The Curcumin Analog CH-5 Exerts Anticancer Effects in Human Osteosarcoma Cells via Modulation of Transcription Factors p53/Sp1
Abstract
1. Introduction
2. Results
2.1. CH-5 Inhibits Cell Viability of Osteosarcoma Cell Lines
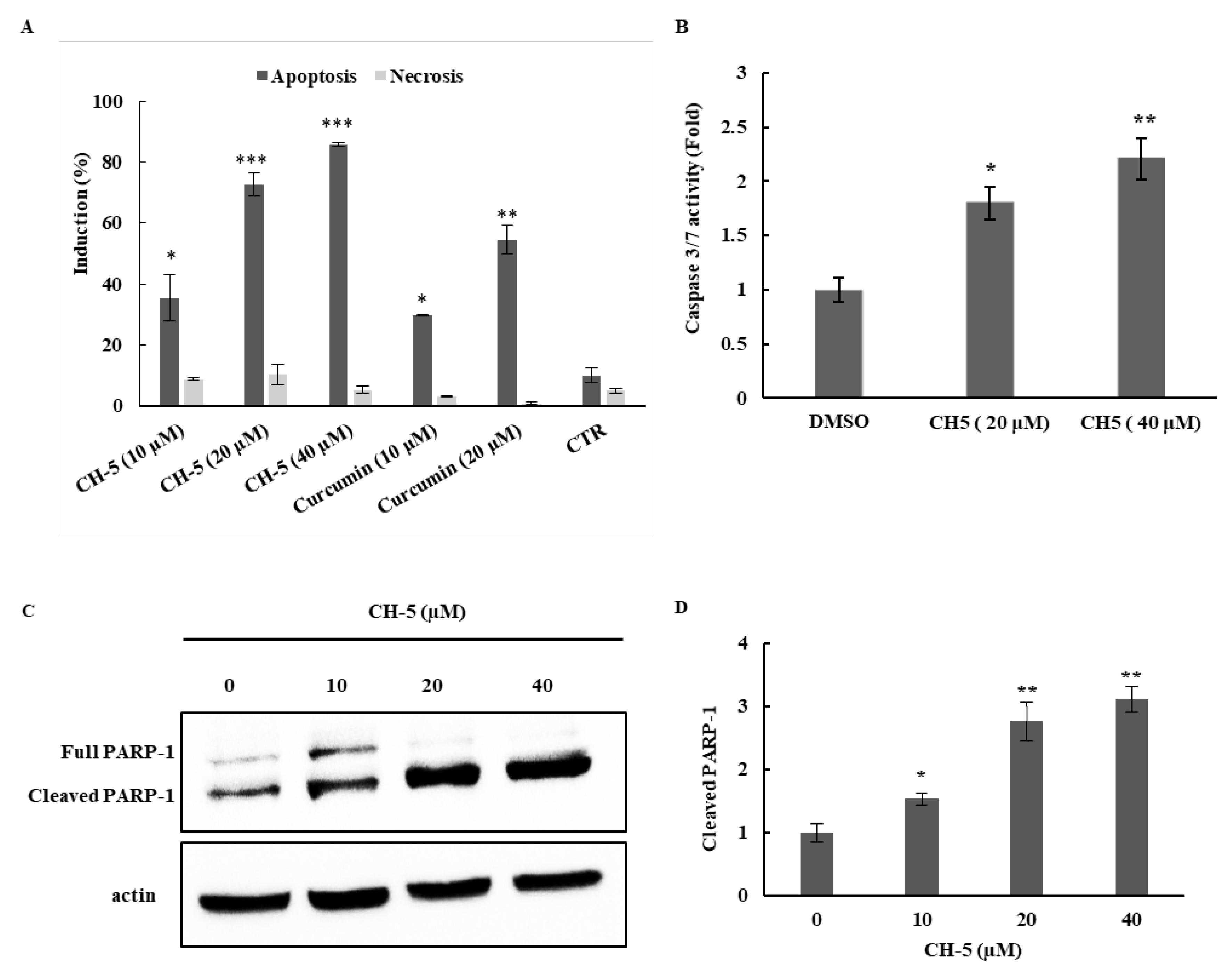
2.2. CH-5 Cytotoxicity Results in Apoptosis Induction
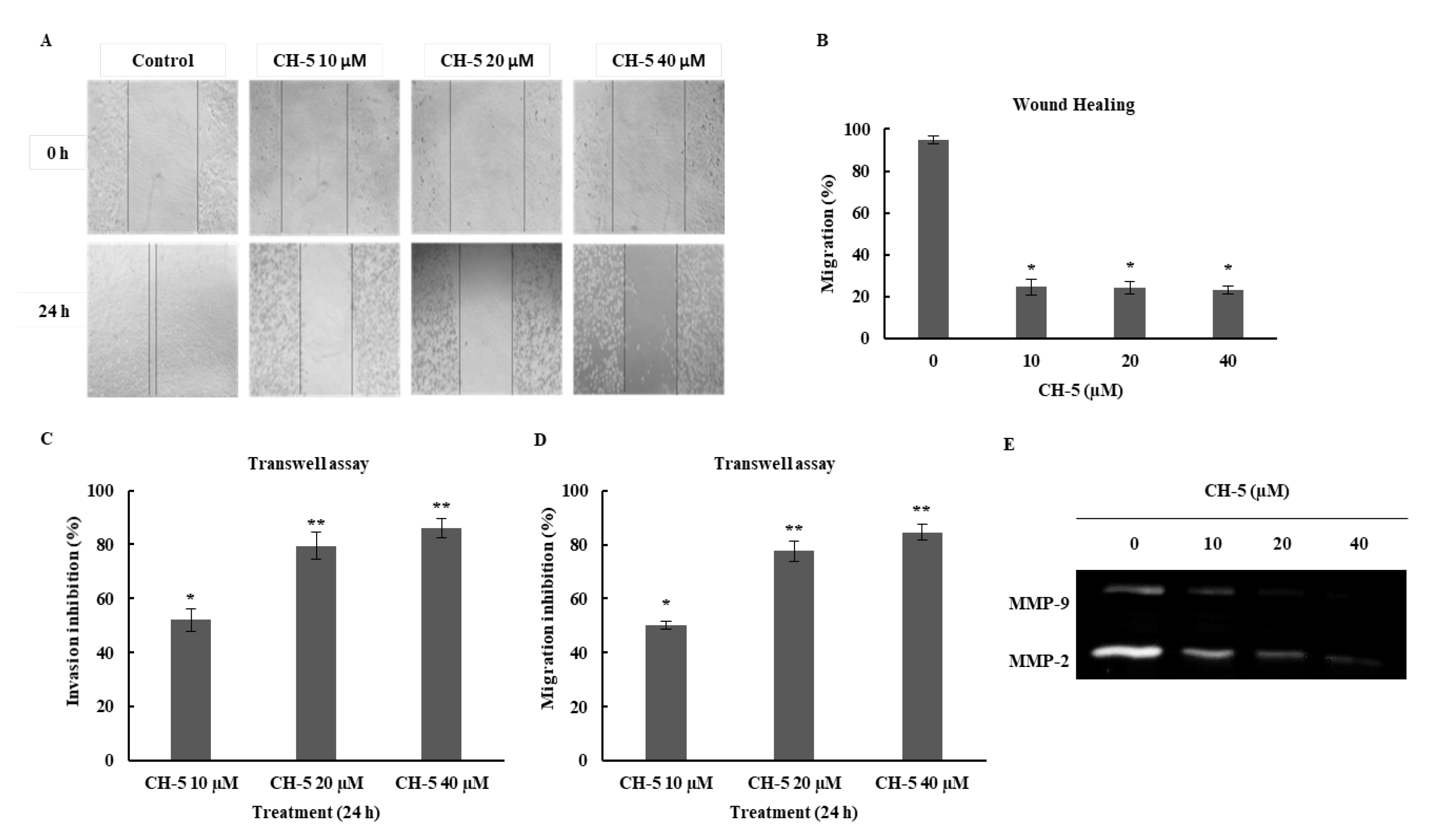
2.3. CH-5 Inhibits Cell Migration and Invasion
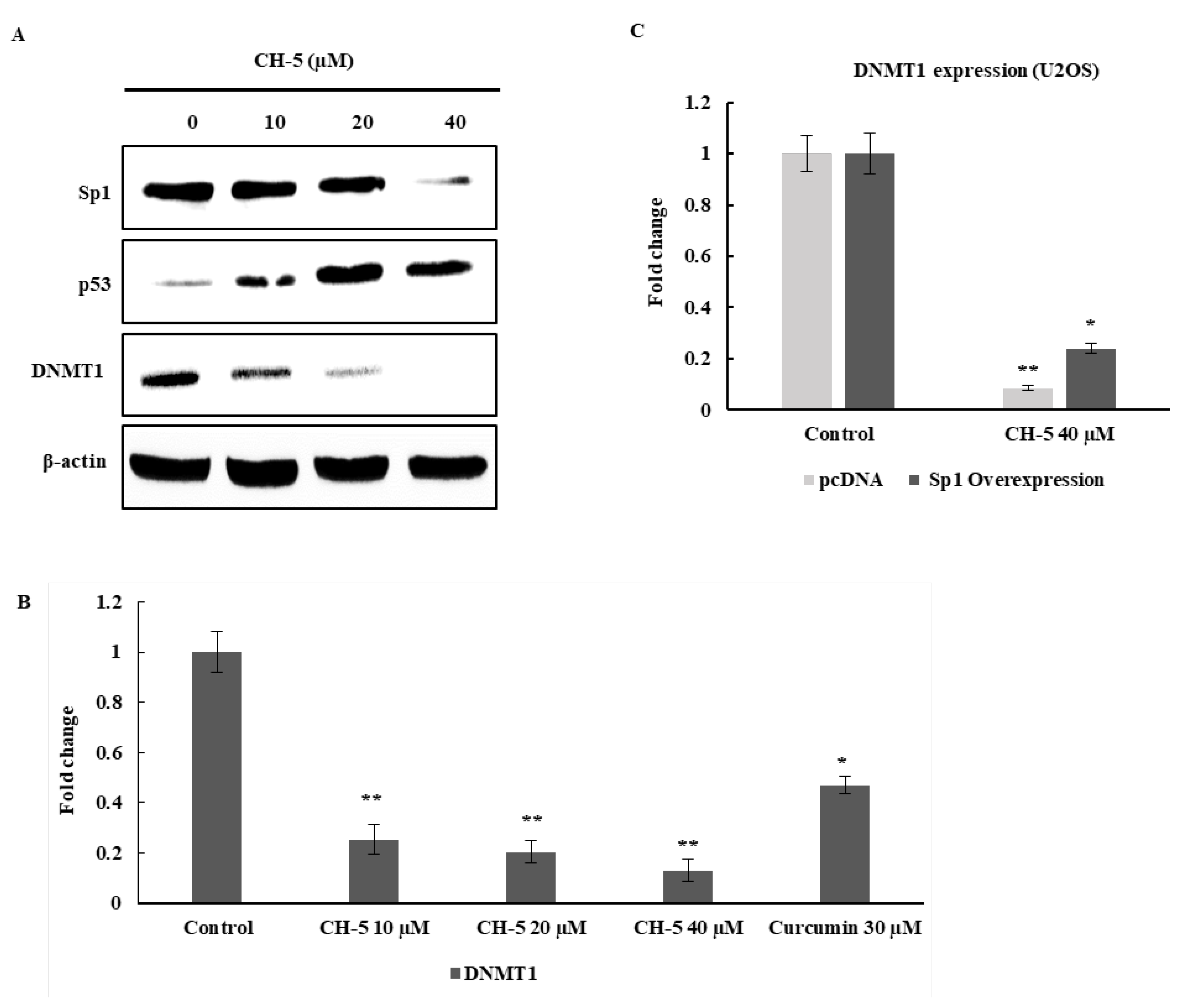
2.4. CH-5 Increases p53 and Reduces Sp1 Protein Levels in U2OS Cells
2.5. The Effect of CH-5 on the p53/Sp1 Axis Affects the Expression of DNA Methyltransferase (DNMT1) Gene
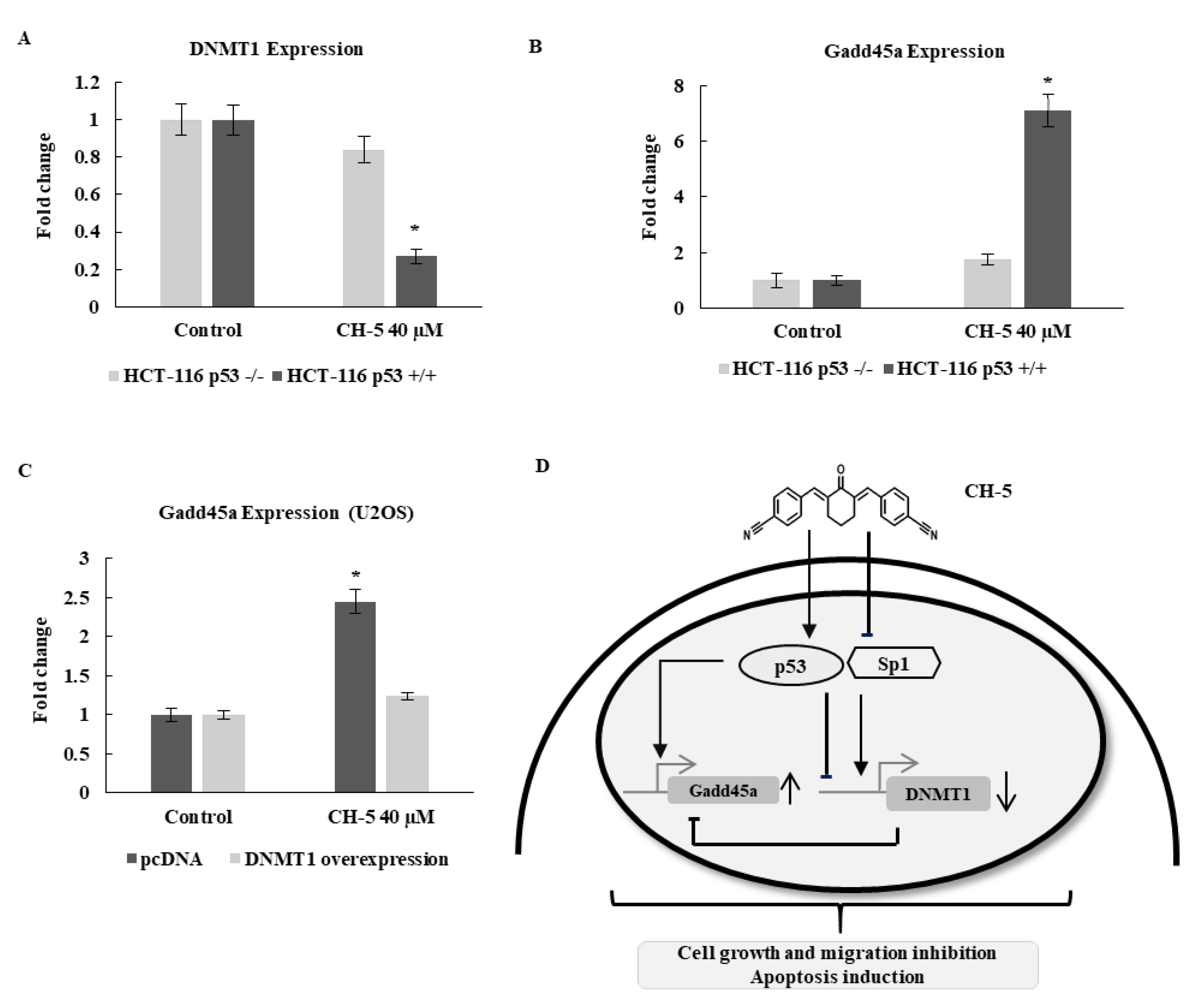
2.6. CH-5 Modulation of Gene Expression is p53-Dependent
2.7. DNMT1 Overexpression Diminishes CH-5-Induced Gene Activation of Gadd45a
3. Discussion
4. Materials and Methods
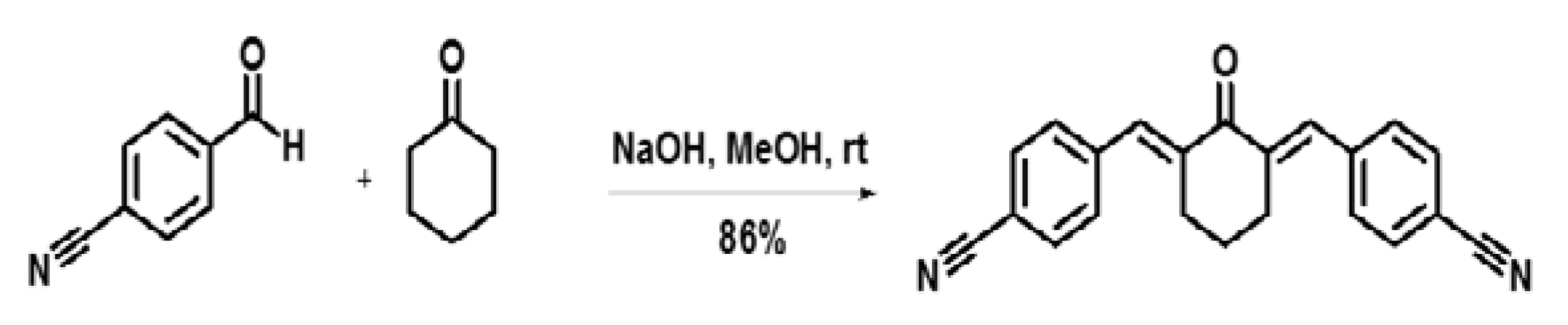
4.1. Synthesis of the Curcumin Analogue CH-5
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. Apoptosis Assay
4.5. Caspase 3/7 Assay
4.6. Wound Healing Assay
4.7. Transwell Assay
4.8. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.9. Gelatin Zymography
4.10. Western Blot
4.11. Expression Vector and Transfection
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, N. Historical Perspective on the Introduction and Use of Chemotherapy for the Treatment of Osteosarcoma. In Current Advances in Osteosarcoma; Kleinerman, E., Ed.; Springer International Publishing: Basel, Switzerland, 2014. [Google Scholar]
- Anderson, M.E. Update on Survival in Osteosarcoma. Orthop. Clin. N. Am. 2016, 47, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, W.; Wang, H.; Zuo, D.; Hua, Y.; Cai, Z. Research progress on the multidrug resistance mechanisms of osteosarcoma chemotherapy and reversal. Tumour Biol. 2015, 36, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Eigner, D.; Scholz, D. Ferula asa-foetida and Curcuma longa in traditional medical treatment and diet in Nepal. J. Ethnopharmacol. 1999, 67, 1–6. [Google Scholar] [CrossRef]
- Anand, P.; Sundaram, C.; Jhurani, S.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin and cancer: An “old-age” disease with an “age-old” solution. Cancer Lett. 2008, 267, 133–164. [Google Scholar] [CrossRef] [PubMed]
- Panahi, Y.; Darvishi, B.; Ghanei, M.; Jowzi, N.; Beiraghdar, F.; Varnamkhasti, B.S. Molecular mechanisms of curcumins suppressing effects on tumorigenesis, angiogenesis and metastasis, focusing on NF-κB pathway. Cytokine Growth Factor Rev. 2016, 28 (Suppl. C), 21–29. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Lee, M.K.; Kim, J.H. Curcumin induces cell cycle arrest and apoptosis in human osteosarcoma (HOS) cells. Anticancer Res. 2009, 29, 5039–5044. [Google Scholar] [PubMed]
- Lin, J.K. Molecular targets of curcumin. Adv. Exp. Med. Biol. 2007, 595, 227–243. [Google Scholar] [PubMed]
- Shishodia, S. Molecular mechanisms of curcumin action: Gene expression. BioFactors 2013, 39, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Bora-Tatar, G.; Dayangac-Erden, D.; Demir, A.S.; Dalkara, S.; Yelekci, K.; Erdem-Yurter, H. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorg. Med. Chem. 2009, 17, 5219–5228. [Google Scholar] [CrossRef] [PubMed]
- Marcu, M.G.; Jung, Y.J.; Lee, S.; Chung, E.J.; Lee, M.J.; Trepel, J.; Neckers, L. Curcumin is an inhibitor of p300 histone acetylatransferase. Med. Chem. 2006, 2, 169–174. [Google Scholar] [PubMed]
- Shu, L.; Khor, T.O.; Lee, J.H.; Boyanapalli, S.S.; Huang, Y.; Wu, T.Y.; Saw, C.L.; Cheung, K.L.; Kong, A.N. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011, 13, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; An, F.; He, X.; Cao, X. Curcumin inhibits the proliferation and invasion of human osteosarcoma cell line MG-63 by regulating miR-138. Int. J. Clin. Exp. Pathol. 2015, 8, 14946–14952. [Google Scholar] [PubMed]
- Ramasamy, T.S.; Ayob, A.Z.; Myint, H.H.; Thiagarajah, S.; Amini, F. Targeting colorectal cancer stem cells using curcumin and curcumin analogues: Insights into the mechanism of the therapeutic efficacy. Cancer Cell Int. 2015, 15, 96. [Google Scholar] [CrossRef] [PubMed]
- Borges, G.A.; Rego, D.F.; Assad, D.X.; Coletta, R.D.; De Luca Canto, G.; Guerra, E.N. In vivo and in vitro effects of curcumin on head and neck carcinoma: A systematic review. J. Oral Pathol. Med. 2017, 46, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, P.; Banerjee, S.; Padhye, S.; Sarkar, F.H.; Iyer, A.K. Parenterally administrable nano-micelles of 3,4-difluorobenzylidene curcumin for treating pancreatic cancer. Colloids Surf. B Biointerfaces 2015, 132, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wang, H.; Yang, F.; Chen, H.; He, W.; Wang, J. Curcumin Promotes Osteosarcoma Cell Death by Activating miR-125a/ERRalpha Signal Pathway. J. Cell. Biochem. 2017, 118, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Tyagi, A.K.; Aggarwal, B.B. Recent developments in delivery, bioavailability, absorption and metabolism of curcumin: The golden pigment from golden spice. Cancer Res. Treat. 2014, 46, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B.C.; Kumar, B.; Thilagavathi, R.; Yadhav, A.; Kumar, P.; Selvam, C. Synthesis, evaluation of cytotoxic properties of promising curcumin analogues and investigation of possible molecular mechanisms. Chem. Biol. Drug Des. 2017, 91, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Teymouri, M.; Barati, N.; Pirro, M.; Sahebkar, A. Biological and pharmacological evaluation of dimethoxycurcumin: A metabolically stable curcumin analogue with a promising therapeutic potential. J. Cell. Physiol. 2018, 233, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Teixeira Lima, F.; Seba, V.; Mendes Lourenco, A.L.; Lucas, T.G.; de Andrade, B.V.; Torrezan, G.S.; Polaquini, C.R.; Garcia, M.E.; Couto, L.B.; et al. Curcumin Analog CH-5 Suppresses the Proliferation, Migration, and Invasion of the Human Gastric Cancer Cell Line HGC-27. Molecules 2018, 23, 279. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Marfell, B.J.; Scott, A.P.; Waterhouse, N.J. Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087288. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.; Desnoyers, S.; Ottaviano, Y.; Davidson, N.E.; Poirier, G.G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: An early marker of chemotherapy-induced apoptosis. Cancer Res. 1993, 53, 3976–3985. [Google Scholar] [PubMed]
- Hwang, C.-I.; Matoso, A.; Corney, D.C.; Flesken-Nikitin, A.; Körner, S.; Wang, W.; Boccaccio, C.; Thorgeirsson, S.S.; Comoglio, P.M.; Hermeking, H.; et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc. Natl. Acad. Sci. USA 2011, 108, 14240–14245. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Jeon, B.N.; Koh, D.I.; Kim, K.S.; Park, S.Y.; Yun, C.O.; Hur, M.W. Regulation of the cyclin-dependent kinase inhibitor 1A gene (CDKN1A) by the repressor BOZF1 through inhibition of p53 acetylation and transcription factor Sp1 binding. J. Biol. Chem. 2013, 288, 7053–7064. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, Y.; Strose, A.; Tedesco, D.; Gurova, K.; Selivanova, G. Integrated high-throughput analysis identifies Sp1 as a crucial determinant of p53-mediated apoptosis. Cell Death Differ. 2014, 21, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Jackson-Grusby, L.; Beard, C.; Possemato, R.; Tudor, M.; Fambrough, D.; Csankovszki, G.; Dausman, J.; Lee, P.; Wilson, C.; Lander, E.; et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001, 27, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.K.; Wu, C.Y.; Chang, J.W.; Juan, L.J.; Hsu, H.S.; Chen, C.Y.; Lu, Y.Y.; Tang, Y.A.; Yang, Y.C.; Yang, P.C.; et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010, 70, 5807–5817. [Google Scholar] [CrossRef] [PubMed]
- Kishikawa, S.; Murata, T.; Kimura, H.; Shiota, K.; Yokoyama, K.K. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur. J. Biochem. 2002, 269, 2961–2970. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Li, G.; Zhan, Q.; Li, D. Gadd45a inhibits cell migration and invasion by altering the global RNA expression. Cancer Boil. Ther. 2012, 13, 1112–1122. [Google Scholar] [CrossRef] [PubMed]
- Asuthkar, S.; Nalla, A.K.; Gondi, C.S.; Dinh, D.H.; Gujrati, M.; Mohanam, S.; Rao, J.S. Gadd45a sensitizes medulloblastoma cells to irradiation and suppresses MMP-9-mediated EMT. Neuro-Oncol. 2011, 13, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Al-Romaih, K.; Sadikovic, B.; Yoshimoto, M.; Wang, Y.; Zielenska, M.; Squire, J.A. Decitabine-induced demethylation of 5′ CpG island in GADD45A leads to apoptosis in osteosarcoma cells. Neoplasia 2008, 10, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.A.; Sang, M.X.; Geng, C.Z.; Wang, S.J.; Shan, B.E. A novel curcumin analogue is a potent chemotherapy candidate for human hepatocellular carcinoma. Oncol. Lett. 2016, 12, 4252–4262. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.K.; Cai, J.; Armstrong, J.; Herold, M.; Lu, Y.J.; Sun, A.; Snyder, J.P.; Liotta, D.C.; Jones, D.P.; Shoji, M. EF24, a novel synthetic curcumin analog, induces apoptosis in cancer cells via a redox-dependent mechanism. Anti-Cancer Drugs 2005, 16, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Shi, Q.; Moore, T.W.; Yoon, Y.; Prussia, A.; Maddox, C.; Liotta, D.C.; Shim, H.; Snyder, J.P. Monocarbonyl Curcumin Analogs: Heterocyclic Pleiotropic Kinase Inhibitors that Mediate Anti-Cancer Properties. J. Med. Chem. 2013, 56, 3456–3466. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Pandya, U.; Singhal, S.S.; Lin, J.T.; Thiviyanathan, V.; Seifert, W.E., Jr.; Awasthi, Y.C.; Ansari, G.A. Curcumin-glutathione interactions and the role of human glutathione S-transferase P1-1. Chem.-Biol. Interact. 2000, 128, 19–38. [Google Scholar] [CrossRef]
- Fang, J.; Lu, J.; Holmgren, A. Thioredoxin reductase is irreversibly modified by curcumin: A novel molecular mechanism for its anticancer activity. J. Biol. Chem. 2005, 280, 25284–25290. [Google Scholar] [CrossRef] [PubMed]
- Addala, E.; Rafiei, H.; Das, S.; Bandy, B.; Das, U.; Karki, S.S.; Dimmock, J.R. 3,5-Bis(3-dimethylaminomethyl-4-hydroxybenzylidene)-4-piperidone and related compounds induce glutathione oxidation and mitochondria-mediated cell death in HCT-116 colon cancer cells. Bioorg. Med. Chem. Lett. 2017, 27, 3669–3673. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Garcia, A.; Hevia, D.; Mayo, J.C.; Gonzalez-Menendez, P.; Coppo, L.; Lu, J.; Holmgren, A.; Sainz, R.M. Thioredoxin 1 modulates apoptosis induced by bioactive compounds in prostate cancer cells. Redox Biol. 2017, 12, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Salah, S.; Ahmad, R.; Sultan, I.; Yaser, S.; Shehadeh, A. Osteosarcoma with metastasis at initial diagnosis: Current outcomes and prognostic factors in the context of a comprehensive cancer center. Mol. Clin. Oncol. 2014, 2, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Erler, J.T.; Bennewith, K.L.; Cox, T.R.; Lang, G.; Bird, D.; Koong, A.; Le, Q.T.; Giaccia, A.J. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 2009, 15, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Kasiappan, R. Natural Products as Mechanism-based Anticancer Agents: Sp Transcription Factors as Targets. Phytother. Res. PTR 2016, 30, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Wang, C.; Yang, D.; Zhang, X.; Wei, Z.; Zhu, Z.; Xu, J.; Hu, Z.; Zhang, Y.; Wang, W.; et al. Curcumin regulates proliferation, autophagy and apoptosis in gastric cancer cells by affecting PI3K and P53 signaling. J. Cell. Physiol. 2017, 233, 4634–4642. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Li, Z.F.; Wang, J.J.; Han, Z.; Liu, Z.; Liu, B.G. Effects of microRNA-374 on proliferation, migration, invasion, and apoptosis of human SCC cells by targeting Gadd45a through P53 signaling pathway. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, L.F.; Wang, Y.; Correa, R.G.; Cho, J.Y.; Libermann, T.A. Blockage of NF-kappaB induces serine 15 phosphorylation of mutant p53 by JNK kinase in prostate cancer cells. Cell Cycle 2005, 4, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Y.K.R.; Kaye, P. An Efficient Catalyst for Aldol Condensation Reactions. J. Turk. Chem. Soc. 2017, 4, 41–48. [Google Scholar] [CrossRef]
- Li, H.; Rauch, T.; Chen, Z.X.; Szabo, P.E.; Riggs, A.D.; Pfeifer, G.P. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Boil. Chem. 2006, 281, 19489–19500. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.J.; Horowitz, J.M.; Eling, T.E. Molecular Cloning and Characterization of Human Nonsteroidal Anti-inflammatory Drug-activated Gene Promoter: BASAL TRANSCRIPTION IS MEDIATED BY Sp1 AND Sp3. J. Biol. Chem. 2001, 276, 33384–33392. [Google Scholar] [CrossRef] [PubMed]

| Cells | CH-5 | Curcumin |
|---|---|---|
| U2OS | 9.0 ± 2.4 | 27.7 ± 3.2 |
| Saos-2 | 11.7 ± 2.4 | 49.7 ± 7.1 |
| MG-63 | 4.4 ± 0.7 | 13.3 ± 1.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima, F.T.; Seba, V.; Silva, G.; Torrezan, G.S.; Polaquini, C.R.; Pinhanelli, V.C.; Baek, S.J.; Fachin, A.L.; Regasini, L.O.; Marins, M. The Curcumin Analog CH-5 Exerts Anticancer Effects in Human Osteosarcoma Cells via Modulation of Transcription Factors p53/Sp1. Int. J. Mol. Sci. 2018, 19, 1909. https://doi.org/10.3390/ijms19071909
Lima FT, Seba V, Silva G, Torrezan GS, Polaquini CR, Pinhanelli VC, Baek SJ, Fachin AL, Regasini LO, Marins M. The Curcumin Analog CH-5 Exerts Anticancer Effects in Human Osteosarcoma Cells via Modulation of Transcription Factors p53/Sp1. International Journal of Molecular Sciences. 2018; 19(7):1909. https://doi.org/10.3390/ijms19071909
Chicago/Turabian StyleLima, Felipe Teixeira, Viviane Seba, Gabriel Silva, Guilherme Silva Torrezan, Carlos Roberto Polaquini, Vitor Caressato Pinhanelli, Seung J. Baek, Ana Lúcia Fachin, Luis Octavio Regasini, and Mozart Marins. 2018. "The Curcumin Analog CH-5 Exerts Anticancer Effects in Human Osteosarcoma Cells via Modulation of Transcription Factors p53/Sp1" International Journal of Molecular Sciences 19, no. 7: 1909. https://doi.org/10.3390/ijms19071909
APA StyleLima, F. T., Seba, V., Silva, G., Torrezan, G. S., Polaquini, C. R., Pinhanelli, V. C., Baek, S. J., Fachin, A. L., Regasini, L. O., & Marins, M. (2018). The Curcumin Analog CH-5 Exerts Anticancer Effects in Human Osteosarcoma Cells via Modulation of Transcription Factors p53/Sp1. International Journal of Molecular Sciences, 19(7), 1909. https://doi.org/10.3390/ijms19071909








