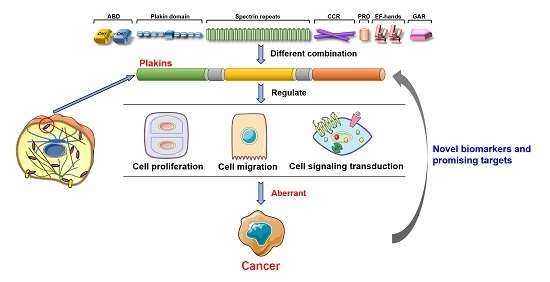
Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer
Abstract
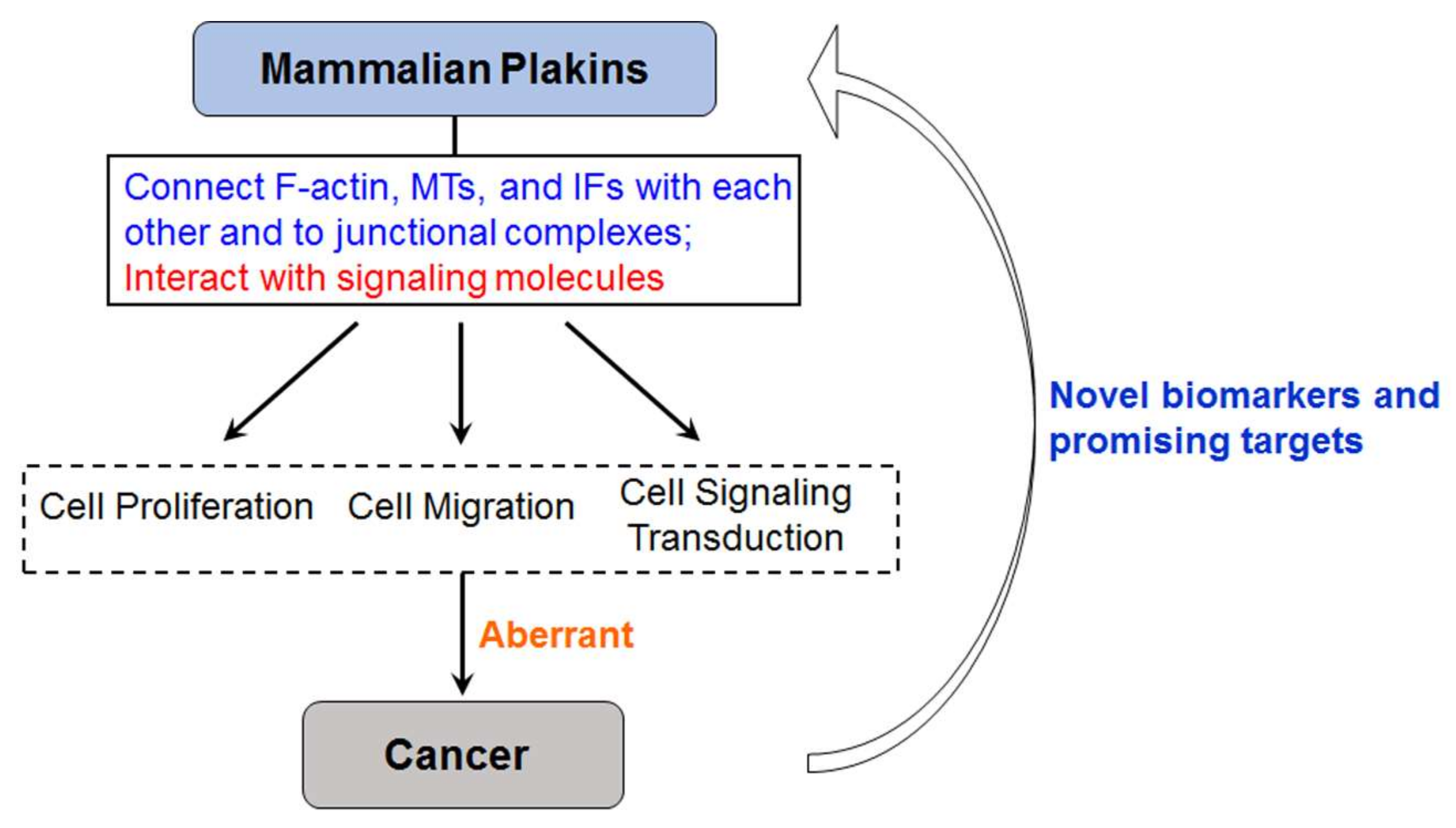
1. Introduction
2. The Mammalian Plakins
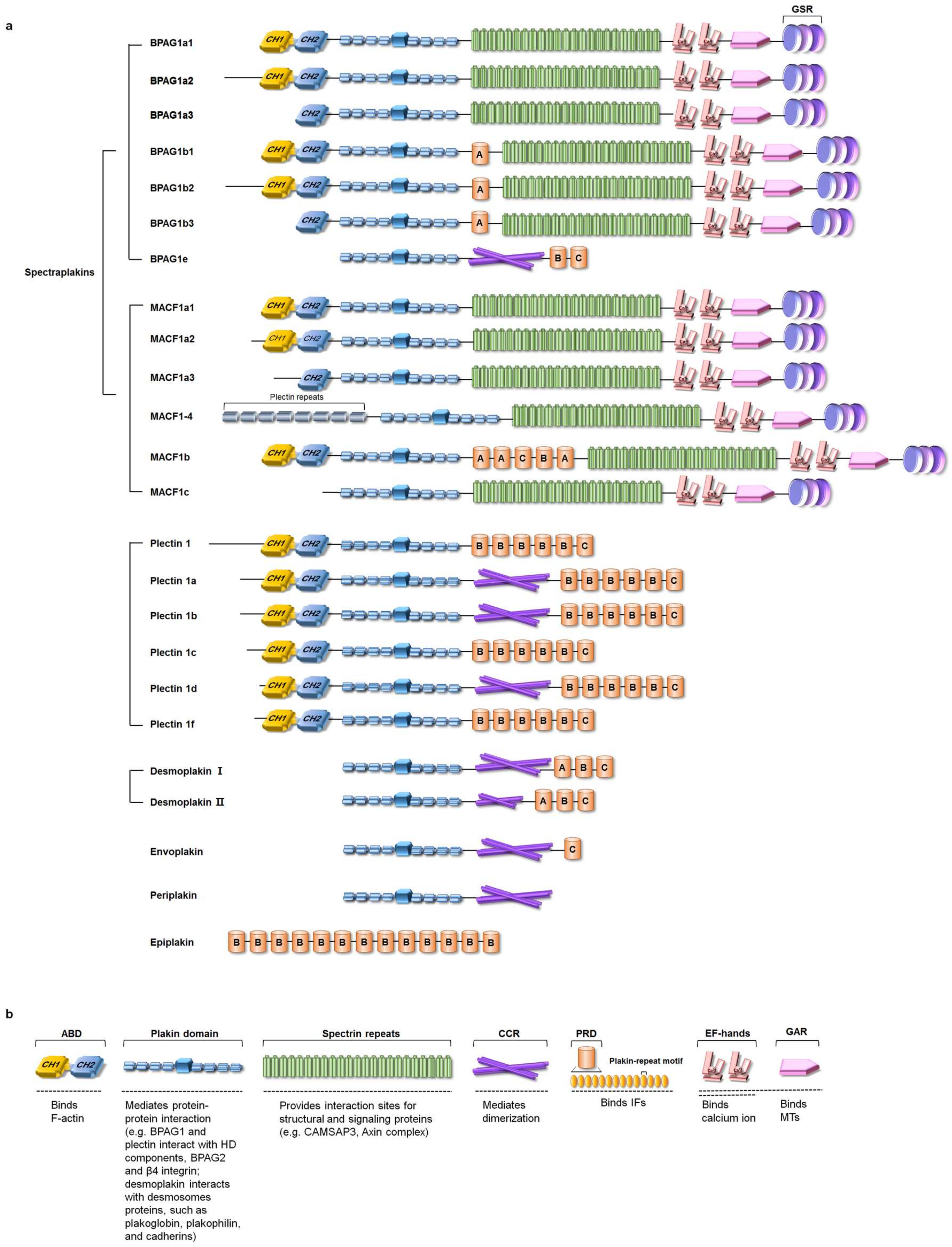
2.1. The Isoforms and Tissue Distribution of Mammalian Plakins
2.2. Domain Structure of Plakins
2.2.1. Actin-Binding Domain (ABD)
2.2.2. Plakin Domain
2.2.3. Spectrin Repeats
2.2.4. Coiled-Coil Rod (CCR)
2.2.5. Plakin Repeat Domain (PRD)
2.2.6. EF-Hand and GAS2-Related Protein (GAR) Domain
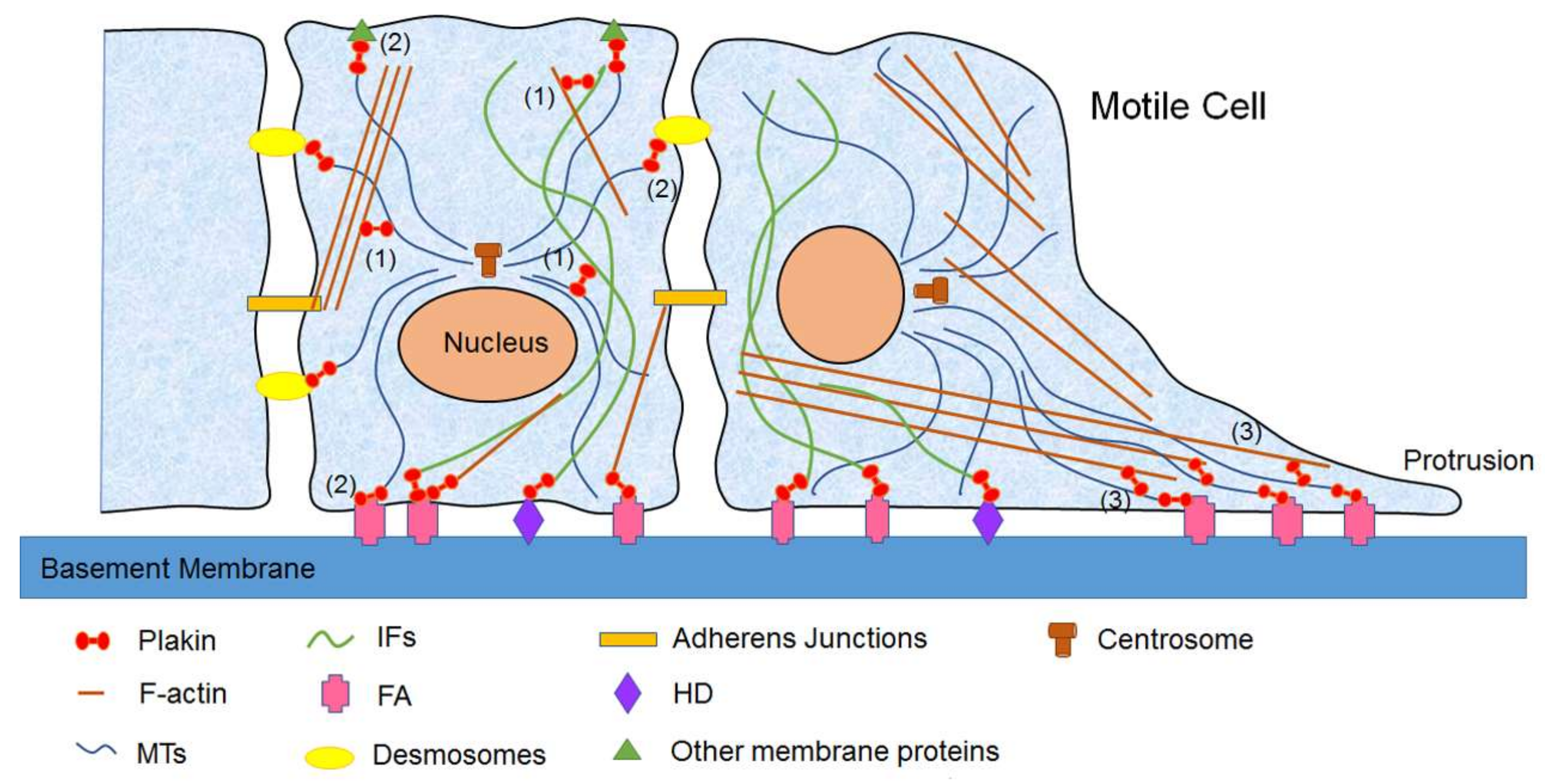
3. Biological Functions of Mammalian Plakins
3.1. Mammalian Plakins in Cell Migration
3.2. Mammalian Plakins in Cell Proliferation
3.3. Mammalian Plakins in Cell Signaling
4. Mammalian Plakins in Human Cancer
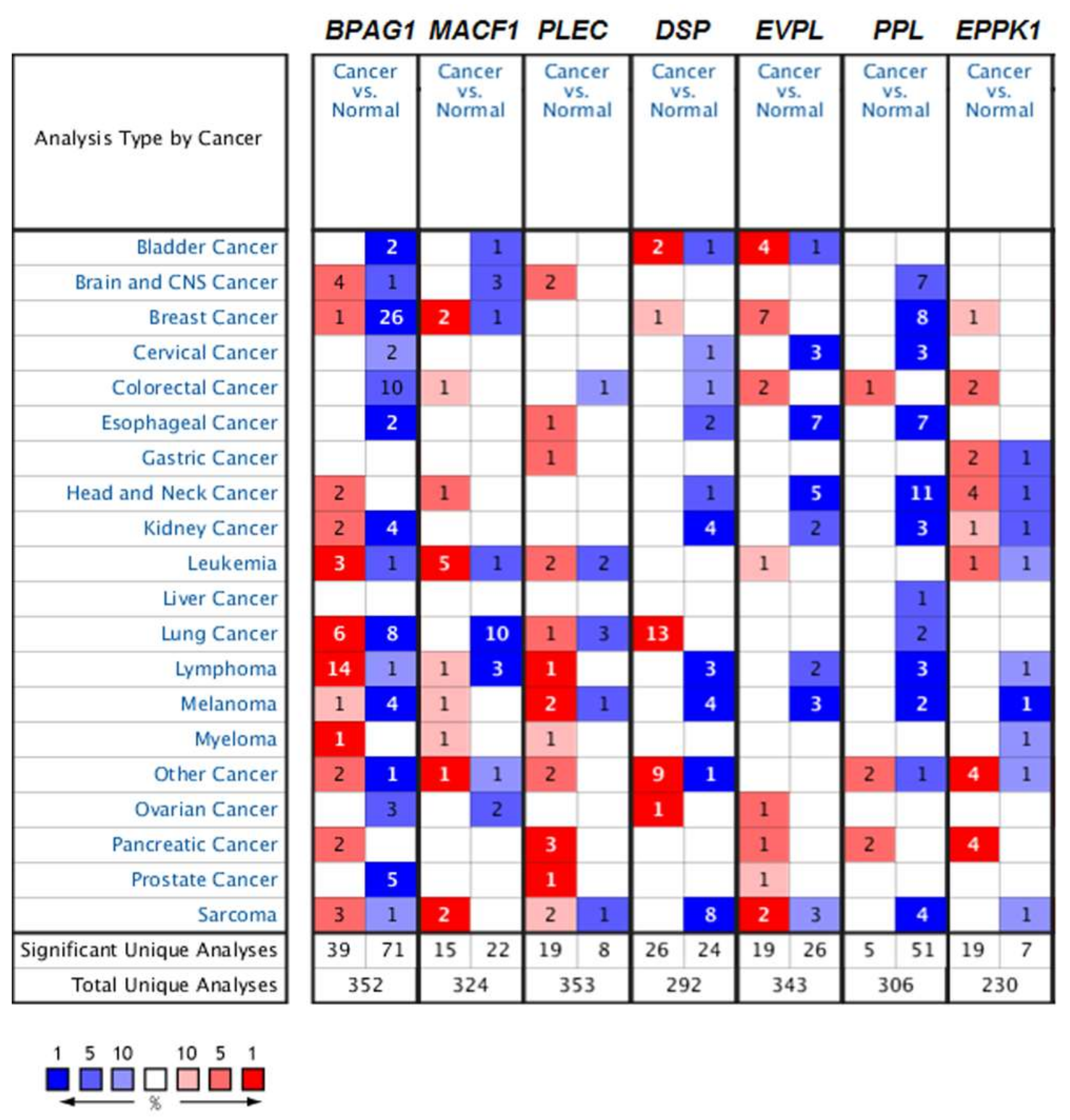
4.1. The Role of Mammalian Plakins in Cancer
4.1.1. Bullous Pemphigoid Antigen 1 (BPAG1) in Cancer
4.1.2. Microtubule Actin Crosslinking Factor 1 (MACF1) in Cancer
4.1.3. Plectin in Cancer
4.1.4. Desmoplakin in Cancer
4.1.5. Other Mammalian Plakins in Cancer
4.2. Mammalian Plakins as Potential Targets for Cancer Therapy
5. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABD | actin-binding domain |
| ACF7 | actin crosslinking factor 7 |
| AMPK | adenosine 5′-monophosphate-activated protein kinase |
| APC | adenomatous polyposis coli |
| BPAG1 | bullous pemphigoid antigen 1 |
| BPAG2 | bullous pemphigoid antigen 2 |
| BP230 | bullous pemphigoid antigen 230 |
| CAMSAP3 | calmodulin regulated spectrin-associated protein 3 |
| CCR | coiled-coil rod |
| CH | calponin homology |
| DSP | desmoplakin |
| ECM | extracellular matrix |
| ELMO | engulfment and motility |
| EPI−/− | triply deficient in envoplakin, periplakin, and involucrin |
| EPPK1 | epiplakin |
| ER | endoplasmic reticulum |
| ERK1/2 | extracellular signal-regulated kinases 1/2 |
| EVPL | envoplakin |
| F-actin | actin filaments |
| FAK | focal adhesion kinase |
| FAs | focal adhesions |
| GAR | GAS2-related protein |
| Gas2 | growth arrest specific 2 |
| GSK3 | glycogen synthase kinase 3 |
| GSK-3β | glycogen synthase kinase 3β |
| GSR | glycine-serine-arginine |
| HD | hemidesmosome |
| HNSCC | head and neck squamous cell carcinoma |
| IFs | intermediate filaments |
| IPMN | intraductal papillary mucinous neoplasms |
| LRP5/6 | low-density lipoprotein receptor 5/6 |
| MACF1 | microtubule actin crosslinking factor 1 |
| MCH-1 | melanin-concentrating hormone-1 |
| MNK2 | mitogen-activated protein kinase-interacting kinase 2 |
| MTOC | microtubule organizing center |
| mTOR | mammalian target of rapamycin |
| MTs | microtubules |
| NDRG1 | N-Myc downstream regulated gene 1 |
| OSCC | oral squamous cell carcinoma |
| PCNA | proliferating cell nuclear antigen |
| PI3K | phosphatidylinositol 3′ kinase |
| PKA | protein kinase A |
| PKB | protein kinase B |
| PKC | protein kinase C |
| PLEC | plecitn |
| PPL | periplakin |
| PRD | plakin repeat domain |
| RCSB | research collaboratory for structural bioinformatics |
| SH3 | src-homology 3 |
| Sp1 | specificity protein 1 |
| TCF | T-cell factor |
| UTRs | untranslated regions |
| Wnt | wingless and int-1 |
References
- Leung, C.L.; Green, K.J.; Liem, R.K. Plakins: A family of versatile cytolinker proteins. Trends. Cell. Biol. 2002, 12, 37–45. [Google Scholar] [CrossRef]
- Jefferson, J.J.; Leung, C.L.; Liem, R.K.H. Plakins: Goliaths that link cell junctions and the cytoskeleton. Nat. Rev. Mol. Cell. Biol. 2004, 5, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, A.; Liem, R.K. Plakins in development and disease. Exp. Cell. Res. 2007, 313, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Lin, C.M.; Lin, C.S.; Perez-Olle, R.; Leung, C.L.; Liem, R.K. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 2006, 20, 1933–1945. [Google Scholar] [CrossRef] [PubMed]
- Takawira, D.; Budinger, G.R.; Hopkinson, S.B.; Jones, J.C. A dystroglycan/plectin scaffold mediates mechanical pathway bifurcation in lung epithelial cells. J. Biol. Chem. 2011, 286, 6301–6310. [Google Scholar] [CrossRef] [PubMed]
- Van den Heuvel, A.P.; de Vries-Smits, A.M.; van Weeren, P.C.; Dijkers, P.F.; de Bruyn, K.M.; Riedl, J.A.; Burgering, B.M. Binding of protein kinase B to the plakin family member periplakin. J. Cell. Sci. 2002, 115, 3957–3966. [Google Scholar] [CrossRef] [PubMed]
- Fife, C.M.; McCarroll, J.A.; Kavallaris, M. Movers and shakers: Cell cytoskeleton in cancer metastasis. Br. J. Pharmacol. 2014, 171, 5507–5523. [Google Scholar] [CrossRef] [PubMed]
- Howe, L.R.; Brown, A.M. Wnt signaling and breast cancer. Cancer Biol. Ther. 2004, 3, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Deschenes-Simard, X.; Kottakis, F.; Meloche, S.; Ferbeyre, G. ERKs in cancer: Friends or foes? Cancer Res. 2014, 74, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.R. Cell adhesion molecules as targets of autoantibodies in pemphigus and pemphigoid, bullous diseases due to defective epidermal cell adhesion. Adv. Immunol. 1993, 53, 291–325. [Google Scholar] [PubMed]
- Kunzli, K.; Favre, B.; Chofflon, M.; Borradori, L. One gene but different proteins and diseases: The complexity of dystonin and bullous pemphigoid antigen 1. Exp. Dermatol. 2016, 25, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Hu, L.; Zhao, F.; Qiu, W.; Wang, P.; Ma, X.; Zhang, Y.; Chen, L.; Qian, A. BPAG1, a distinctive role in skin and neurological diseases. Semin. Cell Dev. Biol. 2017, 69, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, J.J.; Leung, C.L.; Liem, R.K. Dissecting the sequence specific functions of alternative N-terminal isoforms of mouse bullous pemphigoid antigen 1. Exp. Cell Res. 2006, 312, 2712–2725. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.L.; Zheng, M.; Prater, S.M.; Liem, R.K. The BPAG1 locus: Alternative splicing produces multiple isoforms with distinct cytoskeletal linker domains, including predominant isoforms in neurons and muscles. J. Cell Biol. 2001, 154, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Byers, T.J.; Beggs, A.H.; McNally, E.M.; Kunkel, L.M. Novel actin crosslinker superfamily member identified by a two step degenerate PCR procedure. FEBS Lett. 1995, 368, 500–504. [Google Scholar] [PubMed]
- Leung, C.L.; Sun, D.; Zheng, M.; Knowles, D.R.; Liem, R.K. Microtubule actin cross-linking factor (MACF): A hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 1999, 147, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Okuda, T.; Matsuda, S.; Nakatsugawa, S.; Ichigotani, Y.; Iwahashi, N.; Takahashi, M.; Ishigaki, T.; Hamaguchi, M. Molecular cloning of macrophin, a human homologue of Drosophila kakapo with a close structural similarity to plectin and dystrophin. Biochem. Biophys. Res. Commun. 1999, 264, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, J.; Kraeft, S.K.; Auclair, D.; Chang, M.S.; Liu, Y.; Sutherland, R.; Salgia, R.; Griffin, J.D.; Ferland, L.H.; et al. Molecular cloning and characterization of human trabeculin-α, a giant protein defining a new family of actin-binding proteins. J. Biol. Chem. 1999, 274, 33522–33530. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Su, P.; Li, R.; Yin, C.; Zhang, Y.; Shang, P.; Yang, T.; Qian, A. Isoforms, structures, and functions of versatile spectraplakin MACF1. BMB Rep. 2016, 49, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Leung, C.L.; Liem, R.K. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): Identification of a novel group of microtubule associated proteins. J. Cell Sci. 2001, 114, 161–172. [Google Scholar] [PubMed]
- Gong, T.W.; Besirli, C.G.; Lomax, M.I. MACF1 gene structure: A hybrid of plectin and dystrophin. Mamm. Genome 2001, 12, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.M.; Chen, H.J.; Leung, C.L.; Parry, D.A.; Liem, R.K. Microtubule actin crosslinking factor 1b: A novel plakin that localizes to the Golgi complex. J. Cell Sci. 2005, 118, 3727–3738. [Google Scholar] [CrossRef] [PubMed]
- Bernier, G.; Mathieu, M.; De Repentigny, Y.; Vidal, S.M.; Kothary, R. Cloning and characterization of mouse ACF7, a novel member of the dystonin subfamily of actin binding proteins. Genomics 1996, 38, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Bernier, G.; Pool, M.; Kilcup, M.; Alfoldi, J.; De Repentigny, Y.; Kothary, R. Acf7 (MACF) is an actin and microtubule linker protein whose expression predominates in neural, muscle, and lung development. Dev. Dyn. 2000, 219, 216–225. [Google Scholar] [CrossRef]
- Goryunov, D.; He, C.Z.; Lin, C.S.; Leung, C.L.; Liem, R.K. Nervous-tissue-specific elimination of microtubule-actin crosslinking factor 1a results in multiple developmental defects in the mouse brain. Mol. Cell. Neurosci. 2010, 44, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Foisner, R.; Wiche, G. Intermediate filament-associated proteins. Curr. Opin. Cell Biol. 1991, 3, 75–81. [Google Scholar] [CrossRef]
- Svitkina, T.M.; Verkhovsky, A.B.; Borisy, G.G. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J. Cell Biol. 1996, 135, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Wiche, G.; Winter, L. Plectin isoforms as organizers of intermediate filament cytoarchitecture. Bioarchitecture 2011, 1, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Wiche, G.; Krepler, R.; Artlieb, U.; Pytela, R.; Denk, H. Occurrence and immunolocalization of plectin in tissues. J. Cell Biol. 1983, 97, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Wiche, G.; Krepler, R.; Artlieb, U.; Pytela, R.; Aberer, W. Identification of plectin in different human cell types and immunolocalization at epithelial basal cell surface membranes. Exp. Cell Res. 1984, 155, 43–49. [Google Scholar] [CrossRef]
- Fuchs, P.; Zorer, M.; Rezniczek, G.A.; Spazierer, D.; Oehler, S.; Castanon, M.J.; Hauptmann, R.; Wiche, G. Unusual 5′ transcript complexity of plectin isoforms: Novel tissue-specific exons modulate actin binding activity. Hum. Mol. Genet. 1999, 8, 2461–2472. [Google Scholar] [CrossRef] [PubMed]
- Skerrow, C.J. Selective extraction of desmosomal proteins by low ionic strength media. Biochim. Biophys. Acta 1979, 579, 241–245. [Google Scholar] [CrossRef]
- O'Keefe, E.J.; Erickson, H.P.; Bennett, V. Desmoplakin I and desmoplakin II. Purification and characterization. J. Biol. Chem. 1989, 264, 8310–8318. [Google Scholar] [PubMed]
- Green, K.J.; Parry, D.A.; Steinert, P.M.; Virata, M.L.; Wagner, R.M.; Angst, B.D.; Nilles, L.A. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J. Biol. Chem. 1990, 265, 2603–2612. [Google Scholar] [PubMed]
- Angst, B.D.; Nilles, L.A.; Green, K.J. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 1990, 97, 247–257. [Google Scholar] [PubMed]
- Simon, M.; Green, H. Participation of membrane-associated proteins in the formation of the cross-linked envelope of the keratinocyte. Cell 1984, 36, 827–834. [Google Scholar] [CrossRef]
- Ruhrberg, C.; Hajibagheri, M.A.; Simon, M.; Dooley, T.P.; Watt, F.M. Envoplakin, a novel precursor of the cornified envelope that has homology to desmoplakin. J. Cell Biol. 1996, 134, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Ruhrberg, C.; Hajibagheri, M.A.; Parry, D.A.; Watt, F.M. Periplakin, a novel component of cornified envelopes and desmosomes that belongs to the plakin family and forms complexes with envoplakin. J. Cell Biol. 1997, 139, 1835–1849. [Google Scholar] [CrossRef] [PubMed]
- DiColandrea, T.; Karashima, T.; Maatta, A.; Watt, F.M. Subcellular distribution of envoplakin and periplakin: Insights into their role as precursors of the epidermal cornified envelope. J. Cell Biol. 2000, 151, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Kohno, K.; Iwamatsu, A.; Naito, I.; Shinkai, H. Identification of a 450-kDa human epidermal autoantigen as a new member of the plectin family. J. Investig. Dermatol. 1996, 106, 1125–1130. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Takeo, N.; Otani, Y.; Parry, D.A.; Kunimatsu, M.; Lu, R.; Sasaki, M.; Matsuo, N.; Khaleduzzaman, M.; Yoshioka, H. Epiplakin, a novel member of the plakin family originally identified as a 450-kDa human epidermal autoantigen. Structure and tissue localization. J. Biol. Chem. 2001, 276, 13340–13347. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Shiraki, N.; Baba, H.; Goto, M.; Fujiwara, S.; Kume, K.; Kume, S. Expression patterns of epiplakin1 in pancreas, pancreatic cancer and regenerating pancreas. Genes Cells 2008, 13, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Guo, X.; Namekata, K.; Mitamura, Y.; Kume, S.; Harada, T. Expression of Epiplakin1 in the developing and adult mouse retina. Jpn. J. Ophthalmol. 2010, 54, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Roper, K.; Gregory, S.L.; Brown, N.H. The ‘spectraplakins’: Cytoskeletal giants with characteristics of both spectrin and plakin families. J. Cell Sci. 2002, 115, 4215–4225. [Google Scholar] [CrossRef] [PubMed]
- Way, M.; Pope, B.; Weeds, A.G. Evidence for functional homology in the F-actin binding domains of gelsolin and α-actinin: Implications for the requirements of severing and capping. J. Cell Biol. 1992, 119, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Winder, S.J.; Hemmings, L.; Maciver, S.K.; Bolton, S.J.; Tinsley, J.M.; Davies, K.E.; Critchley, D.R.; Kendrick-Jones, J. Utrophin actin binding domain: Analysis of actin binding and cellular targeting. J. Cell Sci. 1995, 108, 63–71. [Google Scholar] [PubMed]
- Sjoblom, B.; Ylanne, J.; Djinovic-Carugo, K. Novel structural insights into F-actin-binding and novel functions of calponin homology domains. Curr. Opin. Struct. Biol. 2008, 18, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Zhang, Y.; Liang, W.G.; Gou, X.; Lee, P.; Liu, H.; Lyu, W.; Tang, W.J.; Chen, S.Y.; Yang, F.; et al. In vivo epidermal migration requires focal adhesion targeting of ACF7. Nat. Commun. 2016, 7, 11692. [Google Scholar] [CrossRef] [PubMed]
- Kodama, A.; Karakesisoglou, I.; Wong, E.; Vaezi, A.; Fuchs, E. ACF7: An essential integrator of microtubule dynamics. Cell 2003, 115, 343–354. [Google Scholar] [CrossRef]
- Hu, L.; Xiao, Y.; Xiong, Z.; Zhao, F.; Yin, C.; Zhang, Y.; Su, P.; Li, D.; Chen, Z.; Ma, X.; et al. MACF1, versatility in tissue-specific function and in human disease. Semin. Cell Dev. Biol. 2017, 69, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Yoshioka, N.; Takebayashi, H. BPAG1 in muscles: Structure and function in skeletal, cardiac and smooth muscle. Semin. Cell Dev. Biol. 2017, 69, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, J.J.; Ciatto, C.; Shapiro, L.; Liem, R.K. Structural analysis of the plakin domain of bullous pemphigoid antigen1 (BPAG1) suggests that plakins are members of the spectrin superfamily. J. Mol. Biol. 2007, 366, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Weis, W.I. Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakin plakin domain. J. Mol. Biol. 2011, 409, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Hopkinson, S.B.; Jones, J.C. The N terminus of the transmembrane protein BP180 interacts with the N-terminal domain of BP230, thereby mediating keratin cytoskeleton anchorage to the cell surface at the site of the hemidesmosome. Mol. Biol. Cell 2000, 11, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Koster, J.; Geerts, D.; Favre, B.; Borradori, L.; Sonnenberg, A. Analysis of the interactions between BP180, BP230, plectin and the integrin α6β4 important for hemidesmosome assembly. J. Cell Sci. 2003, 116, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Rezniczek, G.A.; de Pereda, J.M.; Reipert, S.; Wiche, G. Linking integrin α6β4-based cell adhesion to the intermediate filament cytoskeleton: Direct interaction between the β4 subunit and plectin at multiple molecular sites. J. Cell Biol. 1998, 141, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Getsios, S.; Huen, A.C.; Green, K.J. Working out the strength and flexibility of desmosomes. Nat. Rev. Mol. Cell Biol. 2004, 5, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Winograd, E.; Viel, A.; Cronin, T.; Harrison, S.C.; Branton, D. Crystal structure of the repetitive segments of spectrin. Science 1993, 262, 2027–2030. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Pfuhl, M.; Walther, D.; Saraste, M.; Nilges, M. Solution structure of the spectrin repeat: A left-handed antiparallel triple-helical coiled-coil. J. Mol. Biol. 1997, 273, 740–751. [Google Scholar] [CrossRef] [PubMed]
- Djinovic-Carugo, K.; Gautel, M.; Ylanne, J.; Young, P. The spectrin repeat: A structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002, 513, 119–123. [Google Scholar] [CrossRef]
- Noordstra, I.; Liu, Q.; Nijenhuis, W.; Hua, S.; Jiang, K.; Baars, M.; Remmelzwaal, S.; Martin, M.; Kapitein, L.C.; Akhmanova, A. Control of apico-basal epithelial polarity by the microtubule minus-end-binding protein CAMSAP3 and spectraplakin ACF7. J. Cell Sci. 2016, 129, 4278–4288. [Google Scholar] [CrossRef] [PubMed]
- Manso, J.A.; Garcia Rubio, I.; Gomez-Hernandez, M.; Ortega, E.; Buey, R.M.; Carballido, A.M.; Carabias, A.; Alonso-Garcia, N.; de Pereda, J.M. Purification and structural analysis of plectin and BPAG1e. Method Enzymol. 2016, 569, 177–196. [Google Scholar] [CrossRef]
- Choi, H.J.; Park-Snyder, S.; Pascoe, L.T.; Green, K.J.; Weis, W.I. Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure. Nat. Struct. Biol. 2002, 9, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.L.; Sun, D.; Liem, R.K. The intermediate filament protein peripherin is the specific interaction partner of mouse BPAG1-n (dystonin) in neurons. J. Cell Biol. 1999, 144, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Bouameur, J.E.; Favre, B.; Fontao, L.; Lingasamy, P.; Begre, N.; Borradori, L. Interaction of plectin with keratins 5 and 14: Dependence on several plectin domains and keratin quaternary structure. J. Investig. Dermatol. 2014, 134, 2776–2783. [Google Scholar] [CrossRef] [PubMed]
- Fogl, C.; Mohammed, F.; Al-Jassar, C.; Jeeves, M.; Knowles, T.J.; Rodriguez-Zamora, P.; White, S.A.; Odintsova, E.; Overduin, M.; Chidgey, M. Mechanism of intermediate filament recognition by plakin repeat domains revealed by envoplakin targeting of vimentin. Nat. Commun. 2016, 7, 10827. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Sumiyoshi, H.; Matsuo, N.; Takeo, N.; Goto, M.; Okamoto, O.; Tatsukawa, S.; Kitamura, H.; Fujikura, Y.; Yoshioka, H.; et al. Epiplakin accelerates the lateral organization of keratin filaments during wound healing. J. Dermatol. Sci. 2010, 60, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Suozzi, K.C.; Wu, X.; Fuchs, E. Spectraplakins: Master orchestrators of cytoskeletal dynamics. J. Cell Biol. 2012, 197, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.R.; Fuchs, E.; Slep, K.C. Structure of the ACF7 EF-hand-GAR module and delineation of microtubule binding determinants. Structure 2017, 25, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Kapur, M.; Wang, W.; Maloney, M.T.; Millan, I.; Lundin, V.F.; Tran, T.A.; Yang, Y. Calcium tips the balance: A microtubule plus end to lattice binding switch operates in the carboxyl terminus of BPAG1n4. EMBO Rep. 2012, 13, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Kaverina, I.; Straube, A. Regulation of cell migration by dynamic microtubules. Semin. Cell Dev. Biol. 2011, 22, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, O.C.; Schaefer, A.W.; Mandato, C.A.; Forscher, P.; Bement, W.M.; Waterman-Storer, C.M. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 2003, 5, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.L.; Drubin, D.G.; Barnes, G. Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol. 2000, 12, 63–71. [Google Scholar] [CrossRef]
- Small, J.V.; Kaverina, I. Microtubules meet substrate adhesions to arrange cell polarity. Curr. Opin. Cell Biol. 2003, 15, 40–47. [Google Scholar] [CrossRef]
- Salmon, W.C.; Adams, M.C.; Waterman-Storer, C.M. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 2002, 158, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Degenstein, L.; Dowling, J.; Yu, Q.C.; Wollmann, R.; Perman, B.; Fuchs, E. Gene targeting of BPAG1: Abnormalities in mechanical strength and cell migration in stratified epithelia and neurologic degeneration. Cell 1995, 81, 233–243. [Google Scholar] [CrossRef]
- Poliakova, K.; Adebola, A.; Leung, C.L.; Favre, B.; Liem, R.K.; Schepens, I.; Borradori, L. BPAG1a and b associate with EB1 and EB3 and modulate vesicular transport, Golgi apparatus structure, and cell migration in C2.7 myoblasts. PLoS ONE 2014, 9, e107535. [Google Scholar] [CrossRef] [PubMed]
- Hamill, K.J.; Hopkinson, S.B.; DeBiase, P.; Jones, J.C. BPAG1e maintains keratinocyte polarity through β4 integrin-mediated modulation of Rac1 and cofilin activities. Mol. Biol. Cell 2009, 20, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.; Begum, R.; Fong, K.; Pourreyrone, C.; South, A.P.; McGrath, J.A.; Parsons, M. BPAG1-e restricts keratinocyte migration through control of adhesion stability. J. Investig. Dermatol. 2014, 134, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Kodama, A.; Fuchs, E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell 2008, 135, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shen, Q.T.; Oristian, D.S.; Lu, C.P.; Zheng, Q.; Wang, H.W.; Fuchs, E. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3β. Cell 2011, 144, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Ka, M.; Jung, E.M.; Mueller, U.; Kim, W.Y. MACF1 regulates the migration of pyramidal neurons via microtubule dynamics and GSK-3 signaling. Dev. Biol. 2014, 395, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Ka, M.; Moffat, J.J.; Kim, W.Y. MACF1 controls migration and positioning of cortical GABAergic interneurons in mice. Cereb. Cortex 2017, 27, 5525–5538. [Google Scholar] [CrossRef] [PubMed]
- Zaoui, K.; Benseddik, K.; Daou, P.; Salaun, D.; Badache, A. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc. Natl. Acad. Sci. USA 2010, 107, 18517–18522. [Google Scholar] [CrossRef] [PubMed]
- Margaron, Y.; Fradet, N.; Cote, J.F. ELMO recruits actin cross-linking family 7 (ACF7) at the cell membrane for microtubule capture and stabilization of cellular protrusions. J. Biol. Chem. 2013, 288, 1184–1199. [Google Scholar] [CrossRef] [PubMed]
- Andra, K.; Nikolic, B.; Stocher, M.; Drenckhahn, D.; Wiche, G. Not just scaffolding: Plectin regulates actin dynamics in cultured cells. Genes Dev. 1998, 12, 3442–3451. [Google Scholar] [CrossRef] [PubMed]
- Spurny, R.; Gregor, M.; Castanon, M.J.; Wiche, G. Plectin deficiency affects precursor formation and dynamics of vimentin networks. Exp. Cell Res. 2008, 314, 3570–3580. [Google Scholar] [CrossRef] [PubMed]
- Valencia, R.G.; Walko, G.; Janda, L.; Novacek, J.; Mihailovska, E.; Reipert, S.; Andra-Marobela, K.; Wiche, G. Intermediate filament-associated cytolinker plectin 1c destabilizes microtubules in keratinocytes. Mol. Biol. Cell 2013, 24, 768–784. [Google Scholar] [CrossRef] [PubMed]
- Osmanagic-Myers, S.; Rus, S.; Wolfram, M.; Brunner, D.; Goldmann, W.H.; Bonakdar, N.; Fischer, I.; Reipert, S.; Zuzuarregui, A.; Walko, G.; et al. Plectin reinforces vascular integrity by mediating crosstalk between the vimentin and the actin networks. J. Cell Sci. 2015, 128, 4138–4150. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsberg, C.; Fuchs, P.; Osmanagic-Myers, S.; Fischer, I.; Propst, F.; Elbe-Burger, A.; Wiche, G. Targeted ablation of plectin isoform 1 uncovers role of cytolinker proteins in leukocyte recruitment. Proc. Natl. Acad. Sci. USA 2005, 102, 18449–18454. [Google Scholar] [CrossRef] [PubMed]
- Osmanagic-Myers, S.; Gregor, M.; Walko, G.; Burgstaller, G.; Reipert, S.; Wiche, G. Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J. Cell Biol. 2006, 174, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.C.; Lai, Y.C.; Lai, Y.S.; Hsu, Y.H.; Chao, W.T.; Sia, K.C.; Tseng, Y.H.; Liu, Y.H. Transient knockdown-mediated deficiency in plectin alters hepatocellular motility in association with activated FAK and Rac1-GTPase. Cancer Cell Int. 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Long, H.A.; Boczonadi, V.; McInroy, L.; Goldberg, M.; Maatta, A. Periplakin-dependent re-organisation of keratin cytoskeleton and loss of collective migration in keratin-8-downregulated epithelial sheets. J. Cell Sci. 2006, 119, 5147–5159. [Google Scholar] [CrossRef] [PubMed]
- Kokado, M.; Okada, Y.; Miyamoto, T.; Yamanaka, O.; Saika, S. Effects of epiplakin-knockdown in cultured corneal epithelial cells. BMC Res. Notes 2016, 9, 278. [Google Scholar] [CrossRef] [PubMed]
- Boczonadi, V.; McInroy, L.; Maatta, A. Cytolinker cross-talk: Periplakin N-terminus interacts with plectin to regulate keratin organisation and epithelial migration. Exp. Cell Res 2007, 313, 3579–3591. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Su, P.; Li, R.; Yan, K.; Chen, Z.; Shang, P.; Qian, A. Knockdown of microtubule actin crosslinking factor 1 inhibits cell proliferation in MC3T3-E1 osteoblastic cells. BMB Rep. 2015, 48, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Zhang, Y.; Hu, L.; Tian, Y.; Chen, Z.; Li, D.; Zhao, F.; Su, P.; Ma, X.; Zhang, G.; et al. Mechanical unloading reduces microtubule actin crosslinking factor 1 expression to inhibit β-catenin signaling and osteoblast proliferation. J. Cell. Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Suh, H.N.; Kim, M.O.; Han, H.J. Glucosamine-induced reduction of integrin β4 and plectin complex stimulates migration and proliferation in mouse embryonic stem cells. Stem Cells Dev 2013, 22, 2975–2989. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Su, P.; Yin, C.; Zhang, Y.; Li, R.; Yan, K.; Chen, Z.; Li, D.; Zhang, G.; Wang, L.; et al. Microtubule actin crosslinking factor 1 promotes osteoblast differentiation by promoting β-catenin/TCF1/Runx2 signaling axis. J. Cell. Physiol. 2018, 233, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yin, C.; Hu, L.; Chen, Z.; Zhao, F.; Li, D.; Ma, J.; Ma, X.; Su, P.; Qiu, W.; et al. MACF1 overexpression by transfecting the 21-kbp large plasmid PEGFP-C1A-ACF7 promotes osteoblast differentiation and bone formation. Hum. Gene Ther. 2018. [Google Scholar] [CrossRef] [PubMed]
- Foisner, R.; Malecz, N.; Dressel, N.; Stadler, C.; Wiche, G. M-phase-specific phosphorylation and structural rearrangement of the cytoplasmic cross-linking protein plectin involve p34cdc2 kinase. Mol. Biol. Cell 1996, 7, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Malecz, N.; Foisner, R.; Stadler, C.; Wiche, G. Identification of plectin as a substrate of p34cdc2 kinase and mapping of a single phosphorylation site. J. Biol. Chem. 1996, 271, 8203–8208. [Google Scholar] [CrossRef] [PubMed]
- Bouameur, J.E.; Schneider, Y.; Begre, N.; Hobbs, R.P.; Lingasamy, P.; Fontao, L.; Green, K.J.; Favre, B.; Borradori, L. Phosphorylation of serine 4,642 in the C-terminus of plectin by MNK2 and PKA modulates its interaction with intermediate filaments. J. Cell Sci. 2013, 126, 4195–4207. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Kinbara, M.; Maeda, T.; Yoshizawa, M.; Kokabu, S.; Takano Yamamoto, T. Regulation of osteoclast differentiation and actin ring formation by the cytolinker protein plectin. Biochem. Biophys. Res. Commun. 2017, 489, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Gregor, M.; Zeold, A.; Oehler, S.; Marobela, K.A.; Fuchs, P.; Weigel, G.; Hardie, D.G.; Wiche, G. Plectin scaffolds recruit energy-controlling AMP-activated protein kinase (AMPK) in differentiated myofibres. J. Cell Sci. 2006, 119, 1864–1875. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, L.V.; Zhang, L.; Shabanowitz, J.; Purevjav, E.; Towbin, J.A.; Hunt, D.F.; Green, K.J. GSK3- and PRMT-1-dependent modifications of desmoplakin control desmoplakin-cytoskeleton dynamics. J. Cell Biol. 2015, 208, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Martherus, R.; Jain, R.; Takagi, K.; Mendsaikhan, U.; Turdi, S.; Osinska, H.; James, J.F.; Kramer, K.; Purevjav, E.; Towbin, J.A. Accelerated cardiac remodeling in desmoplakin transgenic mice in response to endurance exercise is associated with perturbed Wnt/β-catenin signaling. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H174–H187. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, H.; Feng, G.J.; Bachner, D.; Ormiston, L.; White, J.H.; Richter, D.; Milligan, G. Periplakin interferes with G protein activation by the melanin-concentrating hormone receptor-1 by binding to the proximal segment of the receptor C-terminal tail. J. Biol. Chem. 2005, 280, 8208–8220. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, C.S.; Bonin, M.; Clare, S.E.; Nieselt, K.; Sotlar, K.; Walter, M.; Fehm, T.; Solomayer, E.; Riess, O.; Wallwiener, D.; et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006, 66, 5278–5286. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.K.; Yuen, P.W.; Liu, Y.; Wang, X.H.; Cheung, A.L.; Wong, Y.C.; Tsao, S.W. Downregulation of hemidesmosomal proteins in nasopharyngeal carcinoma cells. Cancer Lett. 2001, 163, 117–123. [Google Scholar] [CrossRef]
- Vanaja, D.K.; Cheville, J.C.; Iturria, S.J.; Young, C.Y. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003, 63, 3877–3882. [Google Scholar] [PubMed]
- Chaudhari, P.R.; Charles, S.E.; Vaidya, M.M. Role of BPAG1e in neoplastic progression of oral squamous cell carcinoma derived cells. Int. Pharm. Bio Sci. 2017, 8, 519–527. [Google Scholar]
- Chaudhari, P.R.; Charles, S.E.; D’Souza, Z.C.; Vaidya, M.M. Hemidesmosomal linker proteins regulate cell motility, invasion and tumorigenicity in oral squamous cell carcinoma derived cells. Exp. Cell Res. 2017, 360, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ochs, M.F.; Ahn, S.M.; Hennessey, P.; Tan, M.; Soudry, E.; Gaykalova, D.A.; Uemura, M.; Brait, M.; Shao, C.; et al. Expression microarray analysis reveals alternative splicing of LAMA3 and DST genes in head and neck squamous cell carcinoma. PLoS ONE 2014, 9, e91263. [Google Scholar] [CrossRef] [PubMed]
- Bidkhori, G.; Narimani, Z.; Hosseini Ashtiani, S.; Moeini, A.; Nowzari-Dalini, A.; Masoudi-Nejad, A. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS ONE 2013, 8, e67552. [Google Scholar] [CrossRef] [PubMed]
- Afghani, N.; Quick, Q.A. Characterization of the cytoskeletal protein MACF1 in lung cancer. In Proceedings of the 107th Annual Meeting of the American Association for Cancer Research, New Orleans, LA, USA, 16–20 April 2016. [Google Scholar]
- Fleischer, T.; Frigessi, A.; Johnson, K.C.; Edvardsen, H.; Touleimat, N.; Klajic, J.; Riis, M.L.; Haakensen, V.D.; Warnberg, F.; Naume, B.; et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 2014, 15, 435. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, P.R.; Milani, C.; Brentani, M.M.; Katayama, M.L.; de Lyra, E.C.; Carraro, D.M.; Brentani, H.; Puga, R.; Lima, L.A.; Rozenchan, P.B.; et al. Transcriptional profile of fibroblasts obtained from the primary site, lymph node and bone marrow of breast cancer patients. Genet. Mol. Biol. 2014, 37, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Arai, E.; Sakamoto, H.; Ichikawa, H.; Totsuka, H.; Chiku, S.; Gotoh, M.; Mori, T.; Nakatani, T.; Ohnami, S.; Nakagawa, T.; et al. Multilayer-omics analysis of renal cell carcinoma, including the whole exome, methylome and transcriptome. Int. J. Cancer 2014, 135, 1330–1342. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Huang, H.D.; Yeh, K.T.; Chang, J.G. Identification of novel mutations in endometrial cancer patients by whole-exome sequencing. Int. J. Oncol. 2017, 50, 1778–1784. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.; Schell, M.J.; Teer, J.K.; Greenawalt, D.M.; Yang, M.; Yeatman, T.J. Co-evolution of somatic variation in primary and metastatic colorectal cancer may expand biopsy indications in the molecular era. PLoS ONE 2015, 10, e0126670. [Google Scholar] [CrossRef] [PubMed]
- Afghani, N.; Mehta, T.; Wang, J.; Tang, N.; Skalli, O.; Quick, Q.A. Microtubule actin cross-linking factor 1, a novel target in glioblastoma. Int. J. Oncol. 2017, 50, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhou, L.; Xie, Q.F.; Xie, H.Y.; Wei, X.Y.; Gao, F.; Xing, C.Y.; Xu, X.; Li, L.J.; Zheng, S.S. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics 2010, 10, 1557–1572. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Qian, H.; Zhang, R.; Gao, X.; Gou, X. MicroRNA targeting microtubule cross-linked protein (MACF1) would suppress the invasion and metastasis of malignant tumor. Med. Hypotheses 2017, 104, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Ali, A.; Hu, L.; Zhao, F.; Yin, C.; Chen, C.; Yang, T.; Qian, A. Microtubule actin cross-linking factor 1, a novel potential target in cancer. Cancer Sci. 2017, 108, 1953–1958. [Google Scholar] [CrossRef] [PubMed]
- Bausch, D.; Mino-Kenudson, M.; Fernandez-Del Castillo, C.; Warshaw, A.L.; Kelly, K.A.; Thayer, S.P. Plectin-1 is a biomarker of malignant pancreatic intraductal papillary mucinous neoplasms. J. Gastrointest. Surg. 2009, 13, 1948–1954. [Google Scholar] [CrossRef] [PubMed]
- Bausch, D.; Thomas, S.; Mino-Kenudson, M.; Fernandez-del, C.C.; Bauer, T.W.; Williams, M.; Warshaw, A.L.; Thayer, S.P.; Kelly, K.A. Plectin-1 as a novel biomarker for pancreatic cancer. Clin. Cancer Res. 2011, 17, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Katada, K.; Tomonaga, T.; Satoh, M.; Matsushita, K.; Tonoike, Y.; Kodera, Y.; Hanazawa, T.; Nomura, F.; Okamoto, Y. Plectin promotes migration and invasion of cancer cells and is a novel prognostic marker for head and neck squamous cell carcinoma. J. Proteom. 2012, 75, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- McInroy, L.; Maatta, A. Plectin regulates invasiveness of SW480 colon carcinoma cells and is targeted to podosome-like adhesions in an isoform-specific manner. Exp. Cell Res. 2011, 317, 2468–2478. [Google Scholar] [CrossRef] [PubMed]
- Sutoh Yoneyama, M.; Hatakeyama, S.; Habuchi, T.; Inoue, T.; Nakamura, T.; Funyu, T.; Wiche, G.; Ohyama, C.; Tsuboi, S. Vimentin intermediate filament and plectin provide a scaffold for invadopodia, facilitating cancer cell invasion and extravasation for metastasis. Eur. J. Cell Biol. 2014, 93, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.C.; Chao, W.T.; Liao, C.C.; Tseng, Y.H.; Lai, Y.C.; Lai, Y.S.; Hsu, Y.H.; Liu, Y.H. Plectin deficiency in liver cancer cells promotes cell migration and sensitivity to sorafenib treatment. Cell Adh. Migr. 2018, 12, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Narayana, N.; Gist, J.; Smith, T.; Tylka, D.; Trogdon, G.; Wahl, J.K. Desmosomal component expression in normal, dysplastic, and oral squamous cell carcinoma. Dermatol. Res. Pract. 2010, 2010, 649731. [Google Scholar] [CrossRef] [PubMed]
- Papagerakis, S.; Shabana, A.H.; Pollock, B.H.; Papagerakis, P.; Depondt, J.; Berdal, A. Altered desmoplakin expression at transcriptional and protein levels provides prognostic information in human oropharyngeal cancer. Hum. Pathol. 2009, 40, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, Y.; Cui, T.; Knosel, T.; Zhang, Q.; Albring, K.F.; Huber, O.; Petersen, I. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/β-catenin signaling pathway in human lung cancer. Carcinogenesis 2012, 33, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Cipolat, S.; Hoste, E.; Natsuga, K.; Quist, S.R.; Watt, F.M. Epidermal barrier defects link atopic dermatitis with altered skin cancer susceptibility. Elife 2014, 3, e01888. [Google Scholar] [CrossRef] [PubMed]
- Tonoike, Y.; Matsushita, K.; Tomonaga, T.; Katada, K.; Tanaka, N.; Shimada, H.; Nakatani, Y.; Okamoto, Y.; Nomura, F. Adhesion molecule periplakin is involved in cellular movement and attachment in pharyngeal squamous cancer cells. BMC Cell Biol. 2011, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Ikeda, M.; Sato, Y.; Kuruma, H.; Kamata, Y.; Nishimori, T.; Tomonaga, T.; Nomura, F.; Egawa, S.; Iwamura, M. Loss of periplakin expression is associated with pathological stage and cancer-specific survival in patients with urothelial carcinoma of the urinary bladder. Biomed. Res. 2014, 35, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, T.; Tomonaga, T.; Matsushita, K.; Oh-Ishi, M.; Kodera, Y.; Maeda, T.; Nomura, F.; Matsubara, H.; Shimada, H.; Ochiai, T. Proteomic analysis of primary esophageal squamous cell carcinoma reveals downregulation of a cell adhesion protein, periplakin. Proteomics 2006, 6, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, T.; Hagiwara, T.; Tamura-Nakano, M.; Sezaki, T.; Miyake, O.; Hinohara, C.; Shimizu, T.; Yamada, K.; Dohi, T.; Kawamura, Y.I. Aberrant DNA hypermethylation reduces the expression of the desmosome-related molecule periplakin in esophageal squamous cell carcinoma. Cancer Med. 2015, 4, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, G.; Wang, Y.; Elgehama, A.; Sun, Y.; Li, L.; Gu, Y.; Guo, W.; Xu, Q. Loss of periplakin expression is associated with the tumorigenesis of colorectal carcinoma. Biomed. Pharmacother. 2017, 87, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Lorch, J.H.; Thomas, T.O.; Schmoll, H.J. Bortezomib inhibits cell–cell adhesion and cell migration and enhances epidermal growth factor receptor inhibitor-induced cell death in squamous cell cancer. Cancer Res. 2007, 67, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Quick, Q.A. Microtubule-actin crosslinking factor 1 and plakins as therapeutic drug targets. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Ayed, A.O.; Parikh, S.A. Management of patients with chronic lymphocytic leukemia at high risk of relapse on ibrutinib therapy. Leuk. Lymphoma 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hansson, L.; Asklid, A.; Diels, J.; Eketorp-Sylvan, S.; Repits, J.; Soltoft, F.; Jager, U.; Osterborg, A. Ibrutinib versus previous standard of care: An adjusted comparison in patients with relapsed/refractory chronic lymphocytic leukaemia. Ann. Hematol. 2017, 96, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhou, Y.; Cheng, L.; Hu, D.; Zhou, X.; Wang, Z.; Xie, C.; Zhou, F. The anti-esophageal cancer cell activity by a novel tyrosine/phosphoinositide kinase inhibitor PP121. Biochem. Biophys. Res. Commun. 2015, 465, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Che, H.Y.; Guo, H.Y.; Si, X.W.; You, Q.Y.; Lou, W.Y. PP121, a dual inhibitor of tyrosine and phosphoinositide kinases, inhibits anaplastic thyroid carcinoma cell proliferation and migration. Tumour Biol. 2014, 35, 8659–8664. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, H.H.; Berger, D.P.; Kopping, K.; Ottenheijm, H.C.; Zylicz, Z. In vitro and in vivo anticancer activity of mitozolomide and sparsomycin in human tumor xenografts, murine tumors and human bone marrow. J. Cancer Res. Clin. Oncol. 1990, 116, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Guha, G.; Lu, W.; Li, S.; Liang, X.; Kulesz-Martin, M.F.; Mahmud, T.; Indra, A.K.; Ganguli-Indra, G. Novel pactamycin analogs induce p53 dependent cell-cycle arrest at S-Phase in human head and neck squamous cell carcinoma (HNSCC) cells. PLoS ONE 2015, 10, e0125322. [Google Scholar] [CrossRef] [PubMed]
- Prokhorova, I.V.; Akulich, K.A.; Makeeva, D.S.; Osterman, I.A.; Skvortsov, D.A.; Sergiev, P.V.; Dontsova, O.A.; Yusupova, G.; Yusupov, M.M.; Dmitriev, S.E. Amicoumacin A induces cancer cell death by targeting the eukaryotic ribosome. Sci. Rep. 2016, 6, 27720. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Kantarjian, H.M.; Rea, D.; Wetzler, M.; Lipton, J.H.; Akard, L.; Khoury, H.J.; Michallet, M.; Guerci-Bresler, A.; Chuah, C.; et al. Final analysis of the efficacy and safety of omacetaxine mepesuccinate in patients with chronic- or accelerated-phase chronic myeloid leukemia: Results with 24 months of follow-up. Cancer 2015, 121, 1637–1644. [Google Scholar] [CrossRef] [PubMed]
| Plakin Name (Gene) | Gene Locus (Human) | Isoforms | Molecular Weight (kD) | Intracellular Localization | Tissue Distribution |
|---|---|---|---|---|---|
| BPAG1 (BPAG1, DST) | Chromosome 6p12.1 | BPAG1a1 | ~625 | Cortical region, F-actin | Nervous system, brain, liver, spleen, ovary |
| BPAG1a2 | F-actin bundles surrounding the nucleus | Nervous system, brain, kidney, testis, liver, spleen, ovary | |||
| BPAG1a3 | Cortical region | Lung, kidney, testis, liver, spleen, ovary | |||
| BPAG1b1 | ~834 | Cortical region, F-actin | Heart, liver, spleen, ovary | ||
| BPAG1b2 | F-actin bundles surrounding the nucleus | Heart, testis, liver, spleen, ovary | |||
| BPAG1b3 | Cortical region | Lung, heart, testis, liver, spleen, ovary | |||
| BPAG1e | ~300 | HDs, the nucleus | Epidermis, lung, testis, ovary, cornea, bladder | ||
| MACF1 (MACF1) | Chromosome 1p34.3 | MACF1a1 | ~600 | MTs, F-actin | Skin, kidney, stomach |
| MACF1a2 | MTs, F-actin | Brain, heart, lung placenta, liver, kidney, pancreas, spinal cord | |||
| MACF1a3 | MTs, F-actin | Skin, lung, kidney | |||
| MACF1-4 | 670 | Unclear | Heart, lung, pituitary gland, placenta | ||
| MACF1b | ~800 | Golgi complex, MTs | Lung, heart, brain, thymus, liver, spleen, kidney, stomach, small intestine, skeletal muscle, skin, testis | ||
| MACF1c | ~600 | Unclear | Nervous system | ||
| Plectin (PLEC) | Chromosome 8q24.3 | Plectin 1 | ~500 | Nucleus/ER membrane | Connective tissue, vascular, eye lens, skeletal muscle, white blood cells |
| Plectin 1a | HDs | Broad | |||
| Plectin 1b | Mitochondria | Connective tissue, skeletal muscle | |||
| Plectin 1c | MTs | Broad | |||
| Plectin 1d | Z-disks | Skeletal muscle | |||
| Plectin 1f | FAs | Skeletal muscle | |||
| Desmoplakin (DSP) | Chromosome 6p24.3 | Desmoplakin I | ~322 kD | Desmosomes | Epithelia, heart |
| Desmoplakin II | ~259 kD | Desmosomes | Epithelia, heart | ||
| Envoplakin (EVPL) | Chromosome 17q25.1 | 210 kD | Cornified envelope | Stratified squamous epithelia | |
| Periplakin (PPL) | Chromosome 16p13.3 | 195 kD | Cornified envelope, desmosomes | Stratified squamous epithelia | |
| Epiplakin (EPPK1) | Chromosome 8q24.3 | 552 kD | Cornified envelope | Liver, small intestine, colon, salivary glands, stomach, appendix, pancreas, retina |
| Plakin Name | Cancer Type | Plakin’s Alteration and Functions | Mechanisms | References |
|---|---|---|---|---|
| BPAG1 | Breast cancer | Downregulation, related to cancer invasion. | Unclear | [110] |
| Nasopharyngeal carcinoma | Downregulation, related to tumor invasion and metastasis. | Unclear | [111] | |
| Prostate cancer | Downregulation, related to cancer metastasis. | Unclear | [112] | |
| OSCC | Positively regulates cell motility, invasion and tumorigenicity. | Upregulate NDRG1 | [113,114] | |
| HNSCC | Alternative splicing revealed by expression microarray analysis. | Alternative splicing raises cancer-specific BPAG1 isoforms | [115] | |
| MACF1 | Lung cancer | High expression, related to cancer cell migration and metastasis. MACF1 knockdown impairs reproductivity of solid tumors. | Unclear | [116,117] |
| Breast cancer | DNA methylation and altered expression, related to cell motility. | Unclear | [118,119] | |
| Renal cell carcinoma | Mutation. | Associated with Wnt/β-catenin signaling | [120] | |
| Endometrial cancer | Mutation. | Associated with Wnt/β-catenin signaling | [121] | |
| Colon cancer | Mutation. | Associated with Wnt/β-catenin signaling | [122] | |
| Glioblastoma | High expression, promotes cell proliferation and migration. | Activate Wnt signaling | [123] | |
| Hepatocellular carcinoma | A target of microRNA-34a. | Unclear | [124] | |
| Plectin | Pancreatic IPMN | Positive expression. | Unclear | [127] |
| Pancreatic cancer | Positive expression. | Unclear | [128] | |
| HNSCC | High expression, promotes cancer cell migration, invasion and metastasis. | Activates ERK1/2 | [129] | |
| Colon cancer | High expression, promotes cancer cell migration, invasion and metastasis. | Be targeted to podosome-like adhesions in an isoform-specific manner | [130] | |
| Bladder cancer | High expression, promotes cancer cell migration and metastasis. | Anchor invadopodia to IF and stabilizing invadopodia | [131] | |
| OSCC | High expression, promotes cancer cell migration, invasion and tumorigenicity. | Upregulate NDRG1 | [114] | |
| Liver cancer | Low expression, inhibits cancer cell migration. | Unclear | [132] | |
| Desmoplakin | OSCC | Downregulation. | Unclear | [133] |
| Oropharyngeal cancer | Downregulation. | Unclear | [134] | |
| Lung cancer | Downregulation, suppresses cell proliferation, migration and invasion, increases cancer sensitivity to anticancer drug-induced apoptosis. | Inhibits Wnt/β-catenin signaling | [135] | |
| Envoplakin | Skin cancer | Promotes skin cancer development. | Unclear | [136] |
| Periplakin | Pharyngeal squamous cancer | Promotion effect on cell movement and attachment. | Activates the PI3K/Akt axis | [137] |
| Urinary bladder cancer | Downregulation. | Unclear | [138] | |
| Esophageal squamous cancer | Downregulation, inhibits cell migration. | Mislocation of periplakin from cell–cell boundaries to cytoplasm; DNA hypermethylation | [139,140] | |
| Colon cancer | Downregulation, inhibits cell proliferation, migration, and invasion, induces G1/G0 cell cycle arrest. | Inhibits cell proliferation by suppressing ERK1/2 activation and PCNA expression | [141] | |
| Epiplakin | Pancreatic cancer | Downregulation. | Unclear | [43] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Huang, Z.; Wu, Z.; Ali, A.; Qian, A. Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer. Int. J. Mol. Sci. 2018, 19, 974. https://doi.org/10.3390/ijms19040974
Hu L, Huang Z, Wu Z, Ali A, Qian A. Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer. International Journal of Molecular Sciences. 2018; 19(4):974. https://doi.org/10.3390/ijms19040974
Chicago/Turabian StyleHu, Lifang, Zizhan Huang, Zixiang Wu, Arshad Ali, and Airong Qian. 2018. "Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer" International Journal of Molecular Sciences 19, no. 4: 974. https://doi.org/10.3390/ijms19040974
APA StyleHu, L., Huang, Z., Wu, Z., Ali, A., & Qian, A. (2018). Mammalian Plakins, Giant Cytolinkers: Versatile Biological Functions and Roles in Cancer. International Journal of Molecular Sciences, 19(4), 974. https://doi.org/10.3390/ijms19040974