Regulation of Root Development and Architecture by Strigolactones under Optimal and Nutrient Deficiency Conditions
Abstract
:1. Introduction
2. Role of SLs in Root System Development, under Optimal Growth Conditions
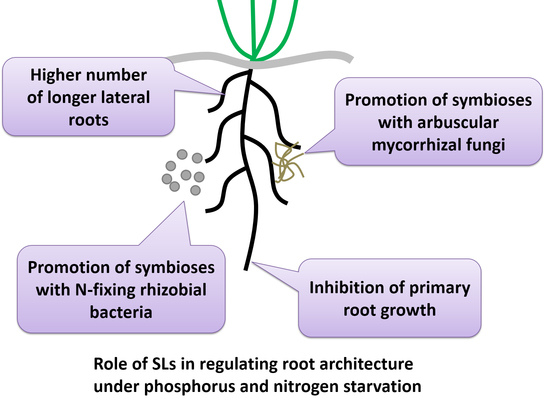
3. Role of SLs in Regulating Root Architecture under Stress Conditions
4. How SLs Influence on Root Development
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pages, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of Witchweed (Striga lutea Lour.): Isolation and Properties of a Potent Stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tao, J.; Gu, P.; Xu, G.; Zhang, Y. The role of strigolactones in root development. Plant Signal. Behav. 2016, 11, e1110662. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Davies, N.W. Strigolactones promote nodulation in pea. Planta 2011, 234, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Furusawa, S.; Nagasaka, S.; Shimomura, K.; Yamaguchi, S.; Umehara, M. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 2014, 240, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Ito, K.; Abeta, N.; Takahashi, R.; Sasaki, Y.; Yajima, S. Effects of strigolactone signaling on Arabidopsis growth under nitrogen deficient stress condition. Plant Signal. Behav. 2016, 11, e1126031. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Muszynska, A.; Gruszka, D. The role of strigolactones in nutrient-stress responses in plants. Int. J. Mol. Sci. 2013, 14, 9286–9304. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Sharma, M.; Pandey, G.K. Emerging Roles of Strigolactones in Plant Responses to Stress and Development. Front. Plant Sci. 2016, 7, 434. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Lv, T.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.; Huang, Z.; Xiao, L.; Engineer, C.; Kim, T.H.; et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.V.; Leyva-Gonzalez, M.A.; Osakabe, Y.; Tran, U.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Yamaguchi, S.; Dong, N.V.; et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA 2014, 111, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; He, H.; Vitali, M.; Visentin, I.; Charnikhova, T.; Haider, I.; Schubert, A.; Ruyter-Spira, C.; Bouwmeester, H.J.; Lovisolo, C.; et al. Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: Exploring the interaction between strigolactones and ABA under abiotic stress. Planta 2015, 241, 1435–1451. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreno, A.M.; Molina, S.; Andreo-Jimenez, B.; Porcel, R.; Garcia-Mina, J.M.; Ruyter-Spira, C.; Lopez-Raez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 2016, 39, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Visentin, I.; Vitali, M.; Ferrero, M.; Zhang, Y.; Ruyter-Spira, C.; Novak, O.; Strnad, M.; Lovisolo, C.; Schubert, A.; Cardinale, F. Low levels of strigolactones in roots as a component of the systemic signal of drought stress in tomato. New Phytol. 2016, 212, 954–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Nguyen, K.H.; Chu, H.D.; Ha, C.V.; Watanabe, Y.; Osakabe, Y.; Leyva-González, M.A.; Sato, M.; Toyooka, K.; Voges, L.; et al. The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1007076. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Muszynska, A. In silico analysis of the genes encoding proteins that are involved in the biosynthesis of the RMS/MAX/D pathway revealed new roles of Strigolactones in plants. Int. J. Mol. Sci. 2015, 16, 6757–6782. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M. Strigolactones as part of the plant defence system. Trends Plant Sci. 2016, 21, 900–903. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.; Rivers, J.; Leon, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and Function of Apocarotenoid Signals in Plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Seto, Y.; Sado, A.; Asami, K.; Hanada, A.; Umehara, M.; Akiyama, K.; Yamaguchi, S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA 2014, 111, 1640–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yoneyama, K.; Kisugi, T.; Uchida, K.; Ito, S.; Akiyama, K.; Hayashi, H.; Yokota, T.; Nomura, T.; Yoneyama, K. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant 2013, 6, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H.; Prandi, C. Strigolactones: Past, present and future. Planta 2016, 243, 1309. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone Signaling and Evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M. Perception and Signaling of Strigolactones. Front. Plant Sci. 2016, 7, 1260. [Google Scholar] [CrossRef] [PubMed]
- Hamiaux, C.; Drummond, R.S.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Xue, Y.L.; Miyakawa, T.; Hou, F.; Qin, H.M.; Fukui, K.; Shi, X.; Ito, E.; Ito, S.; Park, S.H.; et al. Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 2013, 4, 2613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signaling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, T.; Wang, M.; Liu, Y.; Yuan, S.; Gao, Y.; Yin, L.; Sun, W.; Peng, L.; Zhang, W.; et al. DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 2014, 55, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, B.; Jiang, L.; Liu, X.; Li, X.; Lu, Z.; Meng, X.; Wang, Y.; Smith, S.M.; Li, J. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell 2015, 27, 3128–3142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012, 139, 1285–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scaffidi, A.; Waters, M.T.; Sun, Y.K.; Skelton, B.W.; Dixon, K.W.; Ghisalberti, E.L.; Flematti, G.R.; Smith, S.M. Strigolactone Hormones and Their Stereoisomers Signal through Two Related Receptor Proteins to Induce Different Physiological Responses in Arabidopsis. Plant Physiol. 2014, 165, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Delaux, P.M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattacharya, C.; Sejalon-Delmas, N.; Combier, J.P.; Becard, G.; Belausov, E.; et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; van Zeijl, A.; van Bezouwen, L.; de Ruijter, N.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011, 155, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Fang, J.; Lou, L.; Zhao, J.; Yuan, S.; Yin, L.; Sun, W.; Peng, L.; Guo, B.; Li, X. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE 2015, 10, e0118169. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Gruszka, D.; Tylec, P.; Szarejko, I. Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiol. Plant 2016, 158, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Novero, M.; Charnikhova, T.; Ferrandino, A.; Schubert, A.; Ruyter-Spira, C.; Bonfante, P.; Lovisolo, C.; Bouwmeester, H.J.; Cardinale, F. Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J. Exp. Bot. 2013, 64, 1967–1981. [Google Scholar] [CrossRef] [PubMed]
- De Cuyper, C.; Fromentin, J.; Yocgo, R.E.; De Keyser, A.; Guillotin, B.; Kunert, K.; Boyer, F.D.; Goormachtig, S. From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula. J. Exp. Bot. 2015, 66, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Arite, T.; Kameoka, H.; Kyozuka, J. Strigolactone positively controls crown root elongation in rice. J. Plant Growth Regul. 2012, 31, 165–172. [Google Scholar] [CrossRef]
- Koltai, H.; Dor, E.; Hershenhorn, J.; Joel, D.M.; Weininger, S.; Lekalla, S.; Shealtiel, H.; Bhattacharya, C.; Eliahu, E.; Resnick, N.; et al. Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J. Plant Growth Regul. 2010, 29, 129–136. [Google Scholar] [CrossRef]
- Sun, H.; Tao, J.; Liu, S.; Huang, S.; Chen, S.; Xie, X.; Yoneyama, K.; Zhang, Y.; Xu, G. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 2014, 65, 6735–6746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, A.; Mason, M.G.; De Cuyper, C.; Brewer, P.B.; Herold, S.; Agusti, J.; Geelen, D.; Greb, T.; Goormachtig, S.; Beeckman, T.; et al. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 2012, 158, 1976–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Tao, J.; Hou, M.; Huang, S.; Chen, S.; Liang, Z.; Xie, T.; Wei, Y.; Xie, X.; Yoneyama, K.; et al. A strigolactone signal is required for adventitious root formation in rice. Ann. Bot. 2015, 115, 1155–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Bucio, J.; Cruz-Ramirez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Niu, Y.F.; Chai, R.S.; Jin, G.L.; Wang, H.; Tang, C.X.; Zhang, Y.S. Responses of root architecture development to low phosphorus availability: A review. Ann. Bot. 2013, 112, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Asami, T. Target sites for chemical regulation of strigolactone signaling. Front. Plant Sci. 2014, 5, 623. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.; Albertos, P.; Mateos, I.; Sanchez-Vicente, I.; Lechon, T.; Fernandez-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Bi, Y.; Tao, J.; Huang, S.; Hou, M.; Xue, R.; Liang, Z.; Gu, P.; Yoneyama, K.; Xie, X.; et al. Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant Cell Environ. 2016, 39, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Xie, X.; Kusumoto, D.; Sekimoto, H.; Sugimoto, Y.; Takeuchi, Y.; Yoneyama, K. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 2007, 227, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Raez, J.A.; Charnikhova, T.; Fernandez, I.; Bouwmeester, H.; Pozo, M.J. Arbuscular mycorrhizal symbiosis decreases strigolactone production in tomato. J. Plant Physiol. 2011, 168, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Yoneyama, K.; Hugill, C.J.; Quittenden, L.J.; Reid, J.B. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant 2013, 6, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Kohlen, W.; Charnikhova, T.; Liu, Q.; Bours, R.; Domagalska, M.A.; Beguerie, S.; Verstappen, F.; Leyser, O.; Bouwmeester, H.; Ruyter-Spira, C. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011, 155, 974–987. [Google Scholar] [CrossRef] [PubMed]
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010, 51, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, N.; Taylor, C.; Leyser, O. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 2013, 11, e1001474. [Google Scholar] [CrossRef] [PubMed]
- Pandya-Kumar, N.; Shema, R.; Kumar, M.; Mayzlish-Gati, E.; Levy, D.; Zemach, H.; Belausov, E.; Wininger, S.; Abu-Abied, M.; Kapulnik, Y.; et al. Strigolactone analog GR24 triggers changes in PIN2 polarity, vesicle trafficking and actin filament architecture. New Phytol. 2014, 202, 1184–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Matthys, C.; Marquez-Garcia, B.; de Cuyper, C.; Smet, L.; de Keyser, A.; Boyer, F.-D.; Beeckman, T.; Depuydt, S.; Goormachtig, S. Strigolactones spatially influence lateral root development through the cytokinin signaling network. J. Exp. Bot. 2016, 67, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, A.; Hu, Y.; Depaepe, T.; Vandenbussche, F.; Boyer, F.-D.; Straeten, D.V.D.; Geelen, D. Ethylene controls adventitious root initiation sites in Arabidopsis hypocotyls independently of strigolactones. J. Plant Growth Regul. 2017, 36, 897–911. [Google Scholar] [CrossRef]
- Walton, A.; Stes, E.; Goeminne, G.; Braem, L.; Vuylsteke, M.; Matthys, C.; de Cuyper, C.; Staes, A.; Vandenbussche, J.; Boyer, F.D.; et al. The Response of the Root Proteome to the Synthetic Strigolactone GR24 in Arabidopsis. Mol. Cell Proteom. 2016, 15, 2744–2755. [Google Scholar] [CrossRef] [PubMed]
- Maloney, G.S.; DiNapoli, K.T.; Muday, G.K. The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 2014, 166, 614–631. [Google Scholar] [CrossRef] [PubMed]
- Santelia, D.; Henrichs, S.; Vincenzetti, V.; Sauer, M.; Bigler, L.; Klein, M.; Bailly, A.; Lee, Y.; Friml, J.; Geisler, M.; et al. Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J. Biol. Chem. 2008, 283, 31218–31226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.N.; Kim, J.H.; Hyun, W.Y.; Nguyen, N.T.; Hong, S.W.; Lee, H. TTG1-mediated flavonols biosynthesis alleviates root growth inhibition in response to ABA. Plant Cell Rep. 2013, 32, 503–514. [Google Scholar] [CrossRef] [PubMed]
| Investigated Species | Tools | Role of SLs | Reference |
|---|---|---|---|
| Primary root (PR)/seminal root (SR) length | |||
| Solanum lycopersicon L. | racGR24 | Did not influence root length, reversed the suppressive effect of IAA | [42] |
| Arabidopsis thaliana L. | SL biosynthesis (max3-11, max4-1) and signalling (max2-1), racGR24 | No effect under optimal conditions, promotion of PR length under carbohydrate starvation | [36] |
| Oryza sativa L. | SL biosynthesis (d10, d17, d27) and signalling (d3, d14) mutants, racGR24 | Lack of effect on SR length | [41] |
| Oryza sativa L. | racGR24 | Promotion of SR elongation | [43] |
| Hordeum vulgare L. | SL insensitive mutant (hvd14.d), racGR24 | Promotion of SR growth | [38] |
| Lotus japonicus L. | RNAi transgenic line for LjCCD7 | Inhibition of PR | [39] |
| Lateral root (LR) density | |||
| Arabidopsis thaliana L. | SL biosynthesis (max3-11, max4-1) and signalling (max2-1) mutants, racGR24 | Inhibition of LR density | [35] |
| Arabidopsis thaliana L. | SL biosynthesis (max3-11, max4-1) and signalling (max2-1) mutants, racGR24 | Inhibition of LR density | [36] |
| Arabidopsis thaliana L. | SL signalling (max2) mutant, racGR24 | Inhibition of LR initials outgrowth | [37] |
| Oryza sativa L. | SL biosynthesis (d10) and signalling (d14) mutants | No effect in LR density | [41] |
| Oryza sativa L. | racGR24 | Inhibition of LR density | [43] |
| Hordeum vulgare | SL insensitive mutant (hvd14.d), racGR24 | Inhibition of LR density | [38] |
| Lotus japonicus L. | RNAi transgenic line for LjCCD7 | Inhibition of LR density | [39] |
| Medicago truncatula Gaertn. | racGR24 | Inhibition of LR density | [40] |
| Crown root (CR) length | |||
| Oryza sativa L. | SL biosynthesis (d10) and signalling (d14) mutants | Promotion of CR growth | [41] |
| Adventitious root (AR) formation | |||
| Arabidopsis thaliana L. | SL biosynthesis (max1, max3, max4) and signalling (max2), racGR24 | Negative effect of AR formation | [44] |
| Pisum sativum L. | SL biosynthesis (rms1, rms5) and signalling (rms4), racGR24 | Negative effect of AR formation | [44] |
| Oryza sativa L. | SL biosynthesis (d10, d17, d27) and signalling (d3, d14) mutants, racGR24 | Promotion of AR formation | [45] |
| Applied Stress | Investigated Species | Obtained Results | Reference |
|---|---|---|---|
| P-deficiency (20 µM) | Arabidopsis thaliana L. | Increased number of LRs in wild-type under stress conditions, whereas SL mutants contained more LR primordia with arrested outgrowth. | [36] |
| N-deficiency (0.02 mM) | Oryza sativa L. | Under stress promotion of SR elongation was significantly lower in SL mutants in comparison to wild-type; treatment with racGR24 increased SR elongation in SL biosynthesis mutants to the values observed in wild-type, which was not the case in SL signalling mutant. | [43] |
| P-deficiency (2 µM) | |||
| N-deficiency (0.02 mM) | Oryza sativa L. | Treatment with SL biosynthesis inhibitor stopped elongation of SRs in wild-type plants under stress conditions. | [5] |
| P-deficiency (2 µM) | |||
| Phosphate-removed ½ MS | Oryza sativa L. | Elongation of CRs in wild-type under P-deficiency, not observed in SL mutants. | [41] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzec, M.; Melzer, M. Regulation of Root Development and Architecture by Strigolactones under Optimal and Nutrient Deficiency Conditions. Int. J. Mol. Sci. 2018, 19, 1887. https://doi.org/10.3390/ijms19071887
Marzec M, Melzer M. Regulation of Root Development and Architecture by Strigolactones under Optimal and Nutrient Deficiency Conditions. International Journal of Molecular Sciences. 2018; 19(7):1887. https://doi.org/10.3390/ijms19071887
Chicago/Turabian StyleMarzec, Marek, and Michael Melzer. 2018. "Regulation of Root Development and Architecture by Strigolactones under Optimal and Nutrient Deficiency Conditions" International Journal of Molecular Sciences 19, no. 7: 1887. https://doi.org/10.3390/ijms19071887
APA StyleMarzec, M., & Melzer, M. (2018). Regulation of Root Development and Architecture by Strigolactones under Optimal and Nutrient Deficiency Conditions. International Journal of Molecular Sciences, 19(7), 1887. https://doi.org/10.3390/ijms19071887