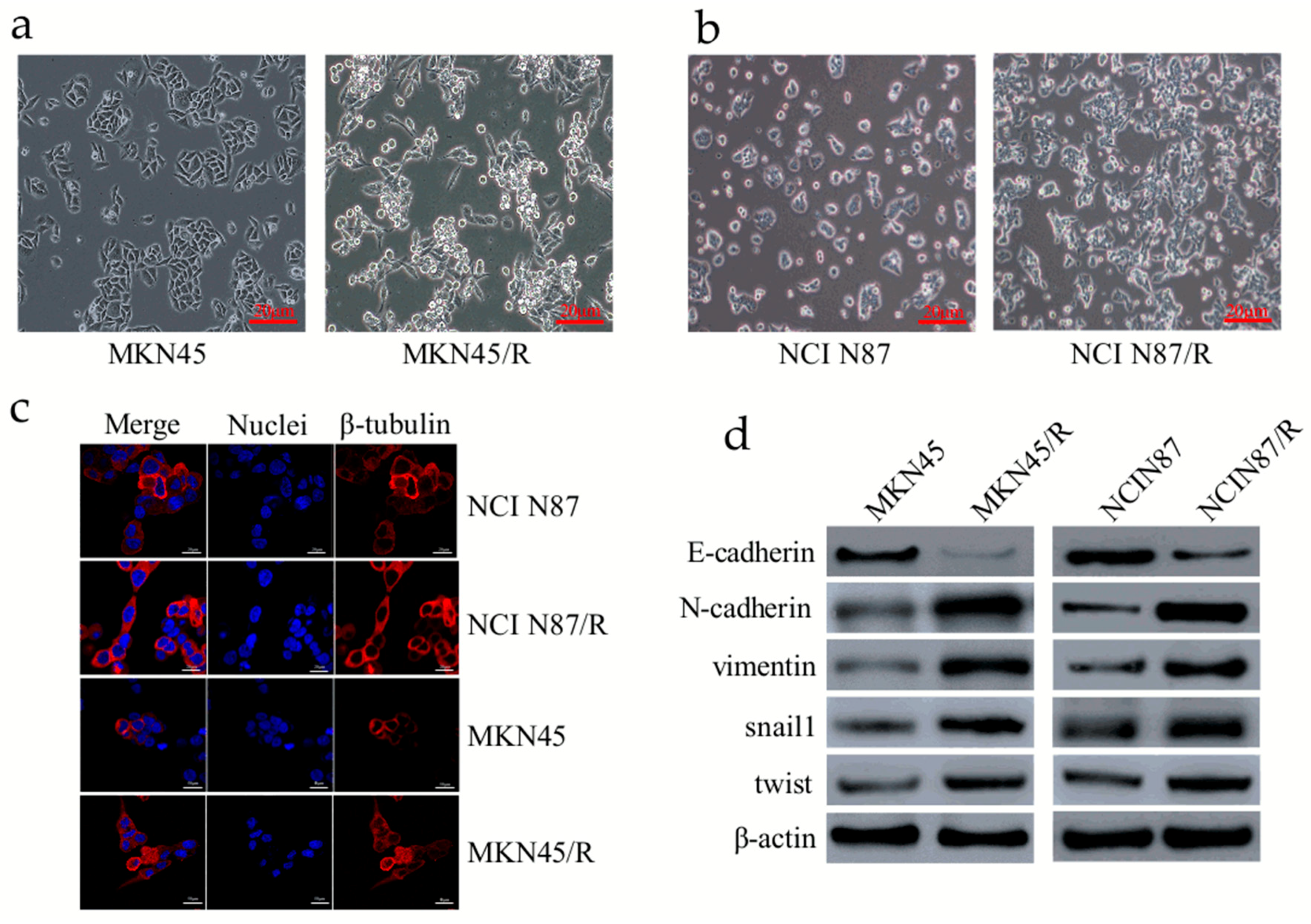
2.1. Establishment of Trastuzumab-Resistant Gastric Cancer Cell Lines
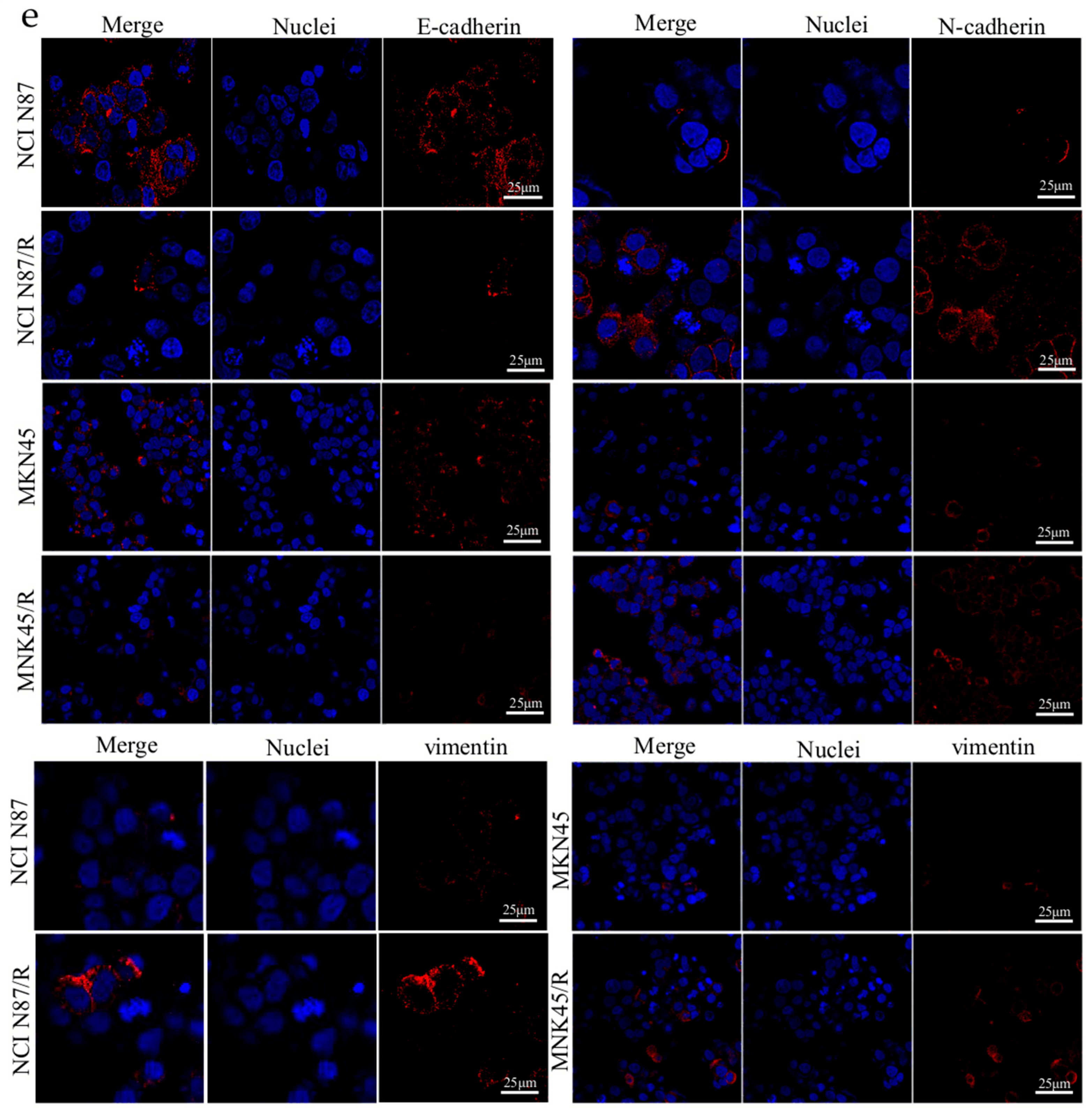
2.2. Trastuzumab-Resistant Gastric Cancer Cells Exhibit an EMT-Like Phenotypic Change
2.3. Trastuzumab Resistant Cells Exhibit Increased Capacity of Migration and Proliferation
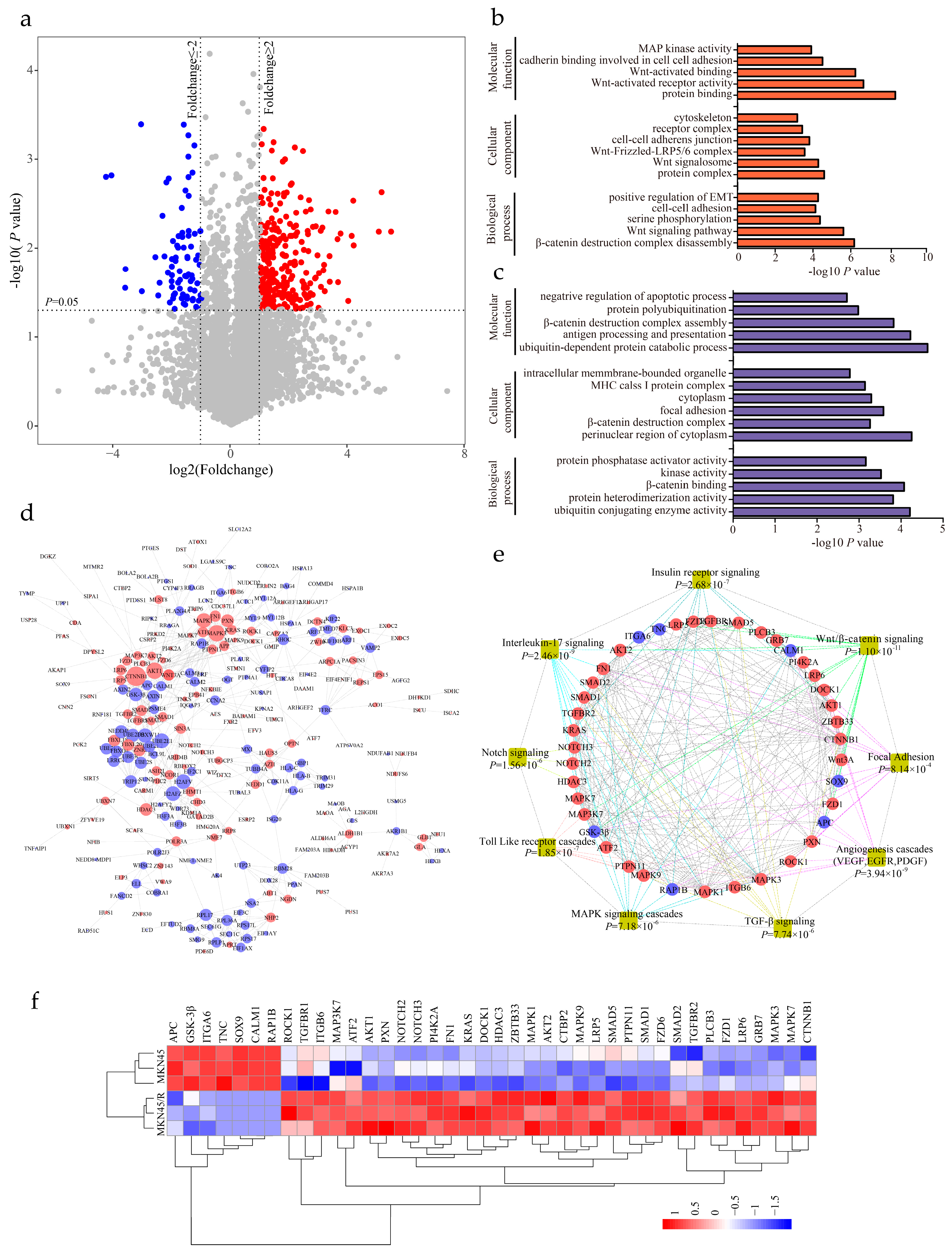
2.4. Label-Free Proteomic Profiling of MKN45 and MKN45/R Cells
2.5. Identification of Differentially Expressed Proteins and Enrichment Analysis
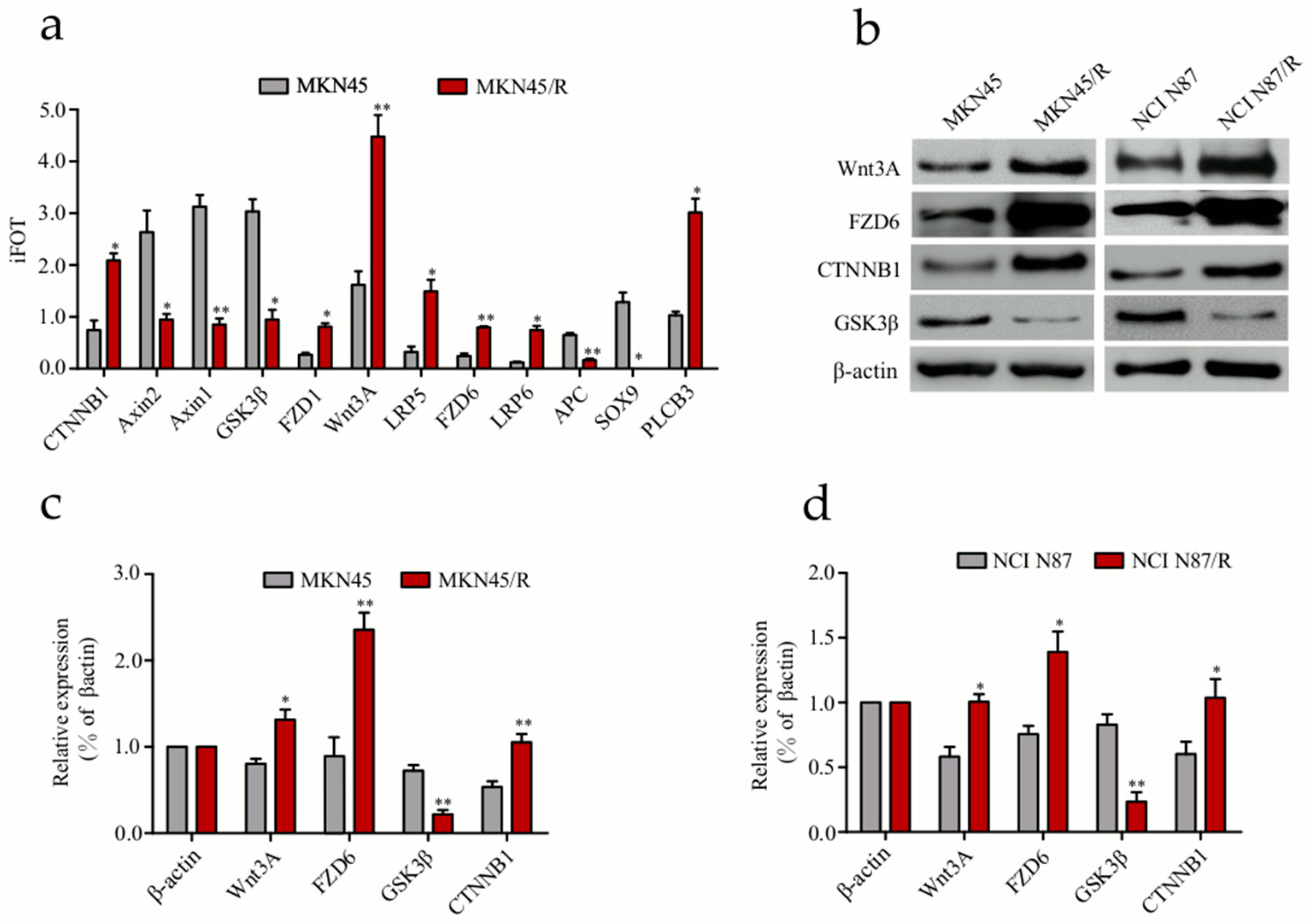
2.6. Label-Free Quantitative Proteomics Profiling Reveals the Activation of Canonical Wnt/β-Catenin Signaling Pathway in Trastuzumab-Resistant Cells
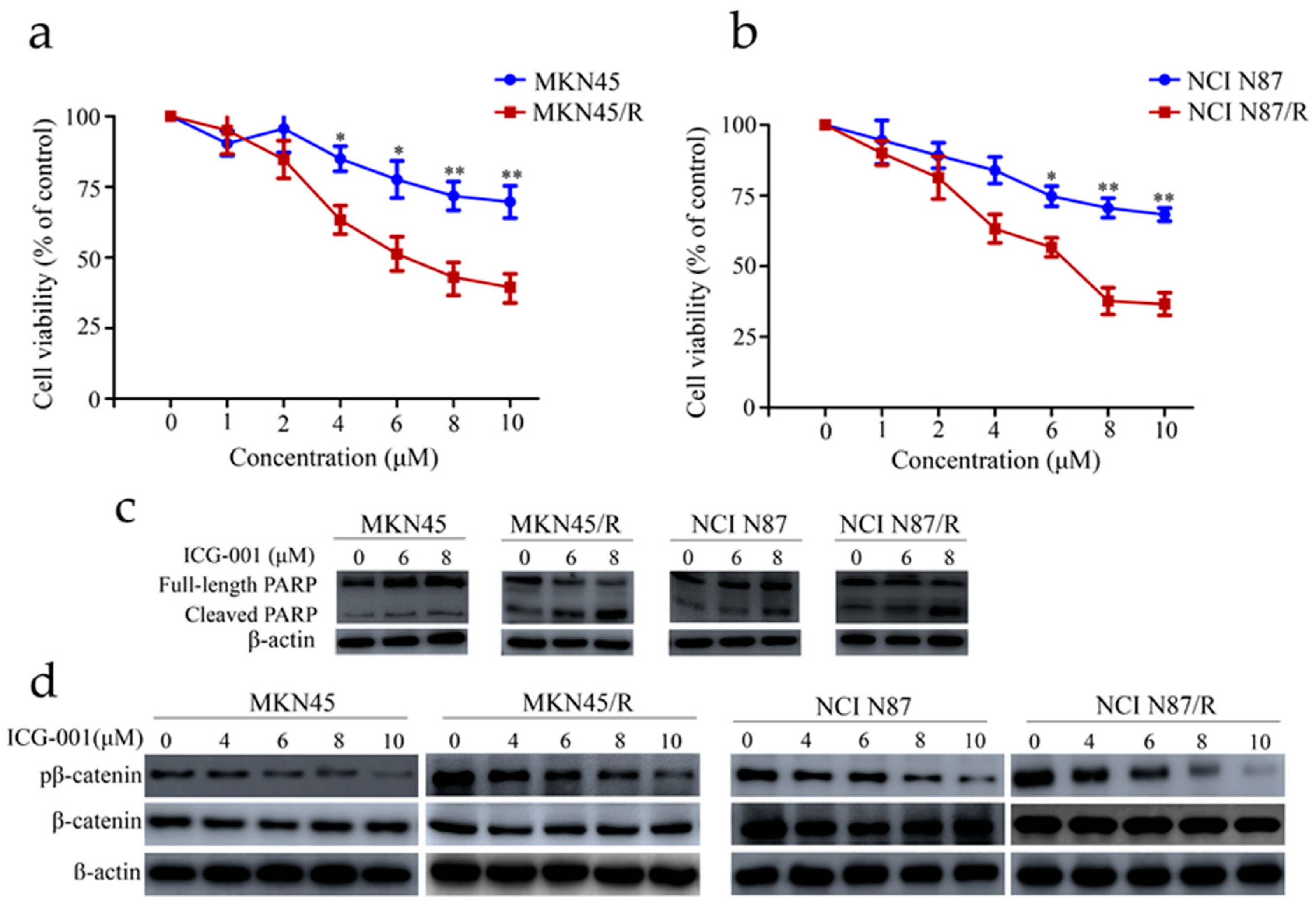
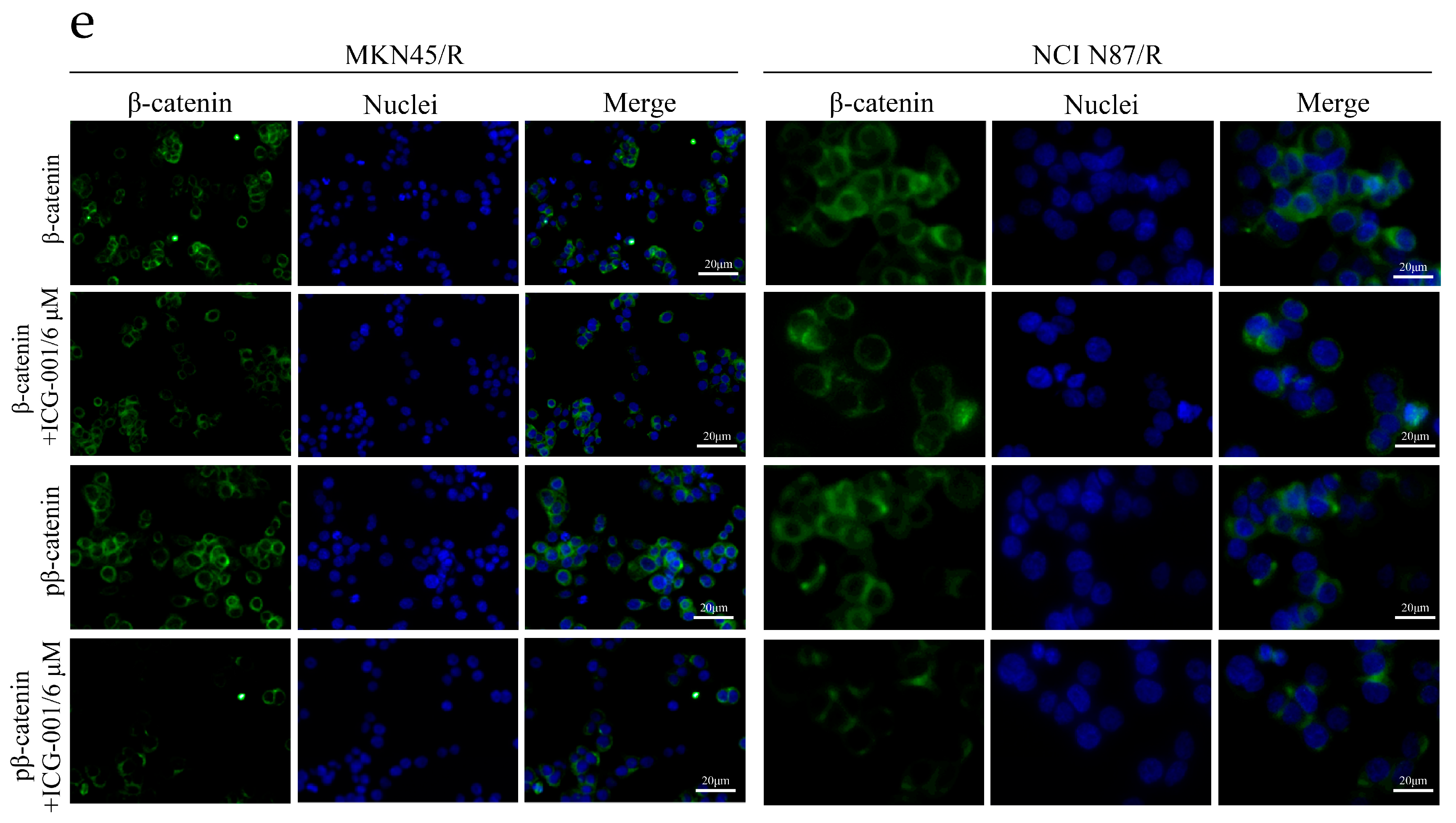
2.7. ICG-001 Reduces Viability, Induces Apoptosis and Inhibits β-Catenin Phosphorylation of MKN45/R and NCI N87/R Cells
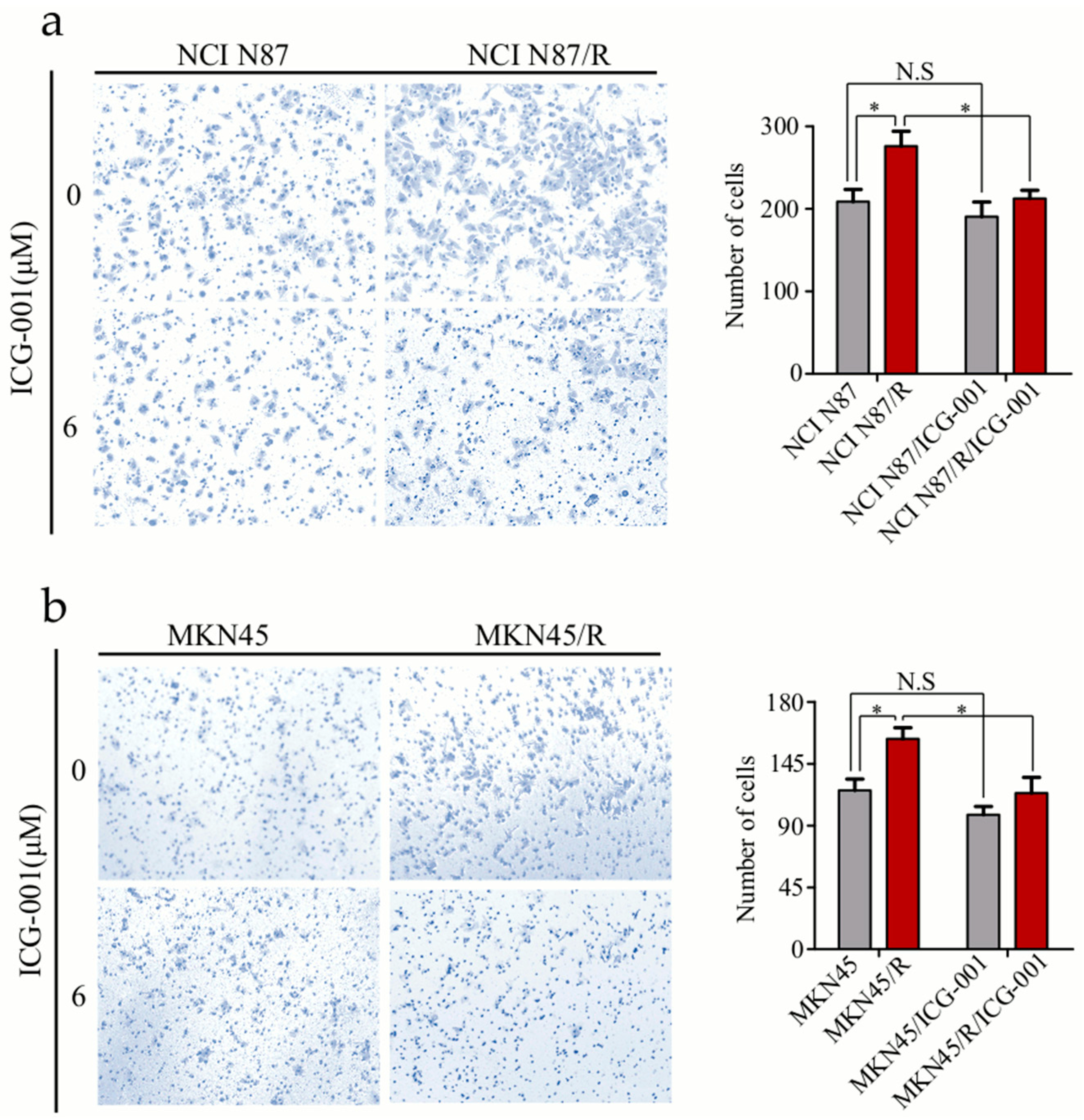
2.8. ICG-001 Inhibits the Invasion of Trastuzumab-Resistant Cells
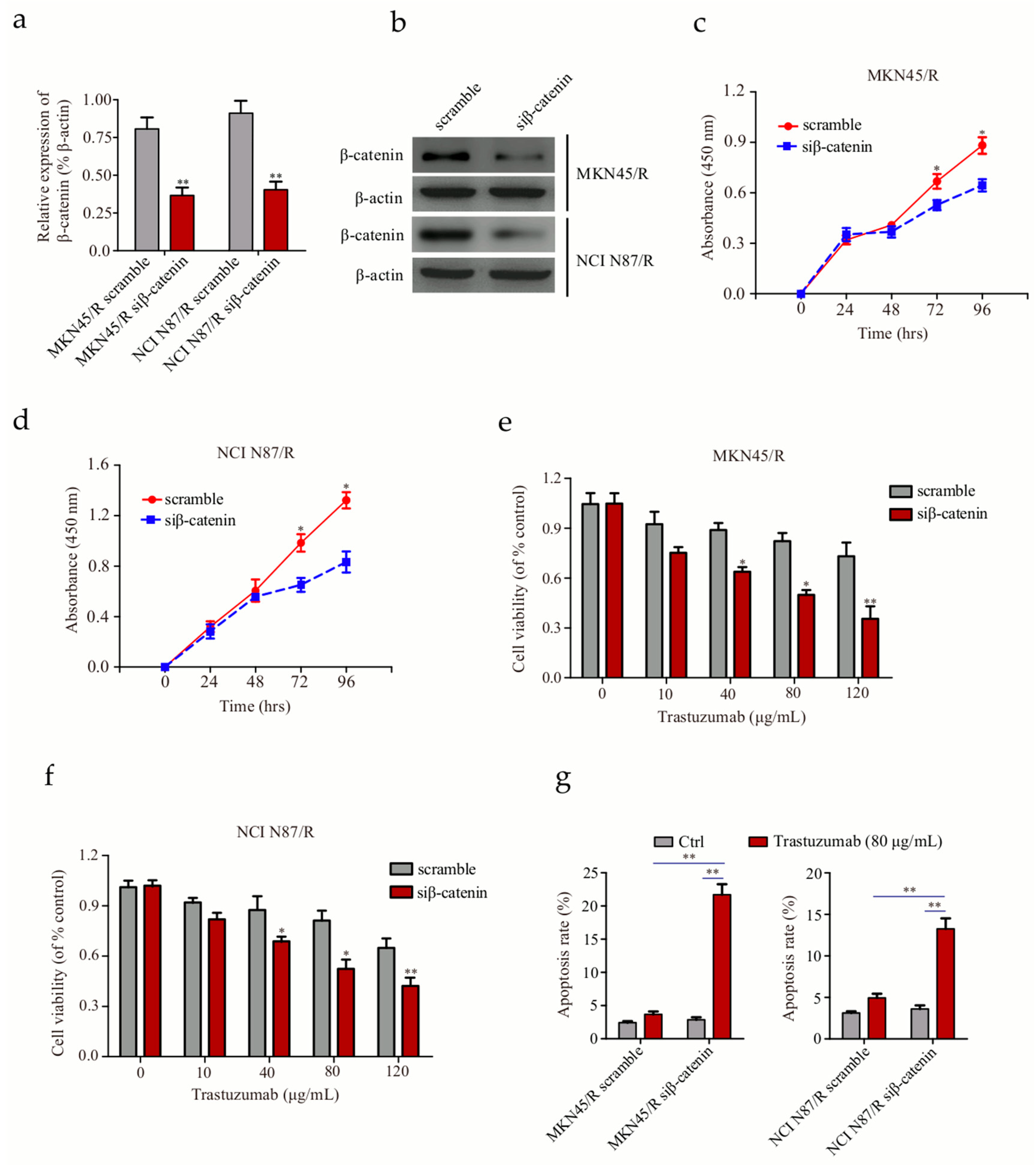
2.9. Knockdown of β-Catenin in Trastuzumab-Resistant Cells Suppresses Proliferation and Enhances Trastuzumab Sensitivity
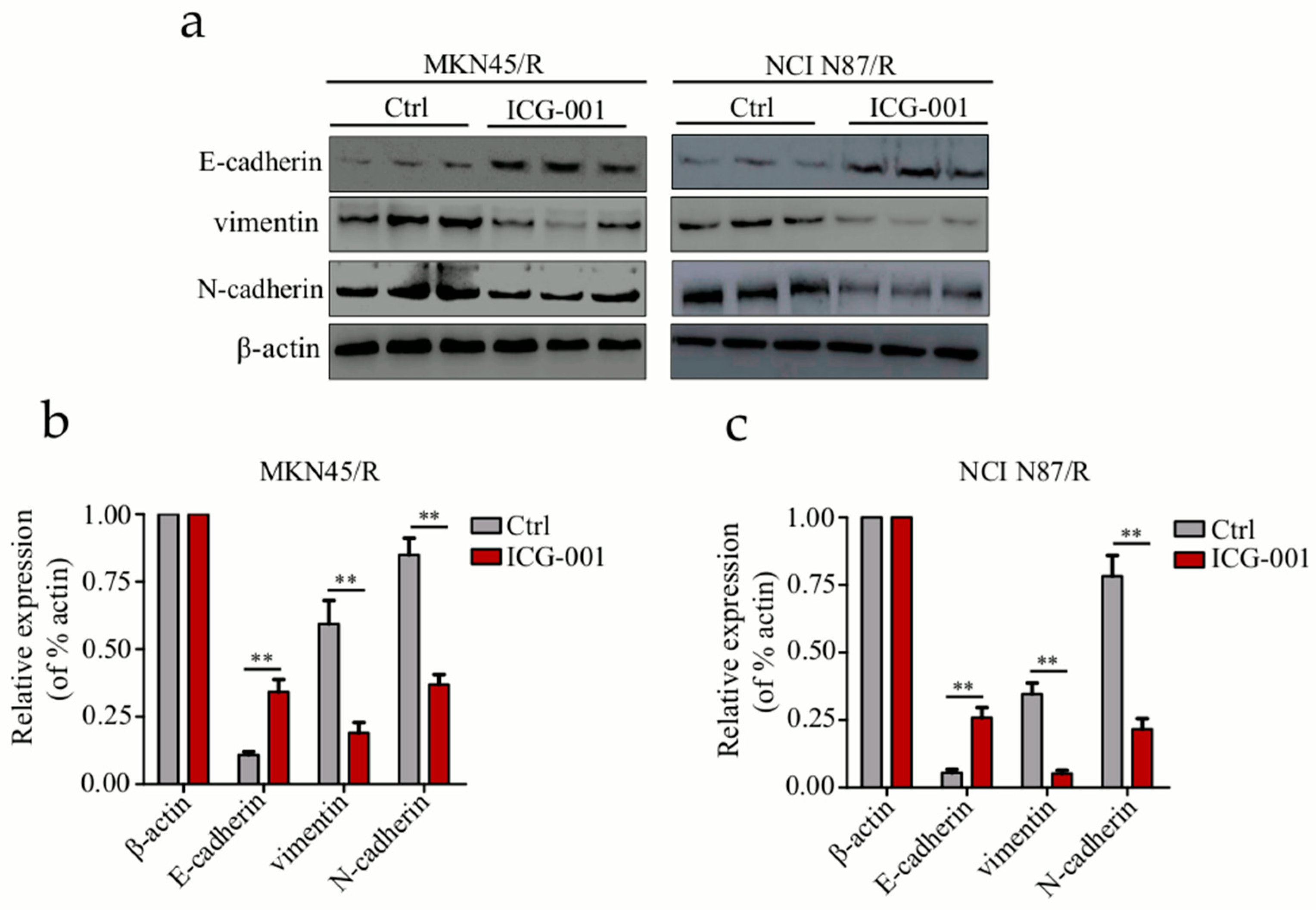
2.10. Inhibition of Wnt/β-Catenin Signaling by ICG-001 Reverses EMT in Trastuzumab Resistant Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Development of Trastuzumab-Resistant MKN45/R and NCI N87/R Gastric Cancer Cell Lines
4.3. Establishment of Stable Trastuzumab Resistant Cell Lines with β-Catenin Knockdown
4.4. Resistance Index Assays
4.5. Wound Healing Assays
4.6. Colony Formation Assays
4.7. Immunofluorescence Staining
4.8. Viability and Apoptosis Assays
4.9. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.10. Western Blotting
4.11. Cell invasion Assays
4.12. Protein Extraction and Peptide Separation
4.13. Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (LC-MS/MS) Analysis
4.14. Protein Identification and Quantification
4.15. Proteome Data Filtering and Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EMT | Epithelial-mesenchymal transition |
| HER-2 | Human epidermal growth factor receptor-2 |
| EGFR | Epidermal growth factor receptor |
| PI3K | Phosphatidylinositide 3-kinase |
| IGFR | Insulin-like growth factor receptor |
| PTEN | Phosphatase and tensin homolog gene |
| FGFR3 | Fibroblast growth factor receptor 3 |
| CBP | cyclic AMP response element-binding protein |
| β2-AR | β2-adrenergic receptor |
| iBAQ | Intensity based absolute quantification |
| FOT | Fraction of total |
| GO | Gene ontology |
| PCA | Principal component analysis |
| SFRP4 | Secreted frizzled related protein 4 |
| CCK-8 | Cell Count kit-8 |
| IC50 | Half maximal inhibitory concentration |
| BSA | Bovine serum albumin |
| RT–PCR | Reverse transcription polymerase chain reaction |
| LC-MS/MS | Liquid chromatography–mass spectrometry/mass spectrometry |
| FDR | False discovery rate |
| FASP | Filter-aided sample preparation |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Martín, C.; Lopez-Rios, F.; Aparicio, J.; Barriuso, J.; García-Carbonero, R.; Pazo, R.; Rivera, F.; Salgado, M.; Salud, A.; Vázquez-Sequeiros, E.; et al. A critical review of HER2-positive gastric cancer evaluation and treatment: From trastuzumab, and beyond. Cancer Lett. 2014, 35, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Boku, N. HER2-positive gastric cancer. Gastric Cancer 2014, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Appert-Collin, A.; Hubert, P.; Cremel, G.; Bennasroune, A. Role of ErbB receptors in cancer cell migration and invasion. Front. Pharmacol. 2015, 6, 283. [Google Scholar] [CrossRef] [PubMed]
- Minner, S.; Jessen, B.; Stiedenroth, L.; Burandt, E.; Köllermann, J.; Mirlacher, M.; Erbersdobler, A.; Eichelberg, C.; Fisch, M.; Brümmendorf, T.H.; et al. Low level HER2 overexpression is associated with rapid tumor cell proliferation and poor prognosis in prostate cancer. Clin. Cancer Res. 2010, 16, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Rimawi, M.F.; Schiff, R.; Osborne, C.K. Targeting HER2 for the treatment of breast cancer. Annu. Rev. Med. 2015, 66, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. The ErbB/HER family of protein–tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef] [PubMed]
- Rexer, B.N.; Arteaga, C.L. Intrinsic and acquired resistance to HER2–targeted therapies in HER2 gene–amplified breast cancer: Mechanisms and clinical implications. Crit. Rev. Oncog. 2012, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, Z.; Lu, N. Molecular targeted therapy for the treatment of gastric cancer. J. Exp. Clin. Cancer Res. 2016, 35, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Shen, A.; Ding, J.; Geng, M. Molecularly targeted cancer therapy: Some lessons from the past decade. Trends Pharmacol. Sci. 2014, 35, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Meza-Junco, J.; Au, H.J.; Sawyer, M.B. Critical appraisal of trastuzumab in treatment of advanced stomach cancer. Cancer Manag. Res. 2011, 3, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.; Eiermann, W.; Robert, N.; Pienkowski, T.; Martin, M.; Press, M.; Mackey, J.; Glaspy, J.; Chan, A.; Pawlicki, M.; et al. Adjuvant trastuzumab in HER2–positive breast cancer. N. Engl. J. Med. 2011, 366, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.; Esteva, F.J. HER2 therapy: Molecular mechanisms of trastuzumab resistance. Breast Cancer Res. 2006, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Lavaud, P.; Andre, F. Strategies to overcome trastuzumab resistance in HER2-over expressing breast cancers: Focus on new data from clinical trials. BMC Med. 2014, 12, 132. [Google Scholar] [CrossRef] [PubMed]
- Chandarlapaty, S.; Sakr, R.A.; Giri, D.; Patil, S.; Heguy, A.; Morrow, M.; Modi, S.; Norton, L.; Rosen, N.; Hudis, C.; et al. Frequent mutational activation of the PI3K–AKT pathway in trastuzumab–resistant breast cancer. Clin. Cancer Res. 2012, 18, 6784–6791. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.; Yuan, L.; Zhang, B.; Kobayashi, R.; Esteva, F.J. Insulin–like growth factor–I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005, 65, 11118–11128. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zi, X.; Zhao, Y.; Mascarenhas, D.; Pollak, M. Insulin–like growth factor–I receptor signaling and resistance to trastuzumab (Herceptin). J. Natl. Cancer Inst. 2001, 93, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Lan, K.H.; Zhou, X.; Tan, M.; Esteva, F.J.; Sahin, A.A.; Klos, K.S.; Li, P.; Monia, B.P.; Nguyen, N.T.; et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 2004, 6, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Arribas, J.; Baselga, J.; Pedersen, K.; Parra–Palau, J.L. p95HER2 and breast cancer. Cancer Res. 2011, 71, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Eto, K.; Iwatsuki, M.; Watanabe, M.; Ishimoto, T.; Ida, S.; Imamura, Y.; Iwagami, S.; Baba, Y.; Sakamoto, Y.; Miyamoto, Y.; et al. The sensitivity of gastric cancer to trastuzumab is regulated by the miR-223/FBXW7 pathway. Int. J. Cancer 2015, 136, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Piro, G.; Carbone, C.; Cataldo, I.; Di Nicolantonio, F.; Giacopuzzi, S.; Aprile, G.; Simionato, F.; Boschi, F.; Zanotto, M.; Mina, M.M.; et al. An FGFR3 autocrine loop sustains acquired resistance to trastuzumab in gastric cancer patients. Clin. Cancer Res. 2016, 22, 6164–6175. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, Z.; Wang, T.; Yang, Z.; Chen, H.; Hu, Y.; Hu, C.; Guo, L.; Deng, Q.; Liu, Y.; et al. β2-AR signaling controls trastuzumab resistance-dependent pathway. Oncogene 2016, 35, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; Caporale, M.; Morano, F.; Scartozzi, M.; Gloghini, A.; De Vita, F.; Giommoni, E.; Fornaro, L.; Aprile, G.; Melisi, D.; et al. HER2 loss in HER2-positive gastric or gastroesophageal cancer after trastuzumab therapy: Implication for further clinical research. Int. J. Cancer 2016, 139, 2859–2864. [Google Scholar] [CrossRef] [PubMed]
- Roepstorff, P. Mass spectrometry based proteomics, background, status and future needs. Protein Cell 2012, 3, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass–spectrometry–based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Jiang, J.; Wei, J.; Liu, W.; Zhang, W.; Liu, M.; Fu, T.; Lu, T.; Song, L.; Ying, W.; et al. A fast workflow for identification and quantification of proteomes. Mol. Cell. Proteom. 2013, 12, 2370–2380. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Liu, J.; Zhang, J.; Wu, M.; Guo, L.; Liao, W. Development of trastuzumab-resistant human gastric carcinoma cell lines and mechanisms of drug resistance. Sci. Rep. 2015, 5, 11634. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. EMT program is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chang, J.; Liu, M.; Yuan, J.; Zhang, J.; Qin, J.; Xia, X.; Wang, Y. Quantitative proteomics profiling reveals activation of mTOR pathway in trastuzumab resistance. Oncotarget 2017, 8, 45793–45806. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, M.; Nguyen, C.; Lee, S.C.; Kahn, M. ICG–001, a novel small molecule regulator of TCF/β–catenin transcription. Med. Chem. 2005, 1, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.; Amerizadeh, F.; ShahidSales, S.; Khazaei, M.; Ghayour-Mobarhan, M.; Sadeghnia, H.R.; Maftouh, M.; Hassanian, S.M.; Avan, A. Therapeutic potential of targeting Wnt/β-catenin pathway in treatment of colorectal cancer: Rational and progress. J. Cell. Biochem. 2017, 119, 1979–1983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, N.; Wisniewski, J.R.; Geiger, T.; Cox, J.; Kircher, M.; Kelso, J.; Pääbo, S.; Mann, M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011, 7, 548. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018, 62, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Ashihara, E.; Maekawa, T. Targeting the Wnt/β-catenin signaling pathway in human cancers. Expert Opin. Ther. Targets 2011, 15, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Jiang, W.; Wang, S.; Wang, L.; Xie, K. Role of Wnt/β-catenin signaling in drug resistance of pancreatic cancer. Curr. Pharm. Des. 2012, 18, 2464–2471. [Google Scholar] [CrossRef] [PubMed]
- Bilir, B.; Kucuk, O.; Moreno, C.S. Wnt signaling blockage inhibits cell proliferation and migration, and induces apoptosis in triple–negative breast cancer cells. J. Transl. Med. 2013, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.K.; Shao, C.; Wang, J.; Wei, Q.; Wang, X.; Collier, Z.; Tang, S.; Liu, H.; Zhang, F.; Huang, J.; et al. Wnt/β–catenin signaling plays an ever–expanding role in stem cell self–renewal, tumorigenesis and cancer chemoresistance. Genes Dis. 2016, 3, 11–40. [Google Scholar] [CrossRef] [PubMed]
- Schade, B.; Lesurf, R.; Sanguin-Gendreau, V.; Bui, T.; Deblois, G.; O’Toole, S.A.; Millar, E.K.; Zardawi, S.J.; Lopez-Knowles, E.; Sutherland, R.L.; et al. β-Catenin signaling is a critical event in ErbB2-mediated mammary tumor progression. Cancer Res. 2013, 73, 4474–4487. [Google Scholar] [CrossRef] [PubMed]
- Arend, R.C.; Londoño-Joshi, A.I.; Straughn, J.M., Jr.; Buchsbaum, D.J. The Wnt/β-catenin pathway in ovarian cancer: A review. Gynecol. Oncol. 2013, 131, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ginther, C.; Kim, J.; Mosher, N.; Chung, S.; Slamon, D.; Vadgama, J.V. Expression of Wnt3 activates Wnt/β-catenin pathway and promotes EMT-like phenotype in trastuzumab-resistant HER2-overexpressing breast cancer cells. Mol. Cancer Res. 2012, 10, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Scholer-Dahirel, A.; Schlabach, M.R.; Loo, A.; Bagdasarian, L.; Meyer, R.; Guo, R.; Woolfenden, S.; Yu, K.K.; Markovits, J.; Killary, K.; et al. Maintenance of adenomatous polyposis coli (APC)-mutant colorectal cancer is dependent on Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 17135–17140. [Google Scholar] [CrossRef] [PubMed]
- Wade, C.A.; Kyprianou, N. Profiling prostate cancer therapeutic resistance. Int. J. Mol. Sci. 2018, 19, 904. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.N.; Bhowmick, N.A. Role of EMT in metastasis and therapy resistance. J. Clin. Med. 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Oliveras-Ferraros, C.; Corominas-Faja, B.; Cufí, S.; Vazquez-Martin, A.; Martin-Castillo, B.; Iglesias, J.M.; López-Bonet, E.; Martin, Á.G.; Menendez, J.A. Epithelial–to–mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin). Cell Cycle 2012, 11, 4020–4032. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.E.; Jary, E.; Ma, S.S.; Nixdorf, S.; Heinzelmann-Schwarz, V.A.; Ward, R.L. The Wnt gatekeeper SFRP4 modulates EMT, cell migration and downstream Wnt signalling in serous ovarian cancer cells. PLoS ONE 2013, 8, e54362. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Liang, L.; Chen, J.; Qiu, N.; Ge, S.; Ji, S.; Shi, T.; Zhen, B.; Liu, M.; Ding, C.; et al. Multidimensional Proteomics Reveals a Role of UHRF2 in the Regulation of Epithelial-Mesenchymal Transition (EMT). Mol. Cell. Proteom. 2016, 15, 2263–2278. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Xia, X.; Ding, C.; Zhen, B.; Zhou, Q.; Feng, J.; Yuan, J.; Chen, R.; Li, Y.; Ge, Z.; et al. A proteomic landscape of diffuse-type gastric cancer. Nat. Commun. 2018, 9, 1012. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Yuan, J.; Liu, Z.; Zhang, J.; Chang, J. Label-Free Quantitative Proteomics Combined with Biological Validation Reveals Activation of Wnt/β-Catenin Pathway Contributing to Trastuzumab Resistance in Gastric Cancer. Int. J. Mol. Sci. 2018, 19, 1981. https://doi.org/10.3390/ijms19071981
Liu W, Yuan J, Liu Z, Zhang J, Chang J. Label-Free Quantitative Proteomics Combined with Biological Validation Reveals Activation of Wnt/β-Catenin Pathway Contributing to Trastuzumab Resistance in Gastric Cancer. International Journal of Molecular Sciences. 2018; 19(7):1981. https://doi.org/10.3390/ijms19071981
Chicago/Turabian StyleLiu, Wenhu, Jiangbei Yuan, Zhenzhong Liu, Jianwu Zhang, and Jinxia Chang. 2018. "Label-Free Quantitative Proteomics Combined with Biological Validation Reveals Activation of Wnt/β-Catenin Pathway Contributing to Trastuzumab Resistance in Gastric Cancer" International Journal of Molecular Sciences 19, no. 7: 1981. https://doi.org/10.3390/ijms19071981
APA StyleLiu, W., Yuan, J., Liu, Z., Zhang, J., & Chang, J. (2018). Label-Free Quantitative Proteomics Combined with Biological Validation Reveals Activation of Wnt/β-Catenin Pathway Contributing to Trastuzumab Resistance in Gastric Cancer. International Journal of Molecular Sciences, 19(7), 1981. https://doi.org/10.3390/ijms19071981





