Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza
Abstract
:1. Introduction
2. Results
2.1. Identification of WRKY Genes in the S. miltiorrhiza Transcriptome
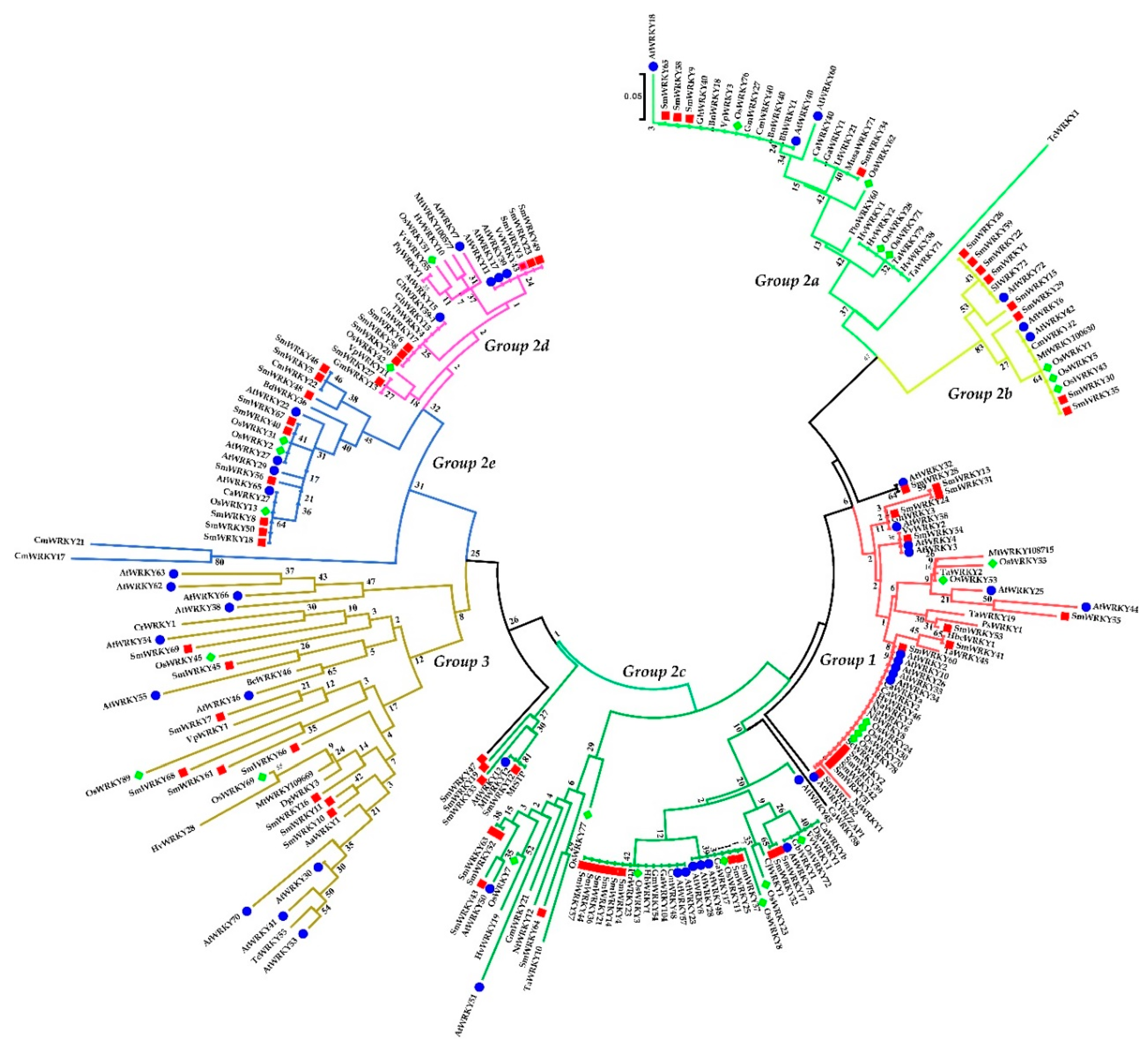
2.2. Phylogenetic Analysis of the S. miltiorrhiza WRKY Family
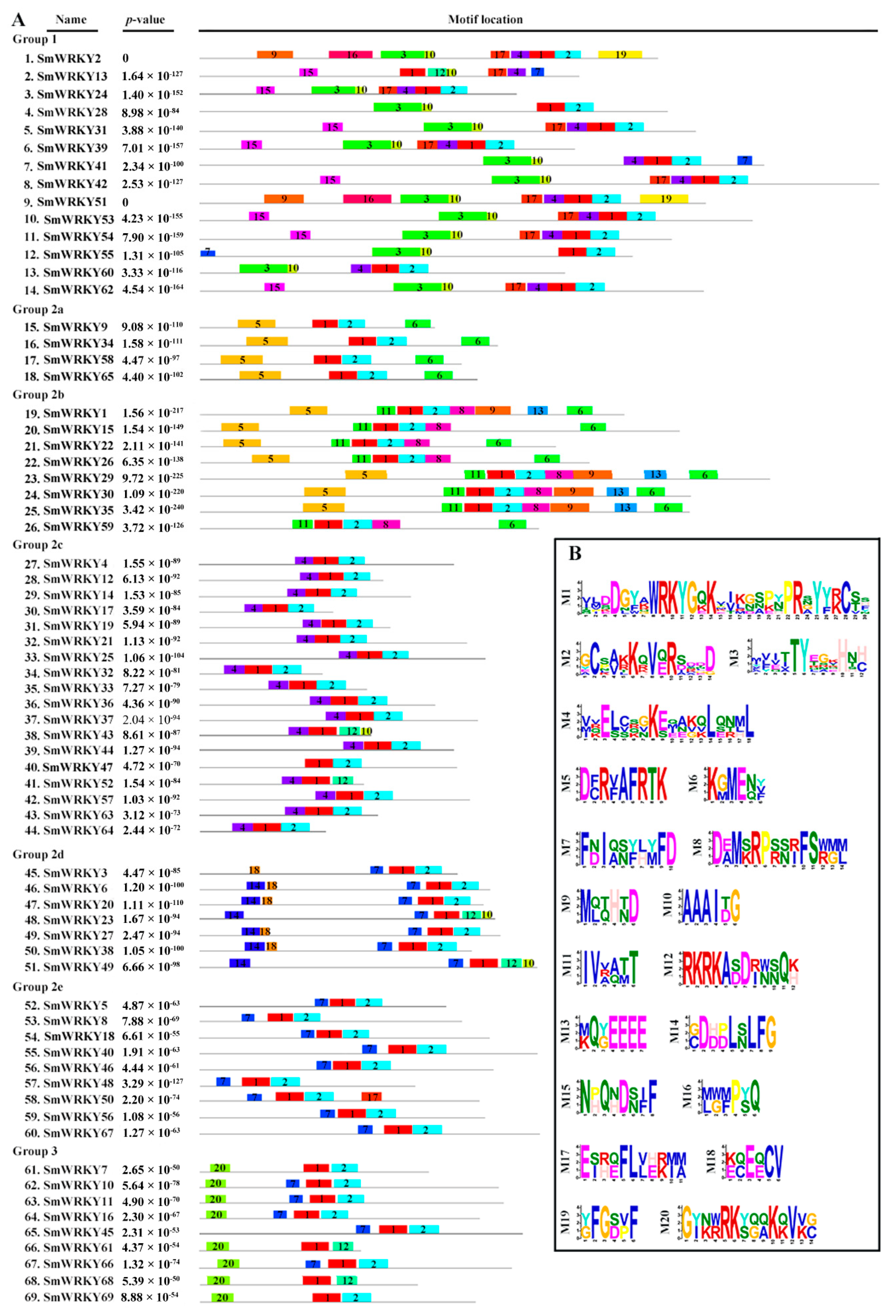
2.3. Motif Composition Analysis of SmWRKY Proteins
2.4. Tissue-Specific Expression of SmWRKYs
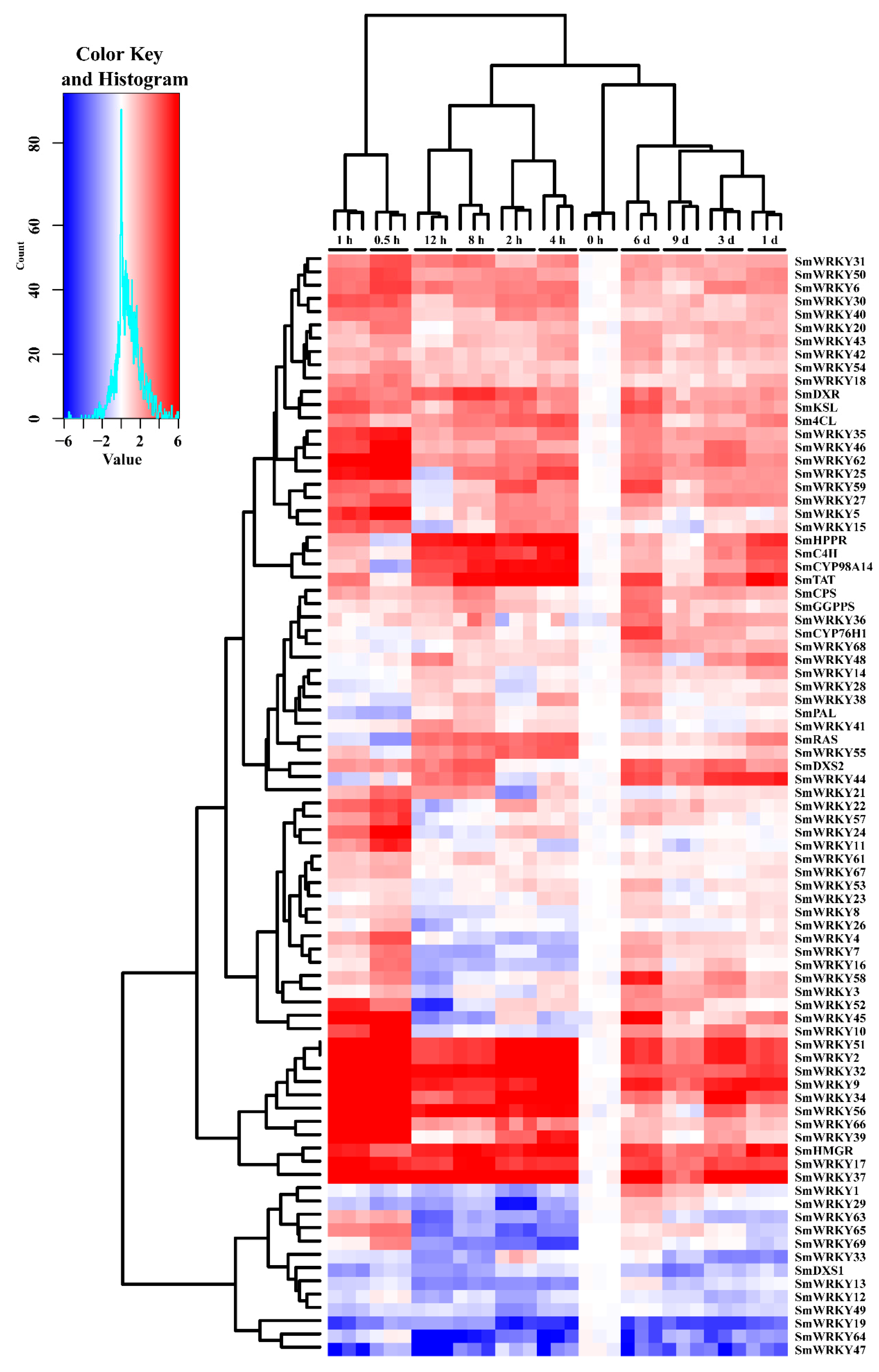
2.5. Differential Expression of SmWRKY Genes in Response to MeJA
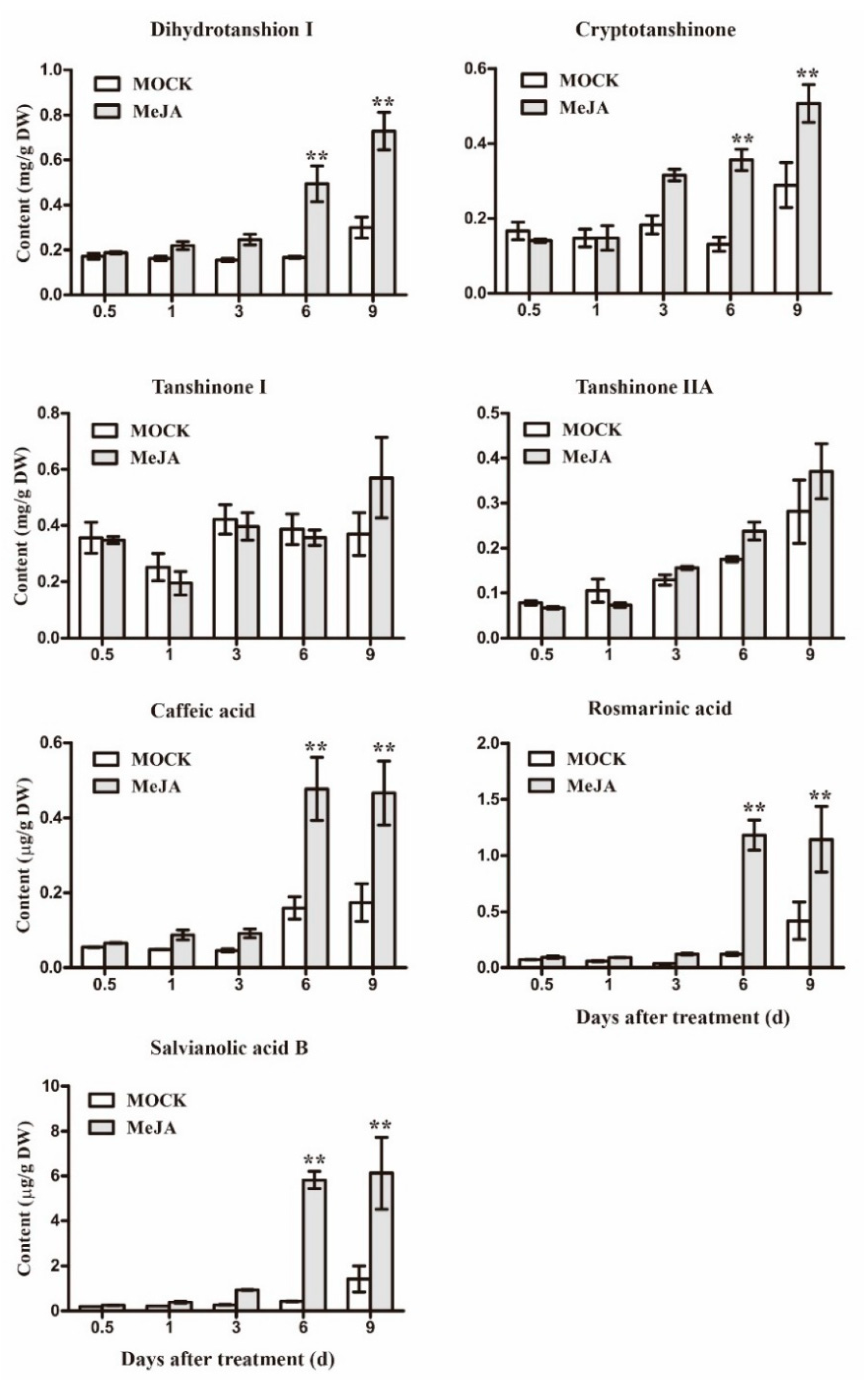
2.6. MeJA-Induced Changes in Secondary Metabolism Biosynthesis
2.7. Integration of Gene Expression Levels and Metabolites Accumulation Analysis
3. Discussion
4. Materials and Methods
4.1. Identification of SmWRKY Genes Family Members
4.2. Multiple Sequence Alignment, Phylogenetic Analysis
4.3. Materials and Hairy Root Culture with MeJA Treatment
4.4. Transcriptional Analysis by Real-Time Quantitative PCR (RT-qPCR)
4.5. Metabolite Extraction and HPLC Analysis
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| C4H | cinnamic acid 4-hydroxylase |
| CPS | copalyl diphosphate synthase |
| CYP98A14 | cytochrome P450-dependent monooxygenase |
| CYP76AH1 | cytochrome P450-dependent monooxygenase |
| DXR | 1-deoxy-d-xylulose 5-phosphate reductoisomerase |
| DXS | 1-deoxy-d-xylulose 5-phosphate synthase |
| GGPP | geranylgeranyl diphosphate |
| GGPPS | geranylgeranyl diphosphate synthase |
| HMGR | 3-hydroxy-3-methylglutaryl CoA reductase |
| HPLC | high performance liquid chromatography |
| HPPR | 4-hydroxyphenylpyruvate reductase |
| KSL | kaurene synthase-like |
| MeJA | methyl jasmonate |
| PAL | phenylalanine ammonia-lyase |
| PDA | photodiode array detector |
| RT-qPCR | quantitative real-time PCR |
| S. miltiorrhiza | Salvia miltiorrhiza |
| TAT | tyrosine aminotransferase |
| 4CL | hydroxycinnamate coenzyme A ligase |
| TFs | transcription factors |
| MVA | mevalonate |
| MEP | methylerythritol phosphate |
References
- Han, J.-Y.; Fan, J.-Y.; Horie, Y.; Miura, S.; Cui, D.-H.; Ishii, H.; Hibi, T.; Tsuneki, H.; Kimura, I. Ameliorating effects of compounds derived from Salvia miltiorrhiza root extract on microcirculatory disturbance and target organ injury by ischemia and reperfusion. Pharmacol. Ther. 2008, 117, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Morris-Natschke, S.L.; Lee, K.-H. New developments in the chemistry and biology of the bioactive constituents of tanshen. Med. Res. Rev. 2007, 27, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S.; et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015, 82, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Di, P.; Zhang, L.; Chen, J.; Tan, H.; Xiao, Y.; Dong, X.; Zhou, X.; Chen, W. 13C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 2013, 8, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, W.; Li, Q.; Li, Y.; Luo, H.; Song, J.; Sun, C.; Qian, J.; Zhu, Y.; Hayward, A.; et al. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 2015, 241, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yuan, L.; Wu, B.; Li, X.; Chen, S.; Lu, S. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 2012, 63, 2809–2823. [Google Scholar] [CrossRef] [PubMed]
- Kai, G.Y.; Xu, H.; Zhou, C.C.; Liao, P.; Xiao, J.B.; Luo, X.Q.; You, L.J.; Zhang, L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.H.; Ma, Y.; Tang, J.F.; He, Y.L.; Liu, Y.C.; Ma, X.J.; Shen, Y.; Cui, G.H.; Lin, H.X.; Rong, Q.X.; et al. The biosynthetic pathways of tanshinones and phenolic acids in Salvia miltiorrhiza. Molecules 2015, 20, 16235–16254. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.M.; Zhu, Y.J.; Song, J.Y.; Xu, L.J.; Sun, C.; Zhang, X.; Xu, Y.H.; He, L.; Sun, W.; Xu, H.B.; et al. Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol. Plant. 2014, 152, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ma, X.H.; Cai, Y.; Ma, Y.; Zhan, Z.L.; Zhou, Y.J.; Liu, W.J.J.; Guan, M.X.; Yang, J.; Cui, G.H.; et al. Cytochrome p450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol. 2016, 210, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, Y.J.J.; Hillwigc, M.L.; Shen, Y.; Yang, L.; Wang, Y.J.; Zhang, X.A.; Liu, W.J.; Peters, R.J.; Chen, X.Y.; et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. USA 2013, 110, 12108–12113. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Løvdal, T.; Olsen, K.M.; Slimestad, R.; Verheul, M.; Lillo, C. Synergetic effects of nitrogen depletion, temperature, and light on the content of phenolic compounds and gene expression in leaves of tomato. Phytochemistry 2010, 71, 605–613. [Google Scholar] [CrossRef] [PubMed]
- D’Angeli, S.; Matteucci, M.; Fattorini, L.; Gismondi, A.; Ludovici, M.; Canini, A.; Altamura, M.M. OeFAD8, OeLIP and OeOSM expression and activity in cold-acclimation of Olea europaea, a perennial dicot without winter-dormancy. Planta 2016, 243, 1279–1296. [Google Scholar] [CrossRef] [PubMed]
- Gismondi, A.; Di Marco, G.; Canuti, L.; Canini, A. Antiradical activity of phenolic metabolites extracted from grapes of white and red Vitis vinifera L. cultivars. Vitis J. Grapevine Res. 2017, 56, 19–26. [Google Scholar]
- Bulgakov, V.P.; Avramenko, T.V.; Tsitsiashvili, G.S. Critical analysis of protein signaling networks involved in the regulation of plant secondary metabolism: Focus on anthocyanins. Crit. Rev. Biotechnol. 2017, 37, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, P.; Yang, D.; Li, W.; Liang, Z.; Liu, Y.; Liu, F. Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. PLoS ONE 2013, 8, e73259. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.P.; Jiang, X.Y.; Feng, L.; Tao, R.; Li, Y.L.; Huang, L.Q. Cloning, molecular characterization and functional analysis of a putative R2R3 MYB transcription factor of the phenolic acid biosynthetic pathway in S. miltiorrhiza bge. F. Alba. Plant Cell Tissue Organ Cult. 2016, 124, 151–168. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, W.; Chen, J.; Tan, H.; Xiao, Y.; Li, Q.; Ji, Q.; Gao, S.; Chen, L.; Chen, S.; et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 2016, 6, 22852. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Okada, K.; Miyamoto, K.; Koga, J.; Shibuya, N.; Nojiri, H.; Yamane, H. Ostgap1, a bzip transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. J. Biol. Chem. 2009, 284, 26510–26518. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Nishizawa, Y.; Minami, E.; Nojiri, H.; Yamane, H.; Okada, K. Overexpression of the bzip transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 2014, 173, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, H.; Li, F.; Chen, Z.; Li, X.; Zhu, L.; Wang, G.; Yu, J.; Huang, D.; Lang, Z. The maize transcription factor ereb58 mediates the jasmonate-induced production of sesquiterpene volatiles. Plant J. 2015, 84, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.P.; Zhang, H.; Kong, L.; Gao, G.; Luo, J.C. Planttfdb 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Yi, S.Y.; Yu, S.H.; Moon, J.S.; Park, J.M.; Choi, D. CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Mol. Cells 2006, 22, 58–64. [Google Scholar] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmüller, R. WRKY transcription factor: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, X.; Yang, F.; Fan, D.; Jiang, Y.Z.; Luo, K.M. PtrWRKY19, a novel WRKY transcription factor, contributes to the regulation of pith secondary wall formation in Populus trichocarpa. Sci. Rep. 2016, 6, 18643. [Google Scholar] [CrossRef] [PubMed]
- Guillaumie, S.; Mzid, R.; Mechin, V.; Leon, C.; Hichri, I.; Destrac-Irvine, A.; Trossat-Magnin, C.; Delrot, S.; Lauvergeat, V. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol. 2010, 72, 215–234. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Miyamoto, K.; Ogawa, S.; Masuda, Y.; Shimizu, T.; Kishi-Kaboshi, M.; Takahashi, A.; Nishizawa, Y.; Minami, E.; Nojiri, H.; et al. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS ONE 2014, 9, e98737. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Z.; Niu, Y.Y.; Xu, J.; Li, Y.; Luo, H.M.; Zhu, Y.J.; Liu, M.Z.; Wu, Q.; Song, J.Y.; Sun, C.; et al. Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tissue Organ Cult. 2013, 114, 269–277. [Google Scholar] [CrossRef]
- Li, C.; Li, D.; Shao, F.; Lu, S. Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genom. 2015, 16, 200. [Google Scholar] [CrossRef] [PubMed]
- Schluttenhofer, C.; Yuan, L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.-L.; Guo, Z.-J.; Wang, H.-H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. Transparent testa glabra2, a trichome and seed coat development gene of arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef] [PubMed]
- Schluttenhofer, C.; Pattanaik, S.; Patra, B.; Yuan, L. Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genom. 2014, 15, 502. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Shao, F.; Ma, Y.; Lu, S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: Genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 2013, 40, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Xing, B.; Yang, D.; Liu, L.; Han, R.; Sun, Y.; Liang, Z. Phenolic acid production is more effectively enhanced than tanshinone production by methyl jasmonate in Salvia miltiorrhiza hairy roots. Plant Cell Tissue Organ Cult. 2018. [Google Scholar] [CrossRef]
- Yan, Q.; Shi, M.; Ng, J.; Wu, J.Y. Elicitor-induced rosmarinic acid accumulation and secondary metabolism enzyme activities in Salvia miltiorrhiza hairy roots. Plant Sci. 2006, 170, 853–858. [Google Scholar] [CrossRef]
- Van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J.M. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean wrky-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Guo, R.; Xu, X.; Gao, M.; Li, X.; Song, J.; Zheng, Y.; Wang, X. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef] [PubMed]
- Rischer, H.; Orešič, M.; Seppänen-Laakso, T.; Katajamaa, M.; Lammertyn, F.; Ardiles-Diaz, W.; Van Montagu, M.C.; Inzé, D.; Oksman-Caldentey, K.M.; Goossens, A. Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc. Natl. Acad. Sci. USA 2006, 103, 5614–5619. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Li, Q.; Tan, H.; Chen, J.; Xiao, Y.; Ma, R.; Gao, S.; Zerbe, P.; Chen, W.; Zhang, L. Gene-to-metabolite network for biosynthesis of lignans in MeJA-elicited Isatis indigotica hairy root cultures. Front. Plant Sci. 2015, 6, 952. [Google Scholar] [CrossRef] [PubMed]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.A.; He, X.; Dixon, R.A. Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol. 2008, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Akagi, A.; Fukushima, S.; Okada, K.; Jiang, C.J.; Yoshida, R.; Nakayama, A.; Shimono, M.; Sugano, S.; Yamane, H.; Takatsuji, H. WRKY45-dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Mol. Biol. 2014, 86, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, V.K.; Negi, N.; Khurana, P. Genome-wide identification and structural, functional and evolutionary analysis of WRKY components of mulberry. Sci. Rep. 2016, 6, 30794. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.X.; Wei, J.B.; Wu, F.L.; Zhang, H.H.; Yang, D.F.; Liang, Z.S.; Jin, W.B. DsTRD: Danshen transcriptional resource database. PLoS ONE 2016, 11, e0149747. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. Expasy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.S.; Yang, D.F.; Liang, X.; Zhang, Y.J.; Liu, Y.; Liu, F.H. Roles of reactive oxygen species in methyl jasmonate and nitric oxide-induced tanshinone production in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2012, 31, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Du, X.; Liang, X.; Han, R.; Liang, Z.; Liu, Y.; Liu, F.; Zhao, J. Different roles of the mevalonate and methylerythritol phosphate pathways in cell growth and tanshinone production of Salvia miltiorrhiza hairy roots. PLoS ONE 2012, 7, e46797. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.F.; Sheng, D.F.; Duan, Q.M.; Liang, X.; Liang, Z.S.; Liu, Y. PEG and ABA trigger the burst of reactive oxygen species to increase tanshinone production in Salvia miltiorrhiza hairy roots. J. Plant Growth Regul. 2012, 31, 579–587. [Google Scholar] [CrossRef]
- Liu, L.; Yang, D.; Liang, T.; Zhang, H.; He, Z.; Liang, Z. Phosphate starvation promoted the accumulation of phenolic acids by inducing the key enzyme genes in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2016, 35, 1933–1942. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Guo, W.; Yang, D.; Hou, Z.; Liang, Z. Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza. Int. J. Mol. Sci. 2018, 19, 1593. https://doi.org/10.3390/ijms19061593
Yu H, Guo W, Yang D, Hou Z, Liang Z. Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza. International Journal of Molecular Sciences. 2018; 19(6):1593. https://doi.org/10.3390/ijms19061593
Chicago/Turabian StyleYu, Haizheng, Wanli Guo, Dongfeng Yang, Zhuoni Hou, and Zongsuo Liang. 2018. "Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza" International Journal of Molecular Sciences 19, no. 6: 1593. https://doi.org/10.3390/ijms19061593
APA StyleYu, H., Guo, W., Yang, D., Hou, Z., & Liang, Z. (2018). Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza. International Journal of Molecular Sciences, 19(6), 1593. https://doi.org/10.3390/ijms19061593



