Abstract
Since their discovery in 1993, numerous microRNAs (miRNAs) have been identified in humans and other eukaryotic organisms, and their role as key regulators of gene expression is still being elucidated. It is now known that miRNAs not only play a central role in the processes that ensure normal development and physiology, but they are often dysregulated in various diseases. In this review, we present an overview of the role of miRNAs in normal renal development and physiology, in maladaptive renal repair after injury, and in the pathogenesis of renal parenchymal diseases. In addition, we describe methods used for their detection and their potential as therapeutic targets. Continued research on renal miRNAs will undoubtedly improve our understanding of diseases affecting the kidneys and may also lead to new therapeutic agents.
1. Introduction
Ribonucleic Acids (RNAs) are a ubiquitous class of unbranched polymeric molecules that serve as intermediates responsible for decoding genetic information from DNA to ribosomes in the form of messenger RNA (mRNAs), transfer of amino acids to ribosomes by transfer RNA (tRNA), and in protein synthesis itself in the form of ribosomal RNA (rRNA). In 1993, a new class of non-coding RNA molecule, microRNAs (miRNAs), was identified. These small molecules play pivotal roles in cell-growth cycle regulation, differentiation, and survival by modulating mRNA stability and translational efficiency. Over the last two decades, the role of miRNAs in various diseases, as well as their role in maladaptive repair, has been elucidated. In addition, miRNAs have been studied for their potential use in disease diagnosis and prognostication, and as therapeutic targets.
In this review, we describe the role of miRNAs in renal physiology and pathology and their putative roles in various renal parenchymal diseases. We also discuss methods of their measurement as well as various strategies for using miRNAs as therapeutic agents.
2. Search Strategy
To identify pertinent studies for this review, we searched Medline for phrases “MicroRNAs” [Mesh] OR “MIRN632 microRNA, human” [Supplementary Concept] AND “Kidney” [Mesh]. We limited our search to papers published in the English language journals from January 2007 to June 2018. We also identified relevant studies from the previously published review papers in this field.
3. miRNA Discovery and Biogenesis
Before 1993, it was thought that mRNAs, transcribed from the coding regions of DNA, are translated by ribosomes into proteins with no other non-coding regions of DNA playing a significant role in the processes of protein synthesis or regulation of gene expression. The processes that lead to miRNA biogenesis have been extensively described and we refer readers to comprehensive reviews for details [1,2,3,4]. At the time of writing this review, more than 28,000 miRNAs have been identified, out of which 2588 are human. A central repository of known miRNA sequences has been established (miRBase) [5].
4. Role of miRNAs in Renal Development
In mammals, renal development is heralded by aggregation of cells in the mesoderm called the urogenital ridge—a portion of which forms the nephrogenic cord. The nephrogenic cord undergoes changes to form the pronephros which regresses, mesonephros which forms the mesonephric duct, and the metanephros which develops as an outgrowth of cells from the mesonephric duct called the ureteric bud. Nephron progenitor cells surround the ureteric bud and form nephrons. The ureteric bud forms the collecting system of the kidneys. These processes involve a series of well-coordinated steps which require expression and suppression of various proteins that have been shown to be regulated by miRNAs.
To better understand the role played by miRNAs in the developing kidney, researchers conditionally deleted Dicer in specific cell populations in the developing kidney. A mouse conditional deletion of Dicer in the nephron progenitor cells resulted in increased apoptosis with a marked reduction of cells and increased expression of pro-apoptotic Bcl-2-Like Protein 11 (BIM) protein in the embryonic kidney [6]. Nephron number was also substantially reduced. Few nephrons that were present exhibited capillary wall abnormalities. Expression of mmu-miR-10a, mmu-miR-17-5p, and mmu-miR-106b was reduced. Suppression of these miRNAs has been associated with increased pro-apoptotic BIM protein activity [7]. Thus, nephron progenitor cell growth is encouraged by miRNA-mediated inhibition of the apoptotic protein, BIM. Similarly, a mouse conditional deletion of Dicer in the ureteric bud caused disruption of ciliogenesis, resulting in small and cystic kidneys and obstruction of urinary flow leading to hydronephrosis [8].
Although these observations highlight the relevance of miRNA regulation in renal development, the identity of the miRNAs and cell populations involved remains unclear. The first question was addressed by targeted deletion of the mmu-miR-17~92 cluster, which has essential roles in development and has been implicated in cancer. Deletion of this family of miRNAs resulted in the preservation of the nephron progenitor population but impaired their proliferation and thus reduced nephron number. Mice lacking the miR-17~92 cluster developed albuminuria by 6 weeks, and focal podocyte effacement and glomerulosclerosis by 3 months [9]. Ablation of Dicer from maturing renal tubular epithelial cells reduced mmu-miR-200 cluster expression levels and upregulated the polycystic kidney disease 1 (PKD1) gene. Predictably, enhanced PKD1 is associated with inhibition of tubulogenesis and cyst formation [10]. In the same study, it was shown that the PKD1 gene was downregulated by miR-mmu-miR-200b/c using a variety of in vitro approaches. Please note that the miR-200 cluster plays a regulatory role in the epithelial-to-mesenchymal transition (EMT) process which is central to fibrotic pathogenesis, suggesting that developmentally relevant miRNAs may also play important roles in the initiation and progression of kidney disease in the adult life. This concept was validated in another series of experiments, which examined the effects of selective inactivation of Dicer in mouse podocytes early in life [11,12]. This highly specific lesion caused proteinuria and death with renal histology showing foot process effacement, collapsing glomerulopathy, podocyte vacuolization, hypertrophy, and apoptosis. These histological features are also observed in severe forms of the nephrotic syndrome in adult animals and humans (e.g., due to focal segmental glomerulopathy). Specifically inactivating Drosha in podocytes led to collapsing glomerulopathy similar to Dicer knock-out mice [13].
Table 1 summarizes data on the role of miRNAs in renal development. Collectively, these results suggest that the establishment of renal structure and maintenance of kidney architecture is dependent on expression of multiple miRNAs expressed in different compartments within the kidney.

Table 1.
Summary of the studies investigating the role of miRNAs in renal development.
5. Role of miRNAs in Renal Physiology
miRNAs play a diverse role in normal renal function, as demonstrated by the elimination of specific miRNAs and/or miRNA-processing enzymes in mouse models. For example, conditional deletion of Dicer in renin-expressing cells in mouse kidneys resulted in a reduced juxtaglomerular cell population, and decreased expression of Ren1 and Ren2 genes, leading to decreased renin concentration, hypotension, abnormal renal function, renal vascular abnormalities and strip fibrosis [14]. Conditional deletion of Dicer in podocytes in the prenatal period not only affects normal renal development but also leads to both structural and functional aberrations after nephrogenesis [11]. A major physiological derangement in progressive renal impairment is the inability to fine tune the balance between the excretion of sodium and conservation of potassium. Such alterations underlie the sodium and potassium retention seen in progressive kidney disease in humans. In that regards, it has been shown that specific miRNAs are involved in fluid and electrolyte handling. A mouse model with selective mmu-miR-192-5p knock-out in the proximal convoluted tubule, the site of the fine regulation of sodium balance in the kidney, exhibits upregulation of the Na+/K+ ATPase β-1 subunit [15]. These animals were unable to increase urine output when fed a high sodium diet [15]. This failure of the adaptive mechanism of sodium natriuresis could contribute to sodium and water retention, which is a common pathophysiological alteration in human kidney disease. microRNAs are also involved in the tight co-regulation of sodium excretion by the kidney in the feed-forward (FF) inhibitory control loops of the with No Lysine kinase system (WNK).
This system is of emerging importance for understanding the development of systemic, volume-sensitive hypertension. Control of the system of miRNAs exemplifies the integration between FF kinase and epigenetic regulatory loops and thus will be examined at some length here (Figure 1). In the normal state, this system ensures renal switching of roles from inter-meal sodium retention to post-meal sodium (natriuresis) and potassium (kaluresis) excretory states. WNK3 upregulates expression of the NaCl cotransporter (NCC) in the distal convoluted tubule of the nephron resulting in sodium retention. On the other hand, natriuresis is mediated by WNK4, which antagonizes WNK3 and decreases NCC expression. WNK4 also increases the expression of renal outer medullary potassium (ROMK) channels in the distal convoluted tubules, thus promoting kaluresis. WNK1 exerts a major regulatory role in switching between the phenotypes of sodium retention and natriuresis by cleaving WNK4, which in turn removes the antagonism on WNK3 mediated sodium retention. It has been shown that mmu-miR-192-5p negatively regulates WNK1, as sodium depletion, aldosterone infusion, and potassium load led to significant kidney-specific WNK1 mRNA expression and reduction in mmu-miR-192-5p expression [16]. This study, in addition to the miR-192 antagonism results presented previously [15], highlights the potential of miRNAs to serve as context-specific regulators: sodium depletion led to a decreased mmu-miR-192-5p level which was associated with decreased urine output. On the other hand, antagonism of mmu-miR-192-5p by a specific antagomir affected urine output only in the setting of high, but not normal salt intake [15]. Hence a single miRNA (mmu-miR-192-5p) appears to play a major regulatory role in one of the most tightly controlled kinase systems in the kidney. Renal potassium handling may be directly controlled by miRNAs independently of effects on the WNK system. High-potassium diet increased mmu-miR-802-5p transcription in the cortical collecting duct in mice, which in turn decreased expression of caveolin-1, which suppresses ROMK activity [17]. mmu-miR-9-5p and mmu-miR-374-5p suppress claudin-14 which in turn suppresses claudin-16 and 19 paracellular cation channels responsible for Ca absorption in the thick ascending limb of the loop of Henle, a major site of sodium, potassium and calcium exchange in the kidney [18]. Extracellular calcium levels also directly regulate mmu-miR-9-5p and mmu-miR-374-5p levels [18].
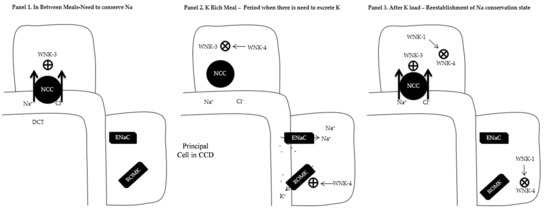
Figure 1.
Overview of the with no Lysine Kinase (WNK) system. Abbreviations: NCC: Sodium/Chloride cotransporter; DCT: Distal Convoluted Tubule; CCD: Cortical Collecting Duct; ENaC; Epithelial Sodium Channel; ROMK: Renal Outer Medullary Potassium Channel; ⊕ Increase expression; ⨂ Decrease expression. (Panel 1) In between meals when the kidney retains Na+ and K+. This is mediated by the presence of WNK3 which increases the expression of NCC in the DCT as well as prevents ROMK expression in the CCD. (Panel 2) K+ rich meal period when there is need to excrete K+. Expression of WNK4 causes suppression of WNK3 which leads to diminished presence of NCC in the DCT and increased Na+ delivery to CCD. In the presence of aldosterone, ENaCs are expressed in the CCD with electrogenic Na absorption making the lumen negative. WNK4 increases the expression of ROMK in the CCD with the removal of K down the electrical gradient. (Panel 3) After K rich meal period. WNK1 antagonizes WNK4 with re-expression to WNK3 phenotype (Panel 1).
Table 2 summarizes key studies investigating the role played by miRNAs in maintaining renal control over sodium and potassium handling. It is evident that miRNAs provide an extra level of complexity and integration of the different systems that maintain the electroneutrality of urine on the one hand while maintaining homeostasis on the other.

Table 2.
Summary of studies investigating the role of miRNAs in renal physiology.
6. Role of miRNAs in Renal Fibrosis and Maladaptive Repair
Renal fibrosis is the final common pathway of various forms of progressive renal disease. TGF-β signaling plays a central role in renal fibrosis. Renal parenchymal cells synthesize TGF-β1 and its isoforms (β2 and β3). Experimental models and human studies have shown that TGF-β1 is upregulated in diseased and fibrotic kidneys [19]. Various stressors inducing stimuli such as hyperglycemia [20], angiotensin II [21], and reactive oxygen species [22] increase TGF-β1 production. It is then activated and exerts its effects in autocrine and paracrine fashion via Smad-dependent and/or Smad-independent pathways [23]. TGF-β1 initiates Smad2 and Smad3 complex formation with Smad4, leading to its activation, translocation to the nucleus, and ultimately transcription of its targets [23]. It is important to note that Smads2 and 3 can also be activated by mediators other than TGF-β [24]. Noting the role of TGF-β in renal fibrosis, we will now discuss the interplay between TGF-β and various miRNAs.
miRNAs regulate TGF-β activity by modulating expression of various components of the TGF-β signaling pathway. In particular, hsa-miR-744-5p has been shown to post-transcriptionally inhibit expression of the TGF-β1 ligand [25]. Similarly, rno-miR-200a-5p has been shown to repress expression of TGF-β2, which prevents renal fibrogenesis [26].
Conversely, TGF-β signaling also influences miRNA expression. TGF-β administration increases expression of miR-192-5p in human, mouse, and rat tubular epithelial, mouse mesangial, and rat proximal tubular epithelial cells, respectively [27,28,29,30]. Low hsa-miR-192-5p is associated with interstitial fibrosis and tubular atrophy [31]. TGF-β is well known to regulate the expression of the miR-200 family of miRNAs, and administration of TGF-β does indeed lead to decreased expression of the miR-200 family in kidney and rat proximal tubular epithelial cells [26].
The final common pathway of TGF-β signaling is the production of extracellular matrix (ECM) proteins and their deposition into the interstitium. Several lines of evidence demonstrate that this process is under miRNA control. In systemic sclerosis—a disease characterized by widespread fibrosis—expression of the mmu-miR-29 family is decreased [32]. mmu-miR-29a-5p suppresses the expression of collagen type I and III [32]. Furthermore, TGF-β suppressed miR-29a-5p which resulted in feedforward upregulation of TGF-β [32]. A mouse model of bleomycin-induced skin fibrosis was associated with decreased mmu-miR-29-5p, which was reversed by the tyrosine-kinase inhibitor imatinib [32], a potent inhibitor of the TGF-β pathway. The role of miR-29 appears not to be limited to systemic sclerosis since a mouse model of mmu-miR-29-5p inhibition demonstrated protection against salt-induced hypertensive renal sclerosis [33]. There was up-regulation of various genes involved with laying of ECM when mmu-miR-29-5p was silenced in the kidneys of these animals [33]. miR-337 was shown to be involved in diabetic nephrosclerosis [34]. It has recently been shown that miR-337 was upregulated when cultured human and mouse mesangial cells were exposed to high glucose and TGF-β to imitate a diabetic milieu [34]. Fibronectin—a key protein involved in fibrosis—was in fact directly induced by miR-377 [34]. Other animal models have been used to study the role of miRNAs in renal fibrosis: hsa-miRNA-449a/b-5p expression was downregulated in hypoxic fibroblasts [35]. Furthermore, hsa-miRNA-449a/b-5p caused upregulation of profibrotic proteins (serine protease inhibitor protein 1-SERPINE1) [35]. Table 3 summarizes some of the studies that investigated the role played by miRNAs in the pathogenesis and maintenance of renal fibrosis. These experiments show that several profibrotic proteins are under the control of miRNAs.

Table 3.
Studies investigating the role of miRNAs in renal fibrosis and maladaptive repair.
7. miRNAs in Select Renal Parenchymal Diseases
miRNA expression profiles have been studied in many renal parenchymal diseases. Specific miRNA expression signatures have been identified for some diseases in both animal models and human studies. We will briefly review some of these associations since they provide the basis for detecting miRNAs as disease-specific biomarkers and potential therapeutic targets.
7.1. Diabetic Nephropathy
miRNAs have been directly implicated in the pathogenesis of diabetic nephropathy. mmu-miR-29c-5p expression—which is associated with podocyte apoptosis—is increased in both the glomeruli and microvascular endothelial cells in a mouse diabetic model [36]. In addition, mmu-miR-29c overexpression promoted activation of the Ras homolog gene family, member A (RhoA)—by suppressing the Sprouty homolog (Spry) 1 gene—which has been shown to play a role in the pathogenesis of diabetic nephropathy [37]. Analysis of kidney biopsy samples from patients with diabetes revealed that hsa-miR-192-5p expression was inversely related to tubulointerstitial fibrosis and directly related to estimated glomerular filtration rate (eGFR) [31].
This association may be causal since the introduction of TGF-β to proximal convoluted tubule cells exposed to high glucose conditions leads to decreased hsa-miR-192-5p expression [31]. Conversely, overexpression of hsa-miR-192-5p ameliorated the TGF-β-mediated fibrosis [31]. Hence, once TGF-β has been activated in high glucose conditions, the decreased expression of hsa-miR-192-5p brought about by TGF-β may further amplify tissue fibrosis.
Several miRNAs may be involved in the expression of the fibrotic renal phenotype. TGF-β increased mmu-miR-216a-5p and collagen type I α1 expression in mouse mesangial models of diabetes [38]. A miRNA circuit has been shown to be directly involved in mediating the autoregulation of TGF-β and the production of ECM [39]. TGF-β induced mmu-miR-192-5p inhibits the expression of the E-box repressors Zeb1/2 which in turn increases the expression of mmu-miR-200b-5p and mmu-miR-200c-5p [39]. These miRNAs further inhibit Zeb1/2 leading to enhanced expression of TGF-β and the ECM components collagen type Iα2 and collagen type IVα1 [39]. Hyperglycemia activates phosphatidylinositol (PI)—3 kinases/Akt pathway leading to cell hypertrophy and increased matrix protein in mouse diabetic models [40]. mmu-miR-21-5p mediates this process by reducing tumor suppressor protein phosphatase and tensin homolog deleted on chromosome 10 (PTEN) [40]. Overexpression of mmu-miR-21-5p is seen to inhibit PTEN expression with an increase in the PI3/Akt pathway, leading to renal cell hypertrophy and fibronectin expression [40]. Overall it is clear that the effect of the miRNAs on these functions and pathologies is significant and important. Many of these miRNAs have been shown to be associated with features of the diabetes phenotype (insulin secretion or sensitivity) and the development of diabetic kidney disease. Many of these miRNAs have the potential to serve as biomarkers in clinical, human disease, an area that was extensively reviewed recently [41,42]. We summarize some of the studies that investigate the role played by miRNAs in the pathogenesis of diabetic nephropathy in Table 4.

Table 4.
Mechanistic, experimental studies investigating the role of miRNAs in diabetic nephropathy.
7.2. Hypertension
Hypertension is a major risk factor for developing coronary artery disease, congestive heart failure, sudden death [43], left ventricular hypertrophy [44], and stroke [45]. Coronary artery disease and stroke are the two major causes of death in the U.S. [46]. Hypertension is more prevalent in patients with chronic kidney disease (CKD) and is thought to be the second most common cause of end-stage renal disease in the U.S. [47]. Genetic, environmental, hemodynamic, renal, and hormonal factors have been implicated in the pathogenesis of hypertension. miRNAs are involved in nearly all pathophysiological alterations that underlie the development of hypertension and its cardiovascular and renal complications.
Oxidative stress due to inhibition of nitric oxide (NO) production and generation of reactive oxygen species could be the final common pathway for hypertension development [48]. Production of reactive oxygen species (ROS) may be influenced by specific miRNAs. In experimental models of oxidative stress (ROS generation, hydrogen peroxide exposure), apoptosis of human umbilical vein endothelial cells (HUVECs) was observed in a dose-dependent manner with concomitant increase in miR-210 levels [49]. Overexpression of hsa-miR-210-5p resulted in inhibition of apoptosis and decreased the concentration of reactive oxygen species. Thus, hsa-miR-210-5p may prevent the deleterious effects of ROS [49]. hsa-miR-155-5p was shown to directly inhibit endothelial nitric oxide synthase (eNOS) production by binding to the 3′ UTR of its mRNA, leading to increased oxidative stress [50]. Furthermore, simvastatin decreased hsa-miR-155-5p expression, thus restoring endothelium-dependent vasorelaxation, an effect that was independent of cholesterol levels [50]. Inhibition of miR-155 may be a therapeutic target for improving endothelial dysfunction and may even underlie some of the non-cholesterol (pleiotropic) effects of statins [50].
miRNAs may also play a role in the development of hypertension, by their effects on vascular smooth muscle cells (VSMCs). Aberrant division of VSMCs leads to vascular luminal hypertrophy and luminal narrowing which causes and propagates hypertension. hsa-miR-143-5p and hsa-miR-145-5p ensure proper development and regulation of VSMCs [51]. VSMCs deficient in hsa-miR-143-5p and hsa-miR-145-5p did not respond to vasocontractile stimuli but had increased synthetic activity [51]. These miRNAs played a critical role in the class switching of VSCMs from a synthetic unit to a vasocontractile unit [51].
Activation of renin-angiotensin-aldosterone systems (RAS) plays a cardinal role in pathophysiology and maintenance of different forms of hypertension. Activation of the angiotensin 1 receptor (AT1R) by Angiotensin II (Ang II) increases blood pressure by vascular smooth muscle cell proliferation, vascular constriction, cardiac remodeling, aldosterone production, and sodium retention, which plays a central role in the pathogenesis of hypertension [52]. These angiotensin mediated processes are under miRNA control. mmu-miR-155-5p inhibits AT1R expression and VSMC proliferation [53]. hsa-miR-29b-5p, hsa-miR-129-3p, hsa-miR-132-5p and hsa-miR-212-5p were upregulated by Ang II in human cell culture (HEK293N) [54]. miR-483-3p expression downregulated angiotensinogen and angiotensin-converting enzyme (ACE) and could be a novel therapeutic agent for hypertension management [55]. Inhibitors of the angiotensin-converting enzyme inhibitors (ACEi) have been shown to decrease renal disease progression in early diabetic nephropathy in type 1 and type 2 diabetes mellitus [43] and in preventing coronary artery disease and strokes [56]. Angiotensin-converting enzyme inhibitors have become a mainstay for the therapy of hypertension [57]. Some of the beneficial effects of ACEi could be mediated by suppression of miR-324-3p. In the Munich Wistar Fromter (MWF) rat model, which develops spontaneous progressive nephropathy, ACEi suppresses rno-miR-324-3p and attenuates the development of hypertensive nephropathy [58].
Sympathetic nervous system overactivity is one of the mechanisms for development and maintenance of hypertension. The role of the sympathetic nervous system (SNS) and RAS in the maintenance of hypertension was studied in mice which were genetically prone to develop hypertension (BPH/2J mice) [59]. Ganglion blocker use (SNS suppressor) in mice that were pre-treated with an ACEi (RAS suppressor) showed that hypertension in the BPH/2J was primarily mediated by the sympathetic nervous system during the active periods and RAS system during the inactive periods [60]. During active periods, BPH/2J mice had higher renal Ren1 (Renin) mRNA and lower miR-181a indicating SNS mediated release of renin [60]. These findings suggest that mmu-miR-181a-5p inversely regulates the Ren1 mRNA. The authors postulated that mmu-miR-181a-5p suppression potentiates sympathetic nervous system-mediated increase in renin production in BPH/2J mice during the active periods [60]. These findings were confirmed by a human study of mRNA and miRNA expression profiles in renal biopsies of hypertensive patients that showed hsa-miRNA-181a inversely regulated the Ren1 mRNA [61].
Various animal models have been developed to study the effects of hypertension on kidneys. Dahl salt-sensitive (Dahl-SS) rats develop hypertension with medullary interstitial fibrosis when exposed to a high salt diet. Consomic SS-13BN rats are genetically modified Dahl-SS rats that have less pronounced blood pressure rise and medullary interstitial fibrosis when exposed to a high salt diet [62]. Liu et al. studied miRNA expression profiles in these two rat models and showed that a high salt diet resulted in upregulation of miR-29b in Consomic SS 13BN rats but not in Dahl-SS rats [33]. Various collagen genes that promote fibrosis were upregulated in Dahl-SS rats but not in Consomic SS 13BN rats—a pattern opposite of miR-29b expression. Furthermore, a miR-29b knockdown Consomic SS 13BN rat model had upregulation of various collagen genes, suggesting that miR-29b expression protects rats from hypertension-associated renal injury [33].
Kidney biopsies in patients with hypertension reveal glomerulosclerosis, tubular atrophy, interstitial fibrosis, and vascular smooth muscle cell hypertrophy. In a recent study, intrarenal miRNA expression profiles of 30 patients with hypertensive nephrosclerosis were compared to 20 normal controls. hsa-miR-200a-5p, hsa-miR-200b-5p, hsa-miR-141-5p, hsa-miR-429-5p, hsa-miR-205-5p, and hsa-miR-192-5p were significantly increased in patients with hypertensive nephrosclerosis [63]. Table 5 summarizes the studies exploring the role of miRNAs in the pathogenesis of hypertension.

Table 5.
Studies investigating the role of miRNAs in hypertension.
7.3. Glomerulonephritis
Glomerulonephritides (inflammation of glomeruli) are a group of diverse disorders that may present as proteinuria and or/hematuria with renal dysfunction. Kidney biopsy findings include podocyte injury, mesangial and endocapillary proliferation, and disruption of basement membranes leading to focal and segmental glomerulosclerosis, tubular atrophy, and interstitial fibrosis. We will now discuss the role of miRNAs in some of the conditions that can cause glomerulonephritis.
7.3.1. Focal Segmental Glomerulosclerosis
Focal segmental glomerulosclerosis (FSGS) is a pattern seen on kidney biopsy characterized by involvement of some of the glomeruli, with part of the involved glomerulus showing obliteration of the capillary lumen and increase in mesangial matrix. FSGS is usually caused by infections, medications, and conditions that cause chronic renal injury. At times, no cause of FSGS is found, and this is labeled as primary FSGS. A molecule that increases the permeability of glomerular basement membranes has been postulated in the pathogenesis of primary FSGS [64,65] but the exact nature of that molecule remains elusive. Podocyte injury is considered an inciting event in the development of FSGS.
In a puromycin-induced FSGS rat model, researchers found diminished rno-miR-30s-5p [66]. Replacement of rno-miR-30s-5p resulted in resolution of podocyte injury and proteinuria [66]. Furthermore, a human cell culture with hsa-miR-30a overexpression had amelioration of the cytoskeletal damage and apoptosis induced by puromycin or TGF-β treatment [66]. Glucocorticoid use caused sustained expression of rno-miR-30s-5p in podocytes [66]. Therefore, miR-30a plays a role in podocyte health and maintenance of cytoskeletal integrity. The histological findings in this model recapitulated the abnormal morphology in the mmu-miR-30a deficient Drosha and Dicer knockout podocyte models that were discussed previously [11,12,13].
Recently, the role of Brain Derived Neurotrophic Factor (BDNF) in maintaining the integrity of podocyte layer was studied [67]. Mice podocyte cell culture exposed to BDNF showed increased actin polymerization and increased number and length of podocyte foot processes with an associated increase in mmu-miR-132-5p and decrease in mmu-miR-134-5p. Overexpression of mmu-miR-132-5p and silencing of mmu-miR-134-5p in a podocyte cell culture also showed podocyte cell growth [67]. Furthermore, BDNF repaired podocyte damage in in vivo and in vitro models [67]. This study showed that BDNF maintains and repairs podocyte layer and this effect is mediated by increased expression of mmu-miR-132-5p and decreased expression of mmu-miR-134-5p. This agent may be used as a therapeutic agent in conditions associated with podocyte damage such as FSGS, minimal change disease and diabetic nephropathy.
miRNAs have also been used as biomarkers—both in serum and urine—to assess FSGS disease activity. In one study, researchers found elevated plasma hsa-miR-125b-5p, hsa-miR-186-50 and hsa-miR-193a-3p in patients with FSGS with area under curve (AUC) of 0.88, 0.78, and 0.91, respectively [68]. Patients in remission had lower hsa-miR-125b-5p and hsa-miR-186-5p concentrations [68]. These miRNA levels remained unchanged in patients that did not achieve remission. hsa-miR-186-5p levels also correlated with proteinuria [68]. Patients with FSGS and minimal change disease had higher urinary hsa-miR-200c-5p levels [69]. hsa-miR-196a-5p, hsa-miR-30a-5p and hsa-miR-490-5p were associated with FSGS disease activity [70]. Urinary hsa-miR-30a-5p was a weak predictor of steroid responsiveness in patients with active FSGS [70]. Table 6 summarizes some of the studies looking at the role of miRNAs in FSGS.

Table 6.
Studies investigating the role of miRNAs in FSGS.
7.3.2. IgA Nephropathy
IgA nephropathy is the most prevalent primary glomerulonephritis in the world. Abnormal O-galactosylation of IgA causes the formation of IgA complexes against which IgG and IgA are formed with deposition in the kidneys and activation of the complement system leading to kidney injury.
Genome-wide analysis has revealed various miRNAs which might play a role in IgA nephropathy. miRNA expression of 6 patients, each with biopsy-proven IgA nephropathy, compared to those with renal cell carcinoma, revealed upregulation of 11 miRNAs and downregulation of 74 miRNAs in the IgA nephropathy group [71]. Members of the hsa-miR-200 and hsa-miR-29 families, which regulate EMT and development of tissue fibrosis, showed prominent expression changes in patients with IgA nephropathy and were associated with tissue fibrosis and proteinuria [71].
miRNA hsa-let-7a-5p and hsa-miR-148b-5p control N-acetylgalactosaminyltransferase 2 (GALNT2) and 1 β 1,3 galactosyltransferase 1 (C1GALT1), respectively—these enzymes play a central role in aberrant IgA galactosylation. It has been shown that these enzymes are overexpressed in the peripheral blood mononuclear cells of IgA nephropathy patients [72,73].
miRNA expression profiles in paraffin embedded kidney biopsy specimens and urine from patients with IgA nephropathy was studied [74]. Both the renal tissue and urine of patients with IgA nephropathy exhibited increased expression of hsa-miR-146a-5p and hsa-miR-155-5p than controls. hsa-miR-146a-5p and hsa-miR-155-5p were inversely related with GFR and positively related with proteinuria. hsa-miR-155-5p expression was also associated with renal fibrosis. The possible mechanistic roles of these miRNAs in the pathogenesis of IgA nephropathy was further studied. Urinary hsa-miR-155-5p was inversely associated with interleukine 1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α) and positively related with expression of Forkehad Box P3 (FOXP3—a transcription factor for regulatory T cell development) and regulated upon activation, normal T-cell expressed and secreted (RANTES—a chemokine). It was postulated that hsa-miR-146a-5p and hsa-miR-155-5p regulate the expression of pro-inflammatory molecules in patients with IgA nephropathy [74]. hsa-miR-155-5p has been shown to influence the regulatory T cell development [75,76]. Decreased regulatory T cell number/function has been implicated in the pathogenesis of IgA nephropathy [77]. Table 7 summarizes studies investigating the role of miRNAs in IgA nephropathy.

Table 7.
Studies investigating the role of miRNAs in IgA nephropathy.
7.3.3. Lupus Nephritis
Systemic lupus erythematosus (SLE) is a systemic disease due to dysregulated immune system activity. Kidney involvement in SLE often leads to chronic kidney disease and eventually kidney failure if left untreated, and it is the major cause of morbidity and mortality. Genetic factors have been implicated in SLE pathogenesis, but the underlying control mechanisms remain poorly defined.
Various lines of evidence point towards the role of miRNAs in SLE. miRNAs regulate 72 genes labeled as “autoimmune genes” that control various aspects of the immune system [78]. hsa-miR-181-5p, hsa-miR-186-5p, and miR-590-3p regulate more than 50% of the genes that are known to be differentially expressed in SLE patients [78]. Epstein-Barr virus (EBV) infection could be one of the initiating agents responsible for dysregulated immune response in SLE. EBV affects SLE patients more commonly with an increased number of infected peripheral white cells than healthy controls [79]. The exact causal link between EBV infection and SLE is not known; however, molecular mimicry is suspected. EBV latent membrane protein 1 activates hsa-miR-155-5p transcription through the nuclear factor κβ (NF-κβ) pathway [80]. miR-155 is expressed in regulatory T cells [81] and macrophages and promotes the development of inflammatory T cells [82]. B6.MRLc1 mice exhibit an immune complex-mediated glomerulonephritis with proliferative lesions that progress to glomerulosclerosis, tubular atrophy, and interstitial fibrosis [83]. These lesions showed expression of mmu-miR-146a-5p which increased with age, suggesting that it plays a role in renal inflammation [83]. Kidney biopsy analysis of patients with lupus nephritis showed upregulation of hsa-miR-146a-5p and hsa-miR-198-5p in the glomerular lesions and hsa-miR-638-5p in tubulointerstitial lesions [84]. In this study, the degree of interstitial miR-638 expression was significantly correlated with clinical markers of kidney damage (proteinuria) and the disease activity score [84]. Conversely, glomerular hsa-miR-146a-5p correlated with clinical markers of renal function (estimated glomerular filtration rate) and the disease activity score [84]. Hence, these two miRNAs may play a pathogenic role in the development of clinical lupus nephritis.
The role of miRNAs in the pathogenesis of proliferative lupus nephritis has been a subject of intense research. A high-throughput analysis of paraffin embedded kidney biopsies of pediatric patients showed differential expression of various miRNAs [85]. hsa-miR-26a-5p, hsa-miR-30b-5p, and hsa-miR-4286-5p, which are involved in cell cycle regulation, were decreased in the kidney biopsy samples and were studied further in a human mesangial cell model. Silencing of hsa-miR-26a-5p, hsa-miR-30b-5p, and hsa-miR-4286-5p resulted in increased expression of genes involved in cell cycle regulation. It has been shown previously that Trastuzumab, a monoclonal antibody against the human epithelial growth factor receptor 2 (HER-2), causes the arrest of the cancer cells in G1 phase with up-regulation of hsa-miR-26a-5p and hsa-miR-30b-5p [86]. The authors studied the effects of Trastuzumab on the human mesangial cell model and found that the mesangial cells treated with this agent have increased hsa-miR-26a-5p and hsa-miR-30b-5p expression. This lead to the hypothesis that HER-2 might be responsible for increased cell proliferation in lupus nephritis. This was validated in paraffin fixed human kidney biopsy specimens from patients with lupus nephritis that showed increased HER-2 expression in tubular, glomerular and mesangial compartments [85]. These findings were replicated in the glomeruli of a mouse lupus model. The factors that lead to increased expression of HER-2 in lupus nephritis were further studied. Human mesangial cells exposed to IFNα had increased expression of HER-2 than control cells. The authors found that urinary HER-2 levels are elevated in patients with lupus nephritis than in healthy controls. HER-2 levels were inversely related to lupus activity and correlated with proteinuria. Therefore, it was shown that INFα increases the HER-2 expression that leads to decreased hsa-miR-26a-5p and hsa-miR-30b-5p expression resulting in activation of the cell cycle leading to proliferative lupus nephritis. Table 8 summarizes the studies that investigate the role of miRNAs in the development of lupus nephritis.

Table 8.
Biopsy studies investigating the role of miRNAs in lupus nephritis.
7.3.4. Anti-Neutrophilic Cytoplasmic Antibodies Associated Vasculitis (ANCA)
ANCA vasculitis is a small vessel vasculitis involving the kidneys as well as other organs and is characterized by the presence of either anti-Proteinase 3 (PR3) or anti-Myeloperoxidase (MPO) (components of neutrophils) antibodies. Currently, it is not known whether these antibodies are pathogenic and what are the inciting factors for production of these antibodies.
Pooled plasma samples from 40 patients who had active ANCA vasculitis or were in remission showed up-regulation of hsa-let-7f-5p and hsa-miR-424-5p, and downregulation of hsa-miR-106b, hsa-miR-9-5p, hsa-miR-125a-50, and hsa-miR-15b-5p [87]. These miRNAs regulate various aspects of the immune system [87], suggesting a direct role in the development of clinical disease.
hsa-miR-155-5p is upregulated in patients with ANCA-associated crescentic GN [88]. Nephrotoxic nephritis is a mouse model of ANCA vasculitis developed by injecting rats with rabbit or duck nephrotoxic sera [89] and has been found to closely correlate with human renal ANCA vasculitis [90]. A mmu-miR-155-5p knockout in this mouse model exhibited less severe lesions [88]. It was noted that miR-155 mediates the TH17 immune response and thus may be a therapeutic option for ANCA associated crescentic GN [88]. Table 9 summarizes the studies that investigate the role of miRNAs in ANCA vasculitis.

Table 9.
Studies investigating the role of miRNAs in ANCA associated nephritis.
7.3.5. Systemic Sclerosis (Scleroderma)
Systemic sclerosis is a condition associated with multiple organ fibrosis. It affects the kidney by causing thickening of the blood vessels, leading to hypertension, endothelial injury, and thrombotic microangiopathy. It has been shown that TGF-β [91] and miR-21 [92] are upregulated in systemic sclerosis. Furthermore, TGF-β regulates the expression of hsa-miR-21-5p and fibrosis-related genes, and hsa-miR-21-5p is inversely associated with Smad7 expression and may, therefore, be a therapeutic target for this condition [93]. Table 10 summarizes the results of a study investigating the role of miR-21-5p in scleroderma.

Table 10.
Study investigating the role of miRNAs in scleroderma-associated renal disease.
7.3.6. Autosomal Dominant Polycystic Kidney Disease (ADPKD)
ADPKD is a disease characterized by impaired ciliary function leading to kidney and liver cyst formation and kidney failure in the vast majority of patients.
In the Sprague-Dawley rat model of ADPKD, 29 miRNAs were downregulated and only 1 miRNA (miR-21) was upregulated. Most of the dysregulated miRNAs control cell-to-cell interaction and crosstalk [94]. Global gene-expression studies in embryonic kidneys in an animal PKD model found differential expression of rno-miRs-10a-5p, rno-miR-30a-5p, rno-miR-96-5p, rno-miR-126-5p, rno-miR-182-5p, rno-miR-200a-5p, rno-miR-204-5p, rno-miR-429-5p and rno-miR-488-5p [95]. The mmu-miR-21-5p expression has also been associated with cyst progression [96]. Inhibition of mmu-miR-21-5p slows cyst growth in a mouse model of ADPKD [96]. The miR-17 cluster of miRNAs is upregulated in mouse models of PKD, and deletion of miR-17 cluster results in resolution of cysts, and better renal and animal survival [96]. We summarize the results of a study investigating the miRNA expression profile in ADPKD in Table 11.

Table 11.
Study investigating the role of miRNAs in autosomal dominant polycystic kidney disease.
7.3.7. Alport Syndrome
Alport syndrome is due to abnormalities in genes encoding α3, α4 or α5 chains of collagen Type IV, resulting in abnormal basement membranes in the kidney, eyes, and inner ear. These changes in the kidney lead to abnormalities of glomerular basement membrane and progressive renal disease. mmu-miR-21-5p is preferentially expressed in the tubulointerstitium instead of glomeruli in normal mice; however, in the Col4 α3−/− mice, mmu-miR-21-5p is expressed equally in both compartments [97]. As described previously, miR-21 has been associated with renal fibrosis. Introduction of anti-miR-21 oligonucleotides in Col4 α3−/− mice resulted in the preservation of renal function, reduction in albuminuria, improved survival, reduced glomerulosclerosis, crescent formation, and tubular injury [97]. Currently, a Phase 1 clinical trial is being conducted to assess the safety, pharmacodynamics, and pharmacokinetics of a molecule that inhibits miR-21 (Table 12) [98].

Table 12.
Study investigating the role of miRNAs in Alport syndrome.
8. miRNA Detection
The preceding section highlights the role of specific miRNAs in normal renal development and physiology, but also the initiation and the progression of the interstitial fibrosis that underlies progressive forms of chronic kidney disease. It follows, that miRNAs detected in either plasma or urine, the two fluidic compartments directly affected by renal processing, may be mechanistically plausible, rational biomarkers for diverse forms of kidney diseases. In fact, miRNA associations found in observational human studies may offer a unique opportunity to “reverse translate” such findings into animal studies, which provide mechanistic insights into novel therapeutics that are tested in rigorous interventional clinical trials in humans. (Figure 2).
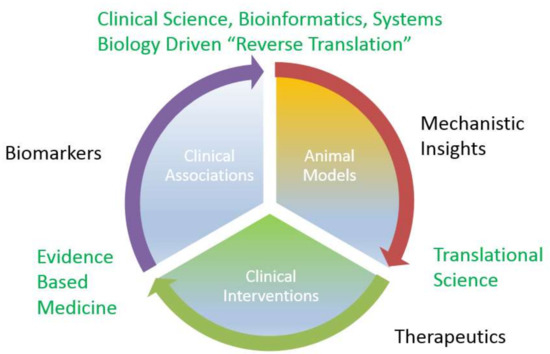
Figure 2.
miRNA discovery cycle from biomarkers to therapeutics.
Nevertheless, detection of miRNAs poses unique challenges because of their short size and the similarity of many sequences to one another. These biochemical features of miRNAs may directly impact the performance of the three methods most commonly used for miRNA detection: quantitative real-time PCR (qPCR), microarrays, and next-generation sequencing (NGS). Each of these approaches comes with its distinct advantages, but also limitations when used as the basis for the development of miRNA biomarker assays.
Of the methods listed above, qPCR has the highest sensitivity, with a theoretical limit of detection of just a few copies per sample [99]. Several commercial kits are available for detection of miRNAs by qPCR, and although the specifics of each kit differ, they generally involve the addition of a known sequence to the 3′ end of the miRNA, followed by reverse transcription and PCR amplification using a miRNA-specific primer. Because each target of interest requires a separate PCR reaction and cannot easily be highly multiplexed, qPCR is less well-suited to high-throughput profiling than either microarrays or NGS. However, for targeted detection of a specific, small set of miRNAs, the cost of qPCR is comparatively low, and the hands-on time required is much less than that of other methods. Nevertheless, the high ionic strength of urine and the presence of urea (a non-specific inhibitor of the polymerase reaction [100]) may pose unique challenges when developing a urine-specific qPCR assay. Furthermore, the development of droplet digital PCR (ddPCR) more recently has improved the precision and reproducibility of qPCR measurements, especially for samples with low target abundance or high contaminant concentrations, and made absolute quantitation more accessible [101].
Microarray-based methods allow for the simultaneous measurement of many miRNAs, making them a better choice than qPCR for profiling a large set of targets. Commercial products are available covering all the mature miRNA sequences in miRBase on a single array. However, the amount of starting material required for microarray analysis is relatively high (~100ng per sample) and it remains difficult to design probes and hybridization conditions that can distinguish between closely related miRNA sequences. In addition, the dynamic range of microarrays is typically lower than either qPCR or NGS.
Unlike qPCR and microarrays, NGS requires no prior knowledge of the target sequences in the sample, and so is ideal for discovery studies. In addition, NGS is not affected by the complications of designing primers or probes with the specificity needed to distinguish between short sequences with high sequence identity. Consequently, as costs for NGS library preparation and sequencing have dropped, small RNA sequencing (sRNA-seq) has become widely used for miRNA profiling because of its ability to comprehensively interrogate the miRNAs (and other types of small non-coding RNAs) in a sample. As opposed to qPCR and microarrays, sRNA-seq allows the analysis of miRNAs with single nucleotide resolution, so not only can the canonical sequences be studied, but also variants arising from RNA editing or imprecise miRNA processing (isomiRs). sRNA-seq is not without drawbacks, however. Compared to qPCR and even microarray analysis, NGS is more expensive and more time intensive, both in sample preparation and data analysis. Most methods for preparing sRNA-seq libraries involve sequential ligation of adapters to the 3′ and 5′ ends of the miRNAs, followed by reverse transcription and PCR amplification to add indexes and other sequences needed for attachment to the flow cell and sequencing. The adapter ligation steps have previously been shown to introduce bias into small RNA libraries, which leads to decreased library diversity and prevents making quantitative comparisons of the expression level of different miRNAs in a sample [102,103,104]. Moreover, because the biases vary depending on the library preparation protocol, comparing data generated by two different sample prep methods is difficult [105,106]. Although attempts have been made to alleviate bias in sRNA-seq through modifications in library preparation methods [104,107,108] or to compensate for it during data analysis [109], it remains a significant issue that will need to be addressed in the future.
Several important issues affect all methods commonly used for measuring miRNAs. For example, whereas several housekeeping genes have been widely used for normalization in mRNA expression profiling studies, there is less consensus on similar invariant transcripts suitable for miRNA expression analysis. Other options include normalization based on sample input (input mass when practical or input volume when the amount of RNA in the sample is too low to reliably measure) or based on the addition of spike-in oligonucleotides added during sample processing. Normalization based on relative read counts (e.g., reads per million total miRNA reads or reads per million genome-mapped reads) is also frequently used to report sRNA-seq data, but this method can yield inaccurate results if samples have different proportions of small RNA species, either because of biological differences or technical differences such as variations in the size selection step of sRNA-seq library preparation.
Another issue affecting miRNA measurement is the lack of correspondence between data generated by different methods. It is not an uncommon practice to identify changes in miRNA expression using high-throughput methods such as microarrays or NGS and subsequently validate those changes by qPCR. However, it has been known for some time that changes in miRNA expression detected by one method are often difficult to corroborate across methods [110]. High among the problems here is that different methods exhibit different sequence-specific biases. Even within a given method, however, results may not be comparable if the data is generated using kits from different vendors, as mentioned above for sRNA-seq, and as previously shown for microarray and qPCR [111,112]. For all these reasons, measurement of miRNAs requires careful planning, care in precisely executing protocols, and repeated measurements when possible.
Measurement of extracellular miRNAs is further complicated by low concentration and inhibition by other macromolecules in the sample. Although there is great interest in profiling RNAs present in biofluids such as blood and urine to identify disease biomarkers, obtaining accurate and reproducible results from these sources remains challenging. The primary reason that measuring miRNA in biofluids is difficult is that the RNA concentration is much lower than in cells or tissues. Typical RNA isolation methods recover only about 10–50 ng of total RNA per milliliter of cell-free plasma, with even lower yields from fluids such as urine and saliva. In addition, the presence of inhibitors derived either from the sample itself or during the collection procedure and co-purified with the RNA, can be problematic for the enzymatic steps in library preparation. Salts present in urine samples and heparin used as an anticoagulant during plasma collection are two examples of inhibitors commonly encountered with biofluid samples. Furthermore, the handling and storage methods of the samples before RNA extraction can have a significant impact on the results. Lysis of cells during collection (hemolysis in plasma, for example) or incomplete removal of intact cells before RNA isolation can also significantly distort the miRNA profile from biofluids since cellular RNA content is much higher than that of the cell-free fluid. Thus, sample collection, RNA isolation, and library protocols are all critical for accurate profiling.
miRNA as Personalized Diagnostics
miRNAs have a unique role among biological regulatory molecules, participating in feed-forward regulatory loops [113,114], whose connectivity (“fan-in” and “fan-out” patterns) differ from other control systems (e.g., transcription factors or kinases). Standard control theory of artificial systems informs us that such layouts are signatures of master controllers that fine-tune the performance of the system being controlled. Translated in biological terms, one should expect miRNAs to be tightly involved when exquisite sensitive control of a critical biological system and the corresponding phenotype(s) is required. In fact, the involvement of miRNAs in sodium and potassium regulation, provide examples of such a role, which is highly context-specific and sensitive to environmental stimuli. A corollary of these lessons from control theory is that miRNAs will likely prove to be successful candidates for the development of personalized diagnostics across the spectrum of kidney diseases (Figure 3). According to this hypothesis, disease processes will perturb the normal organ, but will nonetheless not abolish normal phenotype until very late in the disease process. Such a phenotypic resilience will be due to changes in the normal expression pattern of miRNAs that may be detected in biofluids accessible to the organ in question. The critical challenge for the development of personalized diagnostics is to develop organ-specific signatures whose alterations give clues to the presence or lack thereof of progressive organ dysfunction. Our previous research [115] in the area of diabetic kidney disease, illustrates the potential of miRNAs in the area of kidney disease. In particular, we discovered a signature of miRNAs that can predict the development of overt kidney damage, two years before it is detected by the conventional laboratory tests of serum creatinine and urinary albumin. The miRNAs in this signature were involved in the regulation of normal physiological processes and tissue fibrosis. Furthermore, the signature appeared to be relatively specific for diabetic kidney disease since it predicted less strongly the development of kidney damage in an independent cohort of patients.
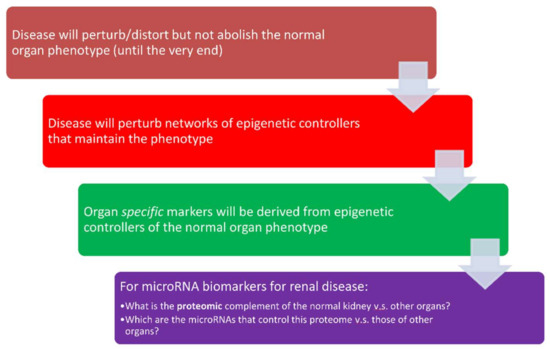
Figure 3.
miRNAs as personalized diagnostics in kidney diseases.
9. Micro RNAs as a Therapeutic Option in Renal Diseases
In the previous sections, we discussed the roles of miRNAs in normal kidney function as well as kidney diseases associated with miRNA dysregulation. There is significant interest in the use of miRNAs as therapeutic agents since they modulate the activity of numerous genes. miRNA-based therapeutics are based on either inhibiting a deleterious miRNA or replacing a deficient beneficial miRNA. miRNA antagonists—also called antagomirs or antimiRs—are single-stranded molecules that are designed to bind directly to a mature miRNA and to block its action. Deficient miRNAs can be replaced by either making small interfering miRNAs (siRNAs), which are small double-stranded RNA molecules encapsulated in nanoparticles and delivered to the target site [116] or by viral vectors that express the desired miRNA.
9.1. miRNA Delivery
Designing an effective miRNA mimic and delivering it to its intended target organ without degradation or causing unintended effects has been the subject of intense research. Designing miRNA mimics that effectively block their targets without affecting any unintended transcripts has proven problematic [117]. Therefore, these molecules must be thoroughly tested to fully understand all their intended and unintended effects.
The process of delivering a miRNA molecule to its intended target is fraught with difficulties. Naked miRNAs are unstable in the blood and are rapidly degraded by the mononuclear macrophage system and removed from circulation by the kidneys and liver. Therefore, effective delivery methods that prevent miRNA degradation must be devised. These delivery vehicles should be non-toxic, have low immunogenicity, and should be able to deliver a large proportion of the miRNAs to their intended target [118]. Some of the techniques used to deliver miRNAs are shown in Table 13.

Table 13.
miRNA delivery methods.
9.2. MicroRNA-Based Renal Therapeutics
Various phase I and phase II trials are underway or have been completed for miRNA-based therapeutic agents for the management of chronic hepatitis C, diabetes mellitus type 2 with fatty liver and cancers [116]. miRNAs are also being used as therapeutic agents in renal diseases. Table 14 shows a summary of miRNA-based therapeutics for the conditions affecting the kidneys.

Table 14.
Trials involving miRNAs in renal parenchymal disease.
Regulus Therapeutics in collaboration with Genzyme has developed a single-stranded molecule RG-012 that inhibits miR-21. In a rat model of Alport syndrome, miR-21 inhibition by this molecule led to milder kidney disease and improved survival than control mice. There was less glomerulosclerosis and tubulointerstitial fibrosis in the treated mice with no adverse events [97]. A phase I randomized, double-blinded, placebo-controlled study is currently being conducted to study the safety and efficacy of RG-012 in male subjects with Alport syndrome [98]. miRagen Therapeutics is developing a molecule MRG-201 that promotes miR-29 activity and thus modulates fibrosis. This molecule has potential roles in preventing progression of CKD in diabetic nephropathy, IgA nephropathy, and scleroderma. A phase 1 study to evaluate the safety and tolerability of this agent has been completed [130].
10. Conclusions
It has been shown in practice over the past decade that extracellular miRNAs can provide informative biomarkers for multiple biological effects and pathologies. The value of understanding miRNA function, however, is much broader. In concert with the multiple factors regulating transcription, miRNAs provide an additional level of control of gene expression, largely at the post-transcriptional level. Their influence on various biological pathways is both widespread and complex and is often subtle. In the last 20 years, tremendous progress has been made in understanding their roles in renal physiology and pathology, and this is beginning to open several new lines of investigation. Research is currently underway to study and modulate miRNAs specifically to control maladaptive repair that leads to fibrosis in various renal diseases. miRNAs also provide us a novel opportunity to develop new ways of studying disease activity and to assess the efficacy of therapeutic agents. Since miRNAs can be targeted directly, although this is sometimes difficult in practice, they provide the opportunity to develop a new class of therapeutic agents. miRNA-based diagnostics and therapeutics, therefore, have the potential to lead medicine into a new era of effectiveness.
Author Contributions
All authors planned the manuscript together; A.E. and D.G. wrote the miRNA detection section; S.K.S wrote all the other sections. All the authors edited, critically reviewed the manuscript, and agreed on the final manuscript.
Funding
This work was supported by grants from the NIH Common Fund Extracellular RNA Communication Consortium (ERCC) to D.G. [U01HL126496 and U54DA036134], the National Institutes of Health, National Center for Advancing Translational Sciences [UL1TR001449 to C.A, in part] Dialysis Clinic Inc. (DCI) [DCI #C-3765 and DCI RF#C-3861.201 to C.A] and by the Pacific Northwest Research Institute. Funding for open access charge: DCI [DCI #C-3765].
Acknowledgments
We thank Elaine Skeffington for careful copy edits.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 3′ UTR | 3′-Untranslated Region |
| ACE | Angiotensin-converting Enzyme |
| ACEi | Angiotensin-converting Enzyme Inhibitors |
| ADPKD | Autosomal Dominant Polycystic Kidney Disease |
| ANCA Vasculitis | Anti-Neutrophilic Cytoplasmic Antibodies associated Vasculitis |
| Ang II | Angiotensin 2 |
| AT1R | Angiotensin 1 Receptor |
| BDNF | Brain Derived Neurotrophic Factor |
| BIM | Bcl-2-Like Protein 11 |
| BP | Blood Pressure |
| C1GALT1 | 1-β-1,3-Galactosyltransferase-1 |
| CCD | Cortical Collecting Duct |
| Col4 α3−/− | Homozygous for Collagen Type 4 alpha 3 Chain Absence |
| Consomic SS-13BN | Consomic Salt Sensitive Rats |
| Dahl-SS | Dahl Salt-Sensitive |
| DCT | Distal Convoluted Tubule |
| ddPCR | Droplet Digital Polymerase Chain Reaction |
| DOPC | 1,2-Dioleoyl-sn-Glycero-3- Phosphocholine |
| ECM | Extra Cellular Matrix |
| eGFR | Estimated Glomerular Filtration Rate |
| EMT | Epithelial to Mesenchymal Transition |
| ENaC | Epithelial Sodium Channel |
| eNOS | Endothelial Nitric Oxide Synthase |
| ESRD | End-stage Renal Disease |
| FF | Feed Forward |
| FOXP3 | Forkhead Box P3 |
| FSGS | Focal Segmental Glomerulosclerosis |
| GALNT2 | N-Acetylgalactosaminyltransferase 2 |
| GN | Glomerulonephritis |
| HUVECs | Human Umbilical Vein Endothelial Cells |
| IL-1β | Interleukin 1 Beta |
| isomiRs | Imprecise miRNA Processing |
| miRNA | Micro RNA |
| MPO | Myeloperoxidase |
| mRNA | Messenger Ribonucleic Acid |
| MWF rat model | Munich Wistar Fromter (MWF) Rat Model |
| NCC | Sodium Chloride Co-Transporter |
| NF-Κβ | Nuclear Factor Kappa Beta |
| NGS | Next-Generation Sequencing |
| NO | Nitric Oxide |
| PI-3/Akt | Phosphatidylinositol-3 Kinase/Akt Pathway |
| PKD1 | Polycystic Kidney Disease 1 |
| PLGA | Poly Lactic-Co-Glycolic Acid |
| PR3 | Proteinase 3 |
| PTEN | Tensin Homolog Deleted on Chromosome 10 |
| qPCR | Quantitative real-time Polymerase Chain Reaction |
| RANTES | Regulated Upon Activation, Normal T-cell Expressed and Secreted |
| RhoA | Ras Homolog Gene Family, Member A |
| RNA | Ribonucleic Acid |
| ROMK | Renal Outer Medullary Potassium Channel |
| ROS | Reactive Oxygen Species |
| rRNA | Ribosomal Ribonucleic Acid |
| SERPINE1 | Serine Protease Inhibitor Protein 1 |
| siRNA | Small Interfering Micro Ribonucleic Acid |
| SLE | Systemic Lupus Erythematosus |
| SPRY1 | Sprouty Homolog 1 |
| sRNA-seq | Small RNA Sequencing |
| TGF-β | Transforming Growth Factor Beta |
| TNF-α | Tumor Necrosis Factor Alpha |
| tRNA | Transfer Ribonucleic Acid |
| VSMCs | Vascular Smooth Muscle Cells |
| WNK | with no Lysine Kinase System |
| ZEB2 | Zinc Finger E-Box-Binding Homeobox 2 |
References
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Creugny, A.; Fender, A.; Pfeffer, S. Regulation of primary microRNA processing. FEBS Lett. 2018. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hung, M.-C. Signaling-mediated regulation of MicroRNA processing. Cancer Res. 2015, 75, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Achkar, N.P.; Cambiagno, D.A.; Manavella, P.A. miRNA Biogenesis: A Dynamic Pathway. Trends Plant Sci. 2016, 21, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- miRBase. Available online: http://www.mirbase.org/ (accessed on 20 November 2017).
- Ho, J.; Pandey, P.; Schatton, T.; Sims-Lucas, S.; Khalid, M.; Frank, M.H.; Hartwig, S.; Kreidberg, J.A. The pro-apoptotic protein Bim is a microRNA target in kidney progenitors. J. Am. Soc. Nephrol. 2011, 22, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, V.K.; Ren, Q.; Pugh, M.M.; Valerius, M.T.; McMahon, A.P.; Yu, J. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011, 79, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Marrone, A.K.; Stolz, D.B.; Bastacky, S.I.; Kostka, D.; Bodnar, A.J.; Ho, J. MicroRNA-17~92 is required for nephrogenesis and renal function. J. Am. Soc. Nephrol. 2014, 25, 1440–1452. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.; Hajarnis, S.; Williams, D.; Hunter, R.; Huynh, D.; Igarashi, P. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J. Am. Soc. Nephrol. 2012, 23, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.J.; Jarad, G.; Cunningham, J.; Goldberg, S.; Schermer, B.; Harfe, B.D.; McManus, M.T.; Benzing, T.; Miner, J.H. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J. Am. Soc. Nephrol. 2008, 19, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Yu, L.; Chiu, C.; Sun, Y.; Chen, J.; Khitrov, G.; Merkenschlager, M.; Holzman, L.B.; Zhang, W.; Mundel, P.; et al. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J. Am. Soc. Nephrol. 2008, 19, 2159–2169. [Google Scholar] [CrossRef] [PubMed]
- Zhdanova, O.; Srivastava, S.; Di, L.; Li, Z.; Tchelebi, L.; Dworkin, S.; Johnstone, D.B.; Zavadil, J.; Chong, M.M.; Littman, D.R.; et al. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 2011, 80, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Sequeira-Lopez, M.L.S.; Weatherford, E.T.; Borges, G.R.; Monteagudo, M.C.; Pentz, E.S.; Harfe, B.D.; Carretero, O.; Sigmund, C.D.; Gomez, R.A. The MicroRNA-Processing Enzyme Dicer Maintains Juxtaglomerular Cells. J. Am. Soc. Nephrol. 2010, 21, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Mladinov, D.; Liu, Y.; Mattson, D.L.; Liang, M. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: MiR-192 targets Na+/K+-ATPase β1. Nucleic Acids Res. 2013, 41, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Elvira-Matelot, E.; Zhou, X.; Farman, N.; Beaurain, G.; Henrion-Caude, A.; Hadchouel, J.; Jeunemaitre, X. Regulation of WNK1 Expression by miR-192 and Aldosterone. J. Am. Soc. Nephrol. 2010, 21, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.-H.; Yue, P.; Pan, C.; Sun, P.; Wang, W.-H. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J. Am. Soc. Nephrol. 2011, 22, 1087–1098. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Renigunta, V.; Himmerkus, N.; Zhang, J.; Renigunta, A.; Bleich, M.; Hou, J. Claudin-14 regulates renal Ca++ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J. 2012, 31, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Böttinger, E.P. TGF-beta in renal injury and disease. Semin. Nephrol. 2007, 27, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Derynck, R. Essential role of TGF-β signaling in glucose-induced cell hypertrophy. Dev. Cell 2009, 17, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Rüster, C.; Wolf, G. Angiotensin II as a morphogenic cytokine stimulating renal fibrogenesis. J. Am. Soc. Nephrol. 2011, 22, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Liu, G.-S.; Dusting, G.J.; Chan, E.C. NADPH oxidase-dependent redox signaling in TGF-β-mediated fibrotic responses. Redox Biol. 2014, 2, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.B. Molecular and cell biology of TGF-β. Miner. Electrolyte Metab. 1998, 24, 111–119. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; Taduri, G.; O’Connell, J.; Teng, Y.; Cooke, V.G.; Woda, C.; Sugimoto, H.; Kalluri, R. Origin and Function of Myofibroblasts in Kidney Fibrosis. Nat. Med. 2013, 19, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Jenkins, R.H.; Bennagi, R.; Krupa, A.; Phillips, A.O.; Bowen, T.; Fraser, D.J. Post-Transcriptional Regulation of Transforming Growth Factor Beta-1 by MicroRNA-744. PLoS ONE 2011, 6, e25044. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Koh, P.; Winbanks, C.; Coughlan, M.T.; McClelland, A.; Watson, A.; Jandeleit-Dahm, K.; Burns, W.C.; Thomas, M.C.; Cooper, M.E.; et al. miR-200a Prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes 2011, 60, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Herman-Edelstein, M.; Koh, P.; Burns, W.; Jandeleit-Dahm, K.; Watson, A.; Saleem, M.; Goodall, G.J.; Twigg, S.M.; Cooper, M.E.; et al. E-Cadherin Expression Is Regulated by miR-192/215 by a Mechanism That Is Independent of the Profibrotic Effects of Transforming Growth Factor-β. Diabetes 2010, 59, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.C.K.; Huang, X.R.; Meng, X.; Lan, H.Y. miR-192 mediates TGF-β/Smad3-driven renal fibrosis. J. Am. Soc. Nephrol. 2010, 21, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Zhang, J.; Wang, M.; Lanting, L.; Yuan, H.; Rossi, J.J.; Natarajan, R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 2007, 104, 3432–3437. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, D.; Liu, F.; Xiang, X.; Ling, G.; Xiao, L.; Liu, Y.; Zhu, X.; Zhan, M.; Yang, Y.; et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J. Pathol. 2011, 225, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Krupa, A.; Jenkins, R.; Luo, D.D.; Lewis, A.; Phillips, A.; Fraser, D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J. Am. Soc. Nephrol. 2010, 21, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Maurer, B.; Stanczyk, J.; Jüngel, A.; Akhmetshina, A.; Trenkmann, M.; Brock, M.; Kowal-Bielecka, O.; Gay, R.E.; Michel, B.A.; Distler, J.H.W.; et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010, 62, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Taylor, N.E.; Lu, L.; Usa, K.; Cowley, A.W.; Ferreri, N.R.; Yeo, N.C.; Liang, M. Renal medullary microRNAs in Dahl salt-sensitive rats: MiR-29b regulates several collagens and related genes. Hypertension 2010, 55, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 4126–4135. [Google Scholar] [CrossRef] [PubMed]
- Muth, M.; Theophile, K.; Hussein, K.; Jacobi, C.; Kreipe, H.; Bock, O. Hypoxia-induced down-regulation of microRNA-449a/b impairs control over targeted SERPINE1 (PAI-1) mRNA—A mechanism involved in SERPINE1 (PAI-1) overexpression. J. Transl. Med. 2010, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, Y.; Wang, W.; Chang, B.H.J.; Danesh, F.R. MicroRNA-29c Is a Signature MicroRNA under High Glucose Conditions That Targets Sprouty Homolog 1, and Its in Vivo Knockdown Prevents Progression of Diabetic Nephropathy. J. Biol. Chem. 2011, 286, 11837–11848. [Google Scholar] [CrossRef] [PubMed]
- Kolavennu, V.; Zeng, L.; Peng, H.; Wang, Y.; Danesh, F.R. Targeting of RhoA/ROCK signaling ameliorates progression of diabetic nephropathy independent of glucose control. Diabetes 2008, 57, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Wang, L.; Putta, S.; Wang, M.; Yuan, H.; Sun, G.; Lanting, L.; Todorov, I.; Rossi, J.J.; Natarajan, R. Post-transcriptional Up-regulation of Tsc-22 by Ybx1, a Target of miR-216a, Mediates TGF-β-induced Collagen Expression in Kidney Cells. J. Biol. Chem. 2010, 285, 34004–34015. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Arce, L.; Wang, M.; Putta, S.; Lanting, L.; Natarajan, R. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011, 80, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Dey, N.; Das, F.; Mariappan, M.M.; Mandal, C.C.; Ghosh-Choudhury, N.; Kasinath, B.S.; Choudhury, G.G. MicroRNA-21 Orchestrates High Glucose-induced Signals to TOR Complex 1, Resulting in Renal Cell Pathology in Diabetes. J. Biol. Chem. 2011, 286, 25586–25603. [Google Scholar] [CrossRef] [PubMed]
- Nassirpour, R.; Raj, D.; Townsend, R.; Argyropoulos, C. MicroRNA biomarkers in clinical renal disease: From diabetic nephropathy renal transplantation and beyond. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2016, 98, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Ghai, V.; Wang, K. Recent progress toward the use of circulating microRNAs as clinical biomarkers. Arch. Toxicol. 2016, 90, 2959–2978. [Google Scholar] [CrossRef] [PubMed]
- Kannel, W.B. Blood pressure as a cardiovascular risk factor: Prevention and treatment. JAMA 1996, 275, 1571–1576. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.E.; Messerli, F.H. Hypertension and the heart. J. Hum. Hypertens. 2000, 14, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Beauchet, O.; Celle, S.; Roche, F.; Bartha, R.; Montero-Odasso, M.; Allali, G.; Annweiler, C. Blood pressure levels and brain volume reduction: A systematic review and meta-analysis. J. Hypertens. 2013, 31, 1502–1516. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Després, J.-P.; Fullerton, H.J.; Howard, V.J.; Huffman, M.D.; et al. Heart Disease and Stroke Statistics—2015 Update: A Report From the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef] [PubMed]
- Saran, R.; Robinson, B.; Abbott, K.C.; Agodoa, L.Y.C.; Albertus, P.; Ayanian, J.; Balkrishnan, R.; Bragg-Gresham, J.; Cao, J.; Chen, J.L.T.; et al. US Renal Data System 2016 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2017, 69, A7–A8. [Google Scholar] [CrossRef] [PubMed]
- Dominiczak, A.F.; Bohr, D.F. Nitric Oxide and Its Putative Role in Hypertension. Hypertension 1995, 25, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Song, X.; Zhang, J.; Zhao, L.; Shi, Y.; Li, Z.; Liu, J.; Liu, N.; Yan, Y.; Xiao, Y.; et al. Protection of Human Umbilical Vein Endothelial Cells against Oxidative Stress by MicroRNA-210. Oxid. Med. Cell. Longev. 2017, 2017, 3565613. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-X.; Zeng, D.-Y.; Li, R.-T.; Pang, R.-P.; Yang, H.; Hu, Y.-L.; Zhang, Q.; Jiang, Y.; Huang, L.-Y.; Tang, Y.-B.; et al. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 1979 2012, 60, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Boettger, T.; Beetz, N.; Kostin, S.; Schneider, J.; Krüger, M.; Hein, L.; Braun, T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Invest. 2009, 119, 2634–2647. [Google Scholar] [CrossRef] [PubMed]
- Crowley, S.D.; Coffman, T.M. Recent advances involving the renin-angiotensin system. Exp. Cell Res. 2012, 318, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, G.; Zhu, G.; Liu, H.; Guo, R.; Qi, F.; Zou, J. MicroRNA-155 inhibits angiotensin II-induced vascular smooth muscle cell proliferation. J. Renin-Angiotensin-Aldosterone Syst. JRAAS 2014, 15, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, P.L.; Christensen, G.L.; Schneider, M.; Nossent, A.Y.; Jensen, H.B.; Andersen, D.C.; Eskildsen, T.; Gammeltoft, S.; Hansen, J.L.; Sheikh, S.P. Angiotensin II Type 1 Receptor Signalling Regulates MicroRNA Differentially in Cardiac Fibroblasts and Myocytes. Br. J. Pharmacol. 2011, 164, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Kemp, J.R.; Unal, H.; Desnoyer, R.; Yue, H.; Bhatnagar, A.; Karnik, S.S. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J. Mol. Cell. Cardiol. 2014, 75, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Law, M.R.; Morris, J.K.; Wald, N.J. Use of blood pressure lowering drugs in the prevention of cardiovascular disease: Meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies. BMJ 2009, 338, b1665. [Google Scholar] [CrossRef] [PubMed]
- 2014 Evidence-Based Guideline for the Management of High Blood Pressure in Adults. The JAMA Network. Available online: https://jamanetwork.com/journals/jama/fullarticle/1791497 (accessed on 26 February 2018).
- Macconi, D.; Tomasoni, S.; Romagnani, P.; Trionfini, P.; Sangalli, F.; Mazzinghi, B.; Rizzo, P.; Lazzeri, E.; Abbate, M.; Remuzzi, G.; et al. MicroRNA-324-3p promotes renal fibrosis and is a target of ACE inhibition. J. Am. Soc. Nephrol. 2012, 23, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- BPH/2J. Mouse Strain Datasheet—003005. Available online: https://www.jax.org/strain/003005 (accessed on 8 February 2018).
- Jackson, K.L.; Marques, F.Z.; Watson, A.M.D.; Palma-Rigo, K.; Nguyen-Huu, T.-P.; Morris, B.J.; Charchar, F.J.; Davern, P.J.; Head, G.A. A novel interaction between sympathetic overactivity and aberrant regulation of renin by miR-181a in BPH/2J genetically hypertensive mice. Hypertension 2013, 62, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.J.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Moreno, C.; Williams, J.M.; Lu, L.; Liang, M.; Lazar, J.; Jacob, H.J.; Cowley, A.W.; Roman, R.J. Narrowing a region on rat chromosome 13 that protects against hypertension in Dahl SS-13BN congenic strains. Am. J. Physiol. Heart Circ. Physiol. 2011, 300, H1530–H1535. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kwan, B.C.-H.; Lai, F.M.-M.; Choi, P.C.-L.; Chow, K.-M.; Li, P.K.-T.; Szeto, C.-C. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am. J. Hypertens. 2010, 23, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Savin, V.J.; Sharma, R.; Sharma, M.; McCarthy, E.T.; Swan, S.K.; Ellis, E.; Lovell, H.; Warady, B.; Gunwar, S.; Chonko, A.M.; et al. Circulating factor associated with increased glomerular permeability to albumin in recurrent focal segmental glomerulosclerosis. N. Engl. J. Med. 1996, 334, 878–883. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, E.T.; Sharma, M.; Savin, V.J. Circulating permeability factors in idiopathic nephrotic syndrome and focal segmental glomerulosclerosis. Clin. J. Am. Soc. Nephrol. CJASN 2010, 5, 2115–2121. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zheng, C.; Fan, Y.; Zeng, C.; Chen, Z.; Qin, W.; Zhang, C.; Zhang, W.; Wang, X.; Zhu, X.; et al. Downregulation of microRNA-30 facilitates podocyte injury and is prevented by glucocorticoids. J. Am. Soc. Nephrol. JASN 2014, 25, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Armelloni, S.; Zennaro, C.; Wei, C.; Corbelli, A.; Ikehata, M.; Berra, S.; Giardino, L.; Mattinzoli, D.; Watanabe, S.; et al. BDNF repairs podocyte damage by microRNA-mediated increase of actin polymerization. J. Pathol. 2015, 235, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, W.; Chen, H.-M.; Liu, C.; Wu, J.; Shi, S.; Liu, Z.-H. Plasma microRNA-186 and proteinuria in focal segmental glomerulosclerosis. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2015, 65, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kwan, B.C.-H.; Lai, F.M.-M.; Chow, K.-M.; Li, P.K.-T.; Szeto, C.-C. Urinary sediment miRNA levels in adult nephrotic syndrome. Clin. Chim. Acta Int. J. Clin. Chem. 2013, 418, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, C.; Chen, H.; Li, L.; Tu, Y.; Liu, C.; Shi, S.; Zen, K.; Liu, Z. Evaluation of microRNAs miR-196a, miR-30a-5P, and miR-490 as biomarkers of disease activity among patients with FSGS. Clin. J. Am. Soc. Nephrol. 2014, 9, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Chen, J.; Li, W.; Chen, Y.; Sui, W.; Zhang, Y.; Dai, Y. Genome-wide analysis of microRNAs expression profiling in patients with primary IgA nephropathy. Genome 2013, 56, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Serino, G.; Sallustio, F.; Curci, C.; Cox, S.N.; Pesce, F.; De Palma, G.; Schena, F.P. Role of let-7b in the regulation of N-acetylgalactosaminyltransferase 2 in IgA nephropathy. Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transpl. Assoc. Eur. Ren. Assoc. 2015, 30, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Serino, G.; Sallustio, F.; Cox, S.N.; Pesce, F.; Schena, F.P. Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J. Am. Soc. Nephrol. 2012, 23, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kwan, B.C.-H.; Lai, F.M.-M.; Chow, K.-M.; Li, P.K.-T.; Szeto, C.-C. Elevated Levels of miR-146a and miR-155 in Kidney Biopsy and Urine from Patients with IgA Nephropathy. Dis. Mark. 2011, 30, 171–179. [Google Scholar] [CrossRef]
- Zheng, Y.; Josefowicz, S.Z.; Kas, A.; Chu, T.-T.; Gavin, M.A.; Rudensky, A.Y. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 2007, 445, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Kohlhaas, S.; Garden, O.A.; Scudamore, C.; Turner, M.; Okkenhaug, K.; Vigorito, E. Cutting edge: The Foxp3 target miR-155 contributes to the development of regulatory T cells. J. Immunol. 2009, 182, 2578–2582. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Peng, Y.; Liu, F.; Lei, H. Is IgA nephropathy induced by abnormalities of CD4+CD25+Treg cells in the tonsils? Med. Hypotheses 2007, 69, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Vinuesa, C.G.; Rigby, R.J.; Yu, D. Logic and Extent of miRNA-Mediated Control of Autoimmune Gene Expression. Int. Rev. Immunol. 2009, 28, 112–138. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.J.; Hochberg, D.; Rand, W.M.; Thorley-Lawson, D.A. EBV and systemic lupus erythematosus: A new perspective. J. Immunol. 1950 2005, 174, 6599–6607. [Google Scholar] [CrossRef]
- Gatto, G.; Rossi, A.; Rossi, D.; Kroening, S.; Bonatti, S.; Mallardo, M. Epstein–Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-κB pathway. Nucleic Acids Res. 2008, 36, 6608–6619. [Google Scholar] [CrossRef] [PubMed]
- Ramkissoon, S.H.; Mainwaring, L.A.; Ogasawara, Y.; Keyvanfar, K.; McCoy, J.P.; Sloand, E.M.; Kajigaya, S.; Young, N.S. Hematopoietic-specific microRNA expression in human cells. Leuk. Res. 2006, 30, 643–647. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.J.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Ichii, O.; Otsuka, S.; Sasaki, N.; Namiki, Y.; Hashimoto, Y.; Kon, Y. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int. 2012, 81, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Kwan, B.C.-H.; Lai, F.M.-M.; Tam, L.-S.; Li, E.K.-M.; Chow, K.-M.; Wang, G.; Li, P.K.-T.; Szeto, C.-C. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrol. Carlton Vic 2012, 17, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Costa-Reis, P.; Russo, P.A.; Zhang, Z.; Colonna, L.; Maurer, K.; Gallucci, S.; Schulz, S.W.; Kiani, A.N.; Petri, M.; Sullivan, K.E. The Role of MicroRNAs and Human Epidermal Growth Factor Receptor 2 in Proliferative Lupus Nephritis. Arthritis Rheumatol. 2015, 67, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Sato, F.; Terasawa, K.; Tsuchiya, S.; Toi, M.; Tsujimoto, G.; Shimizu, K. Trastuzumab Produces Therapeutic Actions by Upregulating miR-26a and miR-30b in Breast Cancer Cells. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.; Harris, S.; Venning, M.; Brenchley, P. Investigating the microRNA signature of ANCA associated vasculitis. In La Press Medicale; Elsevise: New York, NY, USA, 2013; Volume 42, pp. 756–757. [Google Scholar]
- Krebs, C.F.; Kapffer, S.; Paust, H.-J.; Schmidt, T.; Bennstein, S.B.; Peters, A.; Stege, G.; Brix, S.R.; Meyer-Schwesinger, C.; Müller, R.-U.; et al. MicroRNA-155 drives TH17 immune response and tissue injury in experimental crescentic GN. J. Am. Soc. Nephrol. 2013, 24, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Unanue, E.; Dixon, F.J. Experimental glomerulonephritis. IV. participation of complement in nephrotoxic nephritis. J. Exp. Med. 1964, 119, 965–982. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S. Nephrotoxic Nephritis and Glomerulonephritis: Animal Model Versus Human Disease. Br. J. Biomed. Sci. 2014, 71, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Blobe, G.C.; Schiemann, W.P.; Lodish, H.F. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000, 342, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, Y.; Qu, S.; Luo, H.; Zhou, Y.; Wang, Y.; Zhao, H.; You, Y.; Xiao, X.; Zuo, X. MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J. Clin. Immunol. 2012, 32, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Luo, H.; Li, Y.; Zhou, Y.; Jiang, Y.; Chai, J.; Xiao, X.; You, Y.; Zuo, X. MicroRNA-21 in scleroderma fibrosis and its function in TGF-β-regulated fibrosis-related genes expression. J. Clin. Immunol. 2013, 33, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Brors, B.; Srivastava, P.K.; Bott, A.; Boehn, S.N.; Groene, H.-J.; Gretz, N. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics 2008, 9, 624. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Qin, S.; Ho, J.; Zhou, J.; Kreidberg, J.A. Systems biology approach to identify transcriptome reprogramming and candidate microRNA targets during the progression of polycystic kidney disease. BMC Syst. Biol. 2011, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Hajarnis, S.; Lakhia, R.; Patel, V. MicroRNAs and Polycystic Kidney Disease. In Polycystic Kidney Disease; Li, X., Ed.; Codon Publications: Brisbane, Australia, 2015; ISBN 978-0-9944381-0-2. [Google Scholar]
- Gomez, I.G.; MacKenna, D.A.; Johnson, B.G.; Kaimal, V.; Roach, A.M.; Ren, S.; Nakagawa, N.; Xin, C.; Newitt, R.; Pandya, S.; et al. Anti–microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J. Clin. Invest. 2015, 125, 141–156. [Google Scholar] [CrossRef] [PubMed]
- A Study of RG-012 in Subjects With Alport Syndrome. Available online: https://clinicaltrials.gov/ct2/show/NCT03373786 (accessed on 9 February 2018).
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [PubMed]
- Linsen, S.E.V.; de Wit, E.; Janssens, G.; Heater, S.; Chapman, L.; Parkin, R.K.; Fritz, B.; Wyman, S.K.; de Bruijn, E.; Voest, E.E.; Kuersten, S.; Tewari, M.; Cuppen, E. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods 2009, 6, 474–476. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Renwick, N.; Brown, M.; Mihailović, A.; Holoch, D.; Lin, C.; Pena, J.T.G.; Nusbaum, J.D.; Morozov, P.; Ludwig, J.; et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 2011, 17, 1697–1712. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakash, A.D.; Jabado, O.; Brown, B.D.; Sachidanandam, R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011, 39, e141. [Google Scholar] [CrossRef] [PubMed]
- Baran-Gale, J.; Kurtz, C.L.; Erdos, M.R.; Sison, C.; Young, A.; Fannin, E.E.; Chines, P.S.; Sethupathy, P. Addressing Bias in Small RNA Library Preparation for Sequencing: A New Protocol Recovers MicroRNAs that Evade Capture by Current Methods. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Dard-Dascot, C.; Naquin, D.; d’Aubenton-Carafa, Y.; Alix, K.; Thermes, C.; van Dijk, E. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics 2018, 19, 118. [Google Scholar] [CrossRef] [PubMed]
- Sorefan, K.; Pais, H.; Hall, A.E.; Kozomara, A.; Griffiths-Jones, S.; Moulton, V.; Dalmay, T. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence 2012, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lee, J.E.; Riemondy, K.; Anderson, E.M.; Yi, R. High-efficiency RNA cloning enables accurate quantification of miRNA expression by deep sequencing. Genome Biol. 2013, 14, R109. [Google Scholar] [CrossRef] [PubMed]
- Argyropoulos, C.; Etheridge, A.; Sakhanenko, N.; Galas, D. Modeling bias and variation in the stochastic processes of small RNA sequencing. Nucleic Acids Res. 2017, 45, e104. [Google Scholar] [CrossRef] [PubMed]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Tsuchiya, S.; Terasawa, K.; Tsujimoto, G. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS ONE 2009, 4, e5540. [Google Scholar] [CrossRef] [PubMed]
- Redshaw, N.; Wilkes, T.; Whale, A.; Cowen, S.; Huggett, J.; Foy, C.A. A comparison of miRNA isolation and RT-qPCR technologies and their effects on quantification accuracy and repeatability. BioTechniques 2013, 54, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Feala, J.D.; Cortes, J.; Duxbury, P.M.; McCulloch, A.D.; Piermarocchi, C.; Paternostro, G. Statistical properties and robustness of biological controller-target networks. PLoS ONE 2012, 7, e29374. [Google Scholar] [CrossRef] [PubMed]
- Riba, A.; Bosia, C.; Baroudi, M.E.; Ollino, L.; Caselle, M. A Combination of Transcriptional and MicroRNA Regulation Improves the Stability of the Relative Concentrations of Target Genes. PLoS Comput. Biol. 2014, 10, e1003490. [Google Scholar] [CrossRef] [PubMed]
- Argyropoulos, C.; Wang, K.; Bernardo, J.; Ellis, D.; Orchard, T.; Galas, D.; Johnson, J.P. Urinary MicroRNA Profiling Predicts the Development of Microalbuminuria in Patients with Type 1 Diabetes. J. Clin. Med. 2015, 4, 1498–1517. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Conklin, D.S.; Mittal, V. High-throughput selection of effective RNAi probes for gene silencing. Genome Res. 2003, 13, 2333–2340. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Benter, I.F. Nonviral delivery of synthetic siRNAs in vivo. J. Clin. Invest. 2007, 117, 3623–3632. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Carrillo, E.; Liu, Y.P.; Berkhout, B. Improving miRNA Delivery by Optimizing miRNA Expression Cassettes in Diverse Virus Vectors. Hum. Gene Ther. Methods 2017, 28, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Blum, J.S.; Saltzman, W.M. High loading efficiency and tunable release of plasmid DNA encapsulated in submicron particles fabricated from PLGA conjugated with poly-l-lysine. J. Control. Release Off. J. Control. Release Soc. 2008, 129, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Sun, Y.; Hu, L.; Zheng, H.; Ji, P.; Pecot, C.V.; Zhao, Y.; Reynolds, S.; Cheng, H.; Rupaimoole, R.; et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 2013, 23, 186–199. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, J.A.; Mugridge, N.B.; Weiss, J.C.; Phillips, L.; Burn, A.L.; Paulin, R.P.; Haasdyk, J.E.; Dickson, K.-A.; Brahmbhatt, V.N.; Pattison, S.T.; et al. Bacterially Derived 400 nm Particles for Encapsulation and Cancer Cell Targeting of Chemotherapeutics. Cancer Cell 2007, 11, 431–445. [Google Scholar] [CrossRef] [PubMed]
- MesomiR 1: A Phase I Study of TargomiRs as 2nd or 3rd Line Treatment for Patients With Recurrent MPM and NSCLC. Available online: https://clinicaltrials.gov/ct2/show/NCT02369198 (accessed on 27 February 2018).
- Aigner, A. Delivery Systems for the Direct Application of siRNAs to Induce RNA Interference (RNAi) In Vivo. Available online: https://www.hindawi.com/journals/bmri/2006/071659/ref/ (accessed on 1 June 2018).
- Hollins, A.J.; Omidi, Y.; Benter, I.F.; Akhtar, S. Toxicogenomics of drug delivery systems: Exploiting delivery system-induced changes in target gene expression to enhance siRNA activity. J. Drug Target. 2007, 15, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Takei, Y.; Kadomatsu, K.; Yuzawa, Y.; Matsuo, S.; Muramatsu, T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004, 64, 3365–3370. [Google Scholar] [CrossRef] [PubMed]
- Minakuchi, Y.; Takeshita, F.; Kosaka, N.; Sasaki, H.; Yamamoto, Y.; Kouno, M.; Honma, K.; Nagahara, S.; Hanai, K.; Sano, A.; et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004, 32, e109. [Google Scholar] [CrossRef] [PubMed]
- Hu-Lieskovan, S.; Heidel, J.D.; Bartlett, D.W.; Davis, M.E.; Triche, T.J. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005, 65, 8984–8992. [Google Scholar] [CrossRef] [PubMed]
- Safety, Tolerability and Pharmacokinetic Study of MRG-201 in Healthy Volunteers. Available online: https://clinicaltrials.gov/ct2/show/NCT02603224 (accessed on 13 February 2018).
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).