1. Introduction
Many plants accumulate storage reserves such as oil, carbohydrate and protein in seeds, and these reserves are important not only for growth of juvenile seedlings but also for human nutrition and as a source of chemicals for industry. Among these storage reserves, the demand for vegetable oils as fuels and chemical resources has increased because there is a need to increase the use of sustainable energy and chemical resources. However, the yield of plant oil is not enough to meet this demand. Therefore, there is an urgent need to improve the yield of oil crops.
The relative proportions of oil, carbohydrate and protein vary depending on the plant species. The accumulation of each class of storage component requires the coordination of many genes that encode the enzymes in the respective pathways [
1,
2]. Therefore, it is important to reveal the regulation of these genes involved in the metabolism of these storage components to understand how the carbon resources in seeds are allocated to each storage component.
During early embryogenesis, carbon and other nutrients are used mainly for rapid cell division and embryo growth. After cell division ceases, during the maturation phase, the resources are allocated to synthesize storage compounds. In
Arabidopsis, starch accumulates transiently in the early stage of the maturation phase, followed by a major increase in oil and protein contents. The transiently accumulated starch disappears later and presumably is used as a carbon source for fatty acid and protein synthesis. During the subsequent late maturation and desiccation phases, the overall biosynthetic activity decreases as the seed prepares for dormancy [
3].
The temporal patterns of gene expression reflect the accumulation of storage compounds. In the early stage of
Arabidopsis seed development, transcripts for starch synthesis are increased transiently and then reduced to low levels by 8–10 days after flowering (DAF). On the other hand, transcripts for fatty acid synthesis increase gradually from the early stage to maximum level at 10–12 DAF and then start to decrease before those for storage protein synthesis. Transcripts for protein synthesis start to increase at ~8 DAF, and high transcript levels are retained at 13 DAF [
4]. Because carbon resources are limited in developing seeds, there is probably competition for these between fatty acid synthesis and protein synthesis.
Seed storage oils are synthesized in the form of triacylglycerol (TAG). In plants, TAG synthesis occurs in different subcellular compartments. Fatty acids are synthesized in plastids and then exported to the cytosol and activated to form acyl-CoA, which serves as the substrate for the esterification of glycerol-3-phosphate at the endoplasmic reticulum to form glycerolipids. The expression of TAG synthesis genes can be characterized by two main expression patterns in
Arabidopsis [
4]. The expression of many genes for fatty acid synthesis followed a bell-shaped pattern that increased during early developmental stages, peaked between eight and 12 DAF, and then decreased as mentioned previously. The first committed step of fatty acid synthesis is catalyzed by acetyl-CoA carboxylase (ACCase). Most plants, except for Gramineae, have a heteromeric form of ACCase in plastids, and the expression of genes for subunits of the heteromeric ACCase, such as biotin carboxyl carrier protein 1 (BCCP1), BCCP2 and biotin carboxylase, shows a bell-shaped pattern during seed development. On the other hand, the expression patterns of genes for enzymes that function outside of the plastids and are involved in the modification of fatty acids, such as
FAD3 encoding linoleate desaturase and
FAE1 encoding fatty acid elongase 1, show a different profile. Their expression starts to increase at ~8 DAF with maximum expression later in development, and a high level of expression is retained at 13 DAF. Recent research revealed that many of the genes for fatty acid synthesis are regulated by the transcription factor WRINKLED 1 (WRI1) [
5,
6].
To date, there have been several trials to manipulate plant oil content and composition [
7,
8]. In order to increase oil content in seeds, genes for enzymes involved in oil synthesis, such as cytosolic acetyl-CoA carboxylase (homomeric ACCase), lysophosphatidic acid acyltransferase and diacylglycerol acyltransferase 1 (DGAT1), have been overexpressed in seeds and have succeeded in enhancing seed oil content [
9,
10,
11,
12]. Several genes have also been manipulated simultaneously to further increase seed oil content [
13,
14,
15].
In this article, we tried to construct a suitable promoter to express genes involved in oil synthesis to increase seed oil content. We suspect that the bell-shaped expression pattern of genes for fatty acid synthesis is one of the limiting factors for oil content in seeds, because the expression of genes for protein synthesis continued to increase after that of fatty acid synthesis genes was reduced. Therefore, we constructed a promoter that started to express at an early stage of seed development, and high-level expression was kept for later in development by connecting the promoters of the AtBCCP2 gene encoding the BCCP2 subunit of acetyl-CoA carboxylase (ACCase) and the AtFAE1 gene. The constructed promoter was ligated upstream of the AtTAG1 gene encoding DGAT1 and introduced into Arabidopsis. Seeds from the transgenic plants carrying the AtTAG1 gene under the control of the chimeric promoter showed higher oil content (up by 18–73%) compared with that of the wild-type (WT).
3. Discussion
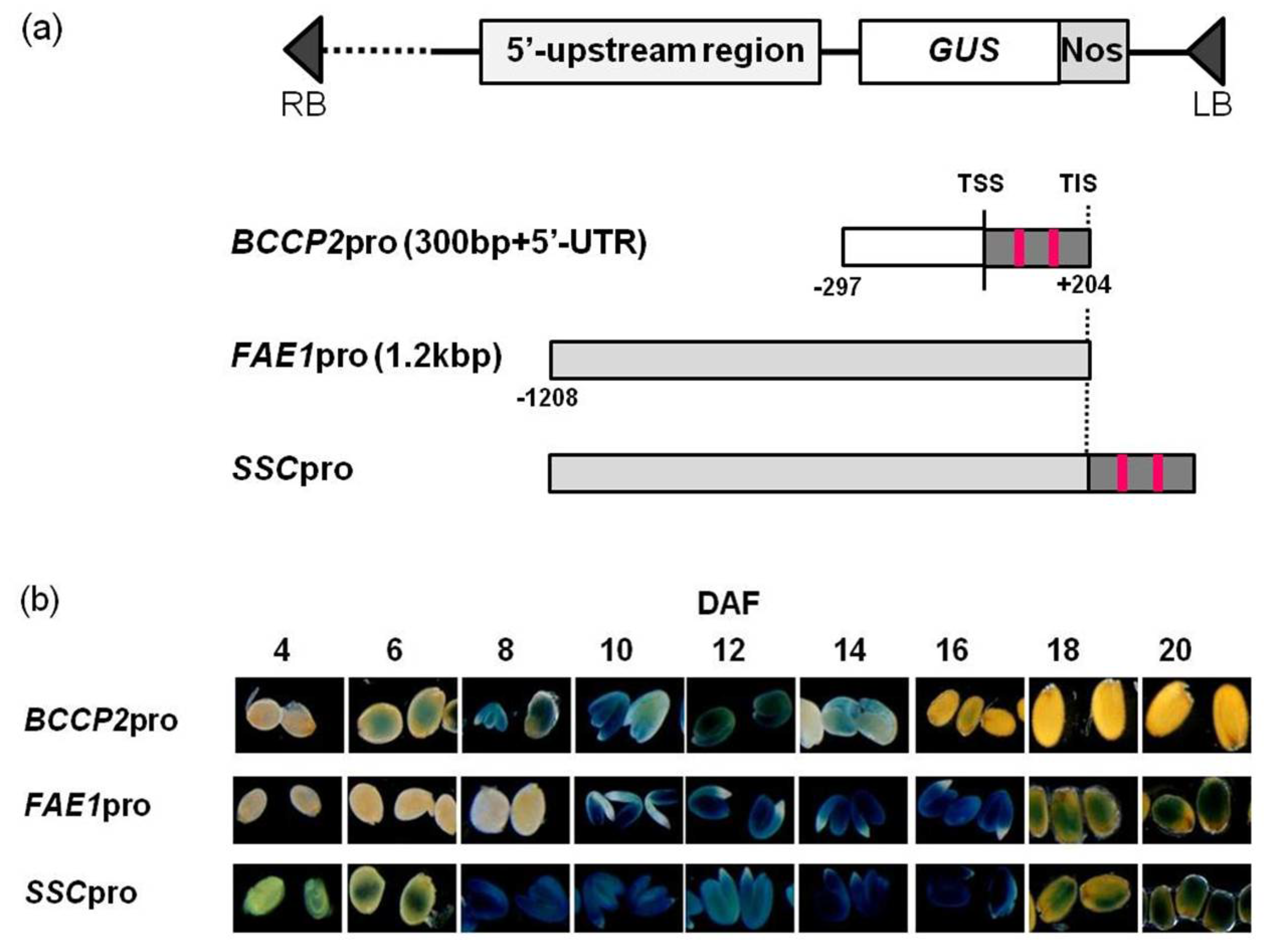
We constructed a chimeric promoter that showed strong activity from early to late stages of seed development by fusing the 5′-UTR of
AtBCCP2 to a 1.2 kb region from the TIS of
AtFAE1 gene (
Figure 1 and
Figure 2).
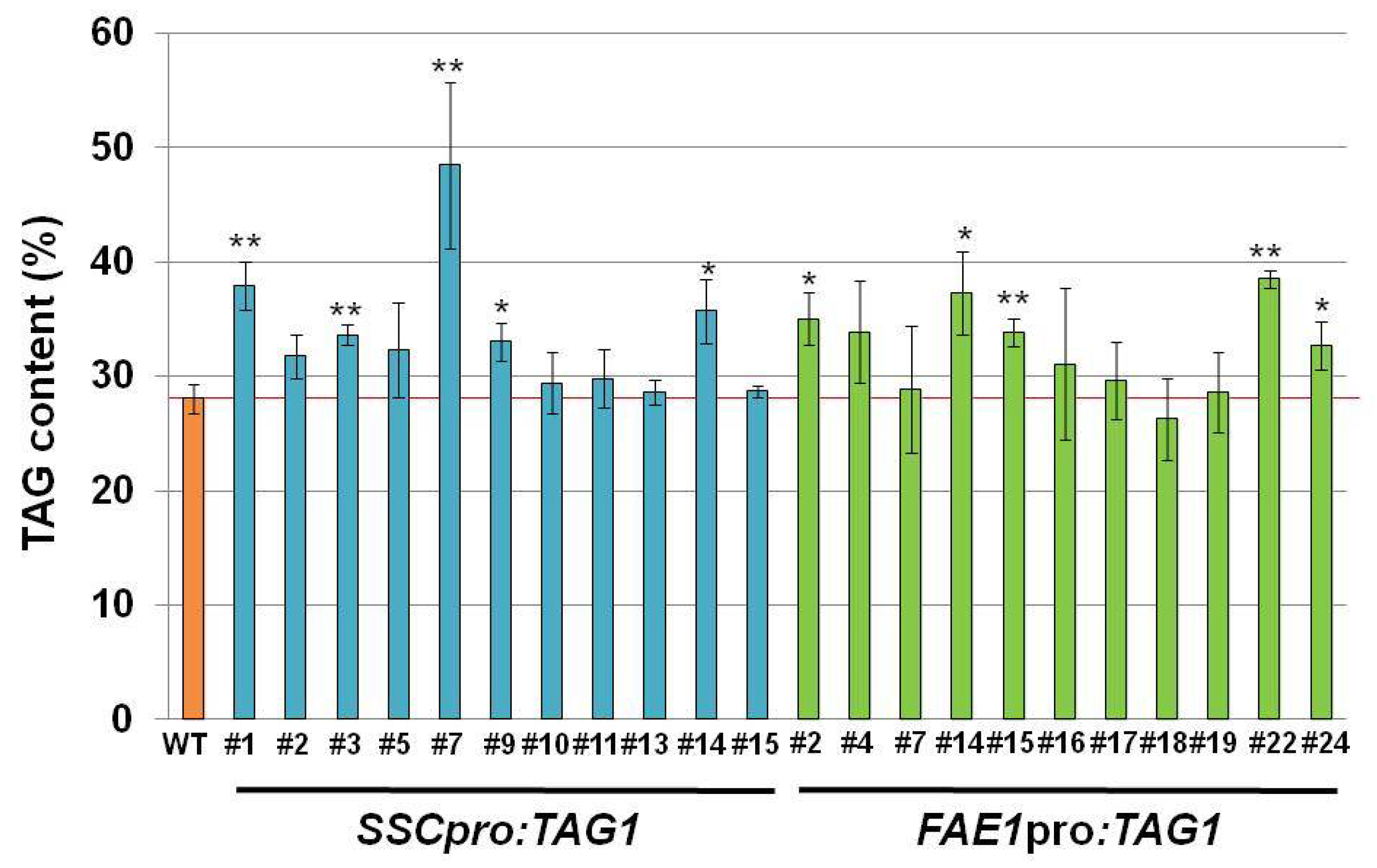
Transgenic plants carrying
SSCpro:TAG1 showed increased amounts of storage oil in seeds compared with WT plants. Compared with plants carrying
FAE1pro:TAG1, similarly increased levels of seed oil were seen in
SSCpro:TAG1 plants (
Figure 3). However, one line containing
SSCpro:TAG1, #7, accumulated much more oil in seeds than the other transgenic plants (
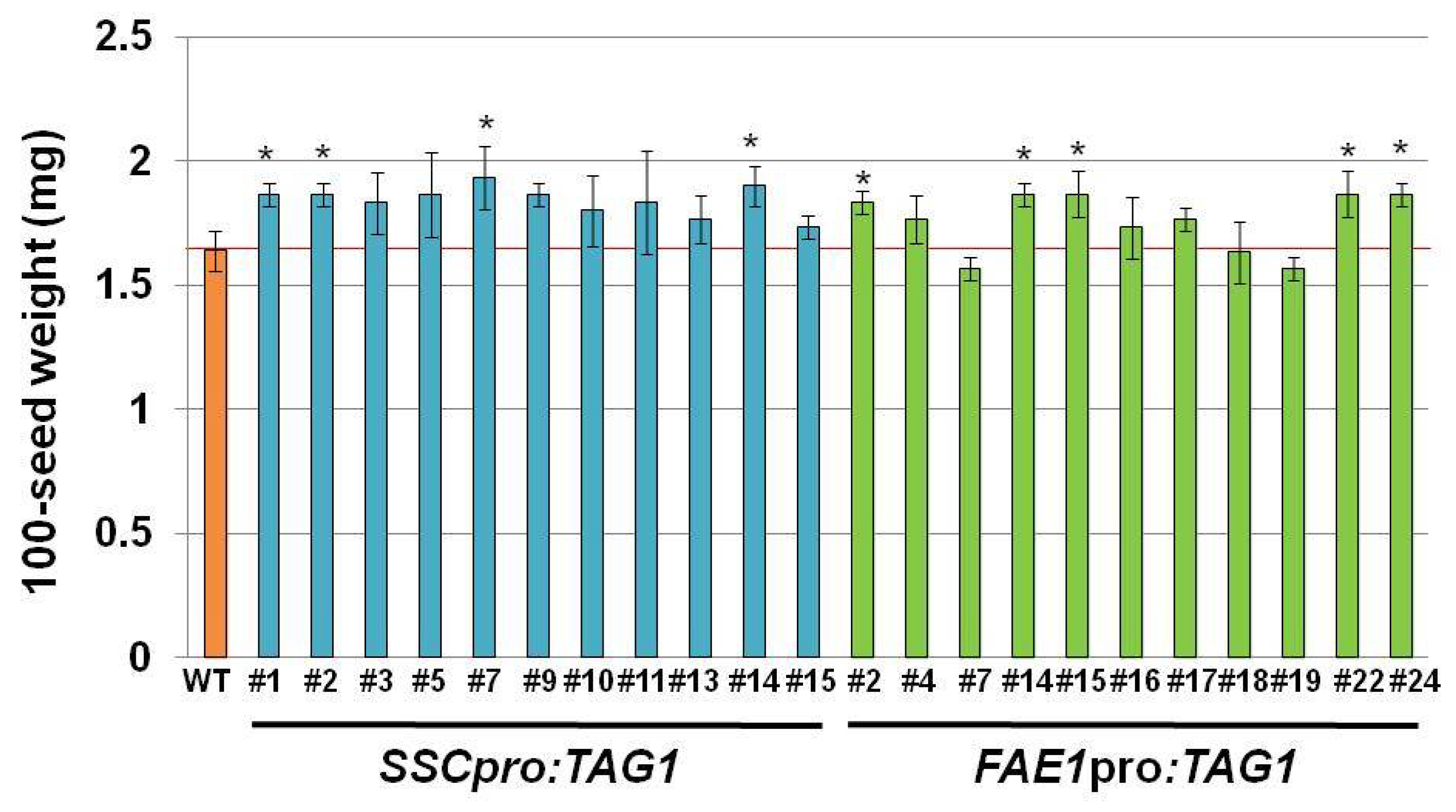
Figure 3). The seed weight and seed size in plant line #7 was also heavier and larger than other seeds. Because we used transgenic plants carrying a single copy transgene, which was confirmed by segregation analysis and quantitative PCR, the effect of copy number on the expression of transgene can be excluded.
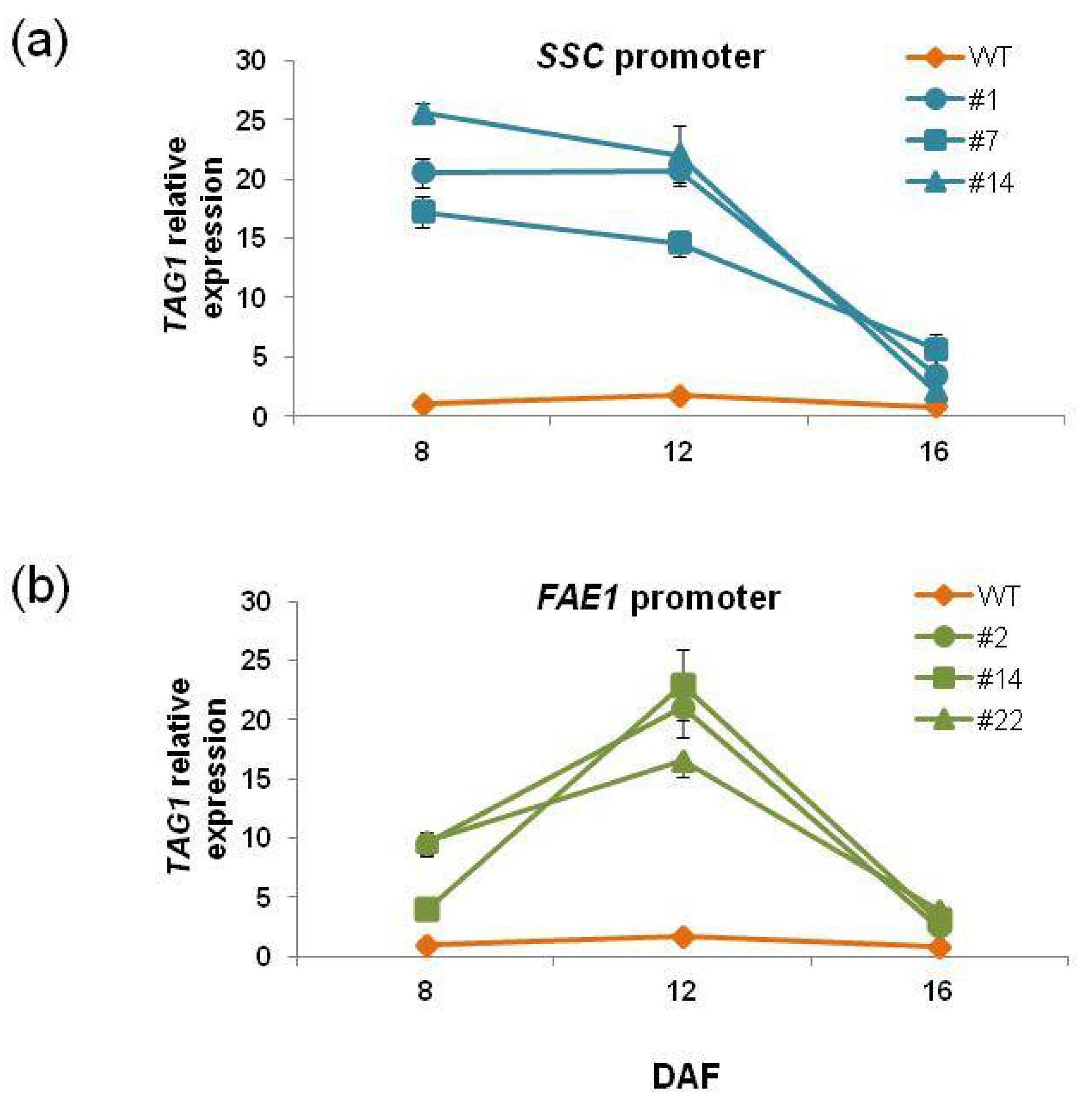
Expression of the
TAG1 gene in transgenic plants showed a unique pattern (
Figure 7).
TAG1 expression in
SSCpro:TAG1 plants at 8 DAF was much higher than that of WT plants. The level was slightly decreased at 12 DAF, and then at 16 DAF, the
TAG1 expression level was lowest among the timepoints measured. On the other hand,
TAG1 expression was highest at 12 DAF in
FAE1pro:TAG1 plants.
We could not find a correlation between oil content and
TAG1 expression level at 8 DAF and 12 DAF. However, it appeared that there was a correlation between oil content and the expression level at 16 DAF. The
SSCpro:TAG1 plant line with highest oil contents (#7) showed the highest expression level of
TAG1 at 16 DAF (relative value 5.6 times), and plant line #14, which accumulated the lowest oil content among the three transgenic lines carrying
SSCpro:TAG1, showed the lowest expression of
TAG1 at 16 DAF (relative value 2.1-fold). Similarly, in plants carrying
FAEpro:TAG1,
TAG1 expression at 16 DAF showed a correlation with oil content; the
TAG1 expression relative value of plant line #22, which had the highest oil content, was 4.0-fold and that in plant line #2, with the lowest oil content, was 2.4-fold. The results indicated that higher expression levels of the
TAG1 gene at 16 DAF (late stage of seed development) were important for higher oil content. In this respect, high expression of the
TAG1 gene in the early stage in
SSCpro:TAG1 plants was not advantageous to accumulate higher content of TAG. However, the chimeric promoter showed high expression in the late stage of seed development, which the
FAE1 promoter could not produce (
Figure 2 and
Figure 7). Therefore, transgenic plants with ideal expression levels of
TAG1, such as plant line #7 (
Figure 7), which accumulate much more oil than other transgenic plants, could be produced by using the chimeric promoter.
The timing of gene expression is important. Kanai et al. (2015) [
15] showed that the
AtWRI1 gene under control of the
FUSCA3 (
FUS3) promoter, which is activated during the middle phase of seed development (peaking at around 8 DAF), enhanced
Arabidopsis seed oil content much more than that under the control of the
35S promoter or the
LEA promoter, which is activated in the late phase of seed development (increases from >8 DAF). Previous studies also showed that activation of
WRI1 during the late phase of seed development was not effective in increasing oil content [
5,
20]. Because fatty acids are essential not only for oil synthesis but also for membrane biogenesis, which is required for embryo development, fatty acid synthesis probably has to be largely activated during the early-to-middle phase of seed development. Moreover, if there is competition between oil synthesis and protein synthesis for carbon resources during seed development, enhancement of fatty acid synthesis should be started before protein synthesis starts to increase. On the other hand, activation of the
TAG1 gene in the late phase of seed development successfully increased oil content [
11,
12].
TAG1 encodes DGAT1, which catalyzes the last step of TAG synthesis and is originally expressed in the later stage of seed development in
Arabidopsis [
21]. Therefore, later expression is probably more effective for the
TAG1 gene to enhance oil production.
In this study, we succeeded in increasing seed oil content using a constructed chimeric promoter. However, expressing genes for enzymes or transcription factors involved in fatty acid synthesis, such as genes for WRI1 or homomeric ACCase (cytosolic ACCase or ACCase from Garmineae), which play a pivotal role in fatty acid synthesis, can take advantage of the properties of the SCC promoter fully because high expression of these genes in the early stage of seed development is required for high accumulation of TAG.
Because the promoter contains the WRI1 binding site and the
FAE1 promoter, which is reported to be regulated by B3 domain factors, LEAFY COTYKEDON 2 and FUS3, through the RY motif [
22], we can use the chimeric promoter in other plants that have similar transcriptional regulation of seed development to
Arabidopsis. Brassicaceae, including
Camelina sativa and
Brassica napus, may be good target plants to be manipulated using the chimeric promoter. Expression of target genes in novel expression patterns could be a promising strategy for substance production in seeds or research on seed development.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The surfaces of seeds were sterilized in 20% bleach for 15 min, rinsed five times with sterile water, and then plated on half-strength Murashige and Skoog (1/2 MS) medium, pH 5.7, 0.8% agar, and 1% sucrose. The transgenic plants were screened on the 1/2 MS with kanamycin (30 μg mL−1). The seeds from homozygous transgenic and WT plants were placed on 1/2 MS without kanamycin. Plates were first placed at 4 °C for 2 days and then transferred to a growth chamber at 23 °C with a light period of 16 h (65 μmol m−2 s−1) and a dark period of 8 h. Seedlings were transferred to soil at 7–10 days after germination.
To collect developmental-stage siliques, colored threads were used to tag flowers on the day of flowering (day 0) when petals just appeared. The siliques that developed from the tagged flowers were collected at specific time points.
For measurement of oil content in seeds, mature seeds were collected from homozygous transgenic T3 plants which were planted at same time and grown under the same condition.
4.2. Construction of Binary Vector and Transformation
Primers used in PCR reactions are described in
Table 1. The 5′-UTR fragment of the
AtBCCP2 gene was amplified using primers BCCP2XF/BCCP2SlR. The promoter of the
AtFAE1 gene was amplified using primers FAEp2kSalF/FAEpSalR. The PCR products were cloned into pCR-Blunt (Invitrogen, Carlsbad, CA, USA). The 5′-UTR fragment of the
AtBCCP2 was digested with
XhoI and
SalI, and the
FAE1 promoter was digested with
SalI and
HindIII whose site is located 1.2 kb upstream of the TIS of the
AtFAE1 gene. Each fragment was collected and ligated to the
HindIII/
SalI site of pBI101 to construct
SSCpro:GUS. To construct
FAE1pro:TAG1,
HindIII/
SalI fragment of the
FAE1 promoter was ligated to the
HindIII/
SalI site of pBI101.
In order to increase the restriction sites to insert the TAG1 gene, the GUS gene was replaced by the GFP gene with several restriction sites on both the 3′ and 5′ sites. The GFP gene was amplified using primer set GFPXXHF/GFPHSSR and the amplified fragment was digested with SalI and SacI. The SSCpro:GUS vector was digested with SalI and SacI and the GUS gene was replaced by the GFP fragment (SSCpro:GFP). The TAG1 fragment was amplified using primer set TAG1XbaSmaF/TAG1EcoRVR and digested with XbaI/EcoRV, and was ligated into the XbaI/SmaI sites of the SSCpro:GFP vector to construct SSCpro:TAG1.
Arabidopsis thaliana (ecotype Col-0) was transformed with
Agrobacterium tumefaciens (strains GV3101) using the floral dip method [
23]. Several transgenic lines were screened on 1/2 MS with kanamycin (30 μg mL
−1) and homozygous transformant lines were established from isolated independent transgenic lines. We confirmed the introduced gene by PCR using a primer set TAG1XbaSmaF/TAG1EcoRVR.
4.3. GUS Staining Assay and GUS Fluorometric Assay
Histochemical staining of GUS activity was conducted using transgenic plants as described previously [
16]. Fresh tissue was incubated in reaction solution (1 mM X-Gluc, 50 mM sodium phosphate buffer pH 7.0, 1 mM potassium ferricyanide/ferrocyanide mixture, 0.01% Triton X-100, 10 mM 2-mercaptoethanol, 20% methanol, 1 mM EDTA) at 37 °C for 12 h. After staining, the tissue was fixed in a mixture of ethanol:acetic acid at a 6:1 ratio and rinsed with 70% ethanol. Tissues were observed and photographed using a dissecting microscope.
GUS activity in protein extracts was measured using the fluorogenic substrate 4-methylumbelliferyl b-
d-glucuronide (MUG) as described previously [
16]. Proteins were extracted from seeds at 8, 12, and 16 DAF (more than five independent plants). Total protein extracts were prepared by grinding the tissues in extraction buffer (50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 10 mM β-mercaptoethanol) containing 0.1% (
w/
v) SDS and 1% Triton X-100, followed by centrifugation at 13,000×
g for 10 min. GUS activity in the supernatants was measured in extraction buffer containing 1 mM MUG and 20% methanol. Reactions were stopped by adding 0.2 M Na
2CO
3, and the amount of 4-methylumbelliferone was calculated by relating relative fluorescence units with those of a standard of known concentration. The protein concentration of extracts was determined using Protein Assay Kits (Bio-Rad, Hercules, CA, USA).
4.4. Measurement of Oil Content
Lipids were extracted in accordance with the Folch method [
24] with some modifications.
A total of 10 mg of seeds was ground in a glass tube, and 2 mL of chloroform:methanol (1:1, v/v) was added. After standing at room temperature for 30 min, 1 mL of 0.9% NaCl solution (w/v) was added and mixed by vortexing. The extract was centrifuged at 845× g for 15 min at 4 °C. The lower layer was transferred to a new tube. Again, 1 mL of 0.9% NaCl solution (w/v) was added to the collected extract and mixed by vortexing. The extract was centrifuged at 845× g for 5 min at 4 °C. The same process was repeated once again. Finally, the lower layer was transferred to a glass vial and the extract was dried and dissolved in 100 μL of chloroform. A 20 μL aliquot of lipid sample was spotted onto a silica gel thin-layer chromatography (TLC) plate (Merck Millipore, Darmstadt, Germany) to separate TAG from total lipid. TLC separation was carried out using petroleum ether:diethyl ether:acetic acid (70:30:1). The TAG spot was transferred to a glass tube, and 1 mL of hydrogen chloride methanol solution was added and heated at 80 °C for 30 min. After cooling, 1 mL hexane and 1 mL 0.9% NaCl solution was added and mixed by vortexing. The sample was centrifuged at 845× g for 5 min at 4 °C and 800 μL of supernatant was transferred to a new tube. The collected solution was dried and the collected fatty acid methyl ester was dissolved in 50 μL of hexane. The fatty acid methyl esters were quantified using gas–liquid chromatography (GC2014, Shimadzu, Kyoto, Japan) using a capillary column (BPX-90, SGE Analytical Science, Ringwood, VIC, Australia).
4.5. Expression Analysis
Total RNA was isolated from seeds at 8, 12, and 16 DAF using TriPure (Roche Diagnostics, Mannheim, Germany) and Fruitmate (TAKARA, Ohtsu, Japan). cDNA was synthesized using 0.5 μg of RNA as a template and ReverTra Ace (TOYOBO, Ohsaka, Japan).
Real-time PCR amplification of cDNAs was conducted using a LightCycler 480 (Roche Diagnostics) in a 384-well PCR plate. The reaction was carried out in a 10 μL reaction volume containing 5 μL SYBR Premix Ex Taq II (TAKARA) with 0.2 μM each of the forward and reverse primers and 1 μL cDNA (10-fold dilution). The primer sets used for real-time PCR are shown in
Table 1. The primers for amplification of
TAG1 were synthesized according to Yang et al. (2011) [
25]. Expression levels of
ELONGATION FACTOR1 αA4 (
EF1αA4) were used for signal normalization of real-time PCR. The
EF1αA4 primers were designed by the Universal ProbeLibrary Assay Design Center (Roche Diagnostics). All reactions were performed independently at least five times, and at least three sets of consistent data were used for analyses. Relative gene expression levels were calculated using the 2
−ΔΔCT method [
26]. To validate the reliability of the data, amplification efficiencies between the target genes and the house-keeping genes of all the real-time PCR reactions were compared, and dissociation curves of all PCR products were examined to ensure the quality of the PCR.