Isolation and Identification of Putative Protein Substrates of the AAA+ Molecular Chaperone ClpB from the Pathogenic Spirochaete Leptospira interrogans
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
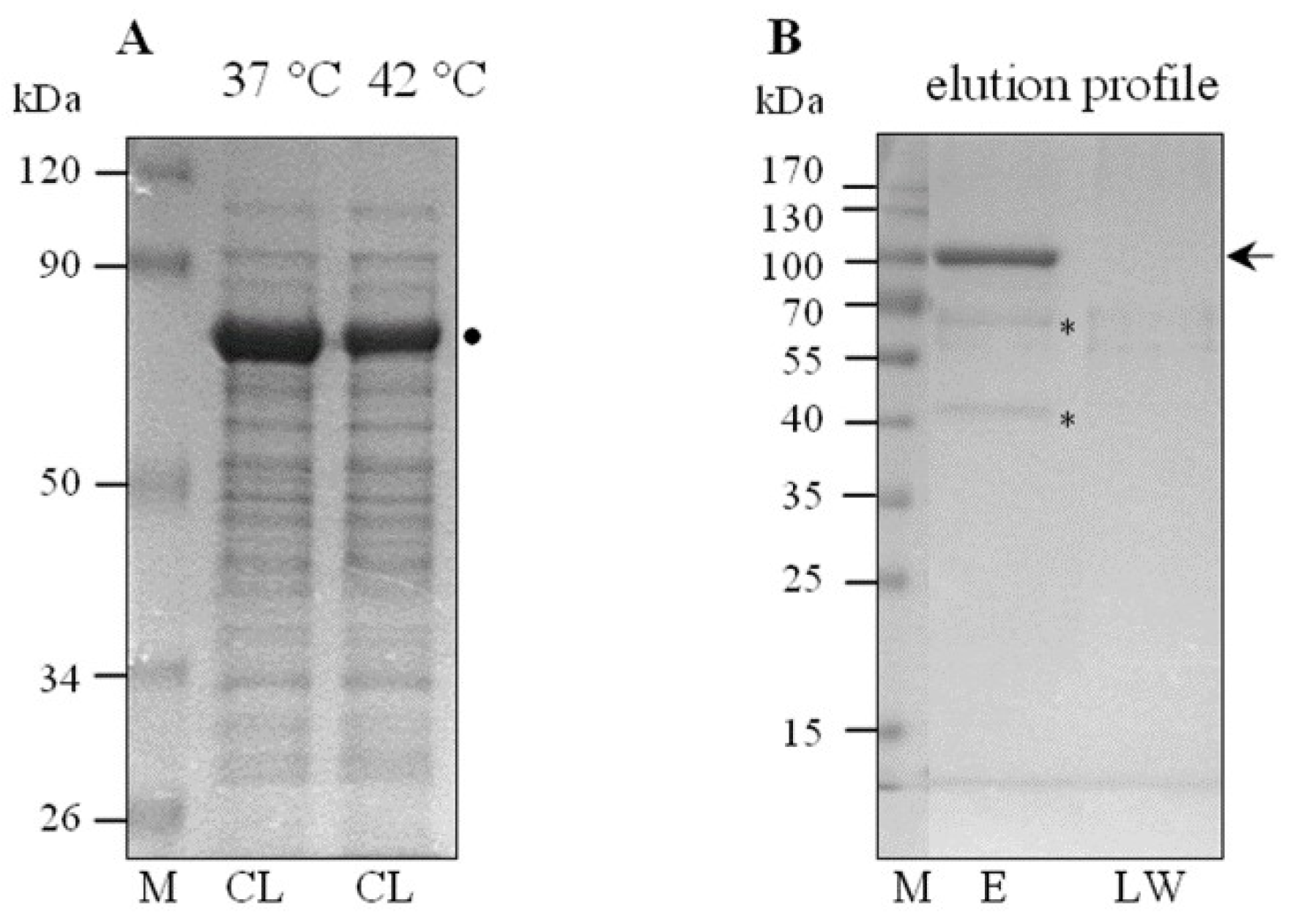
3.1. Leptospira Strain, Growth Conditions, and Cell Lysate Preparation
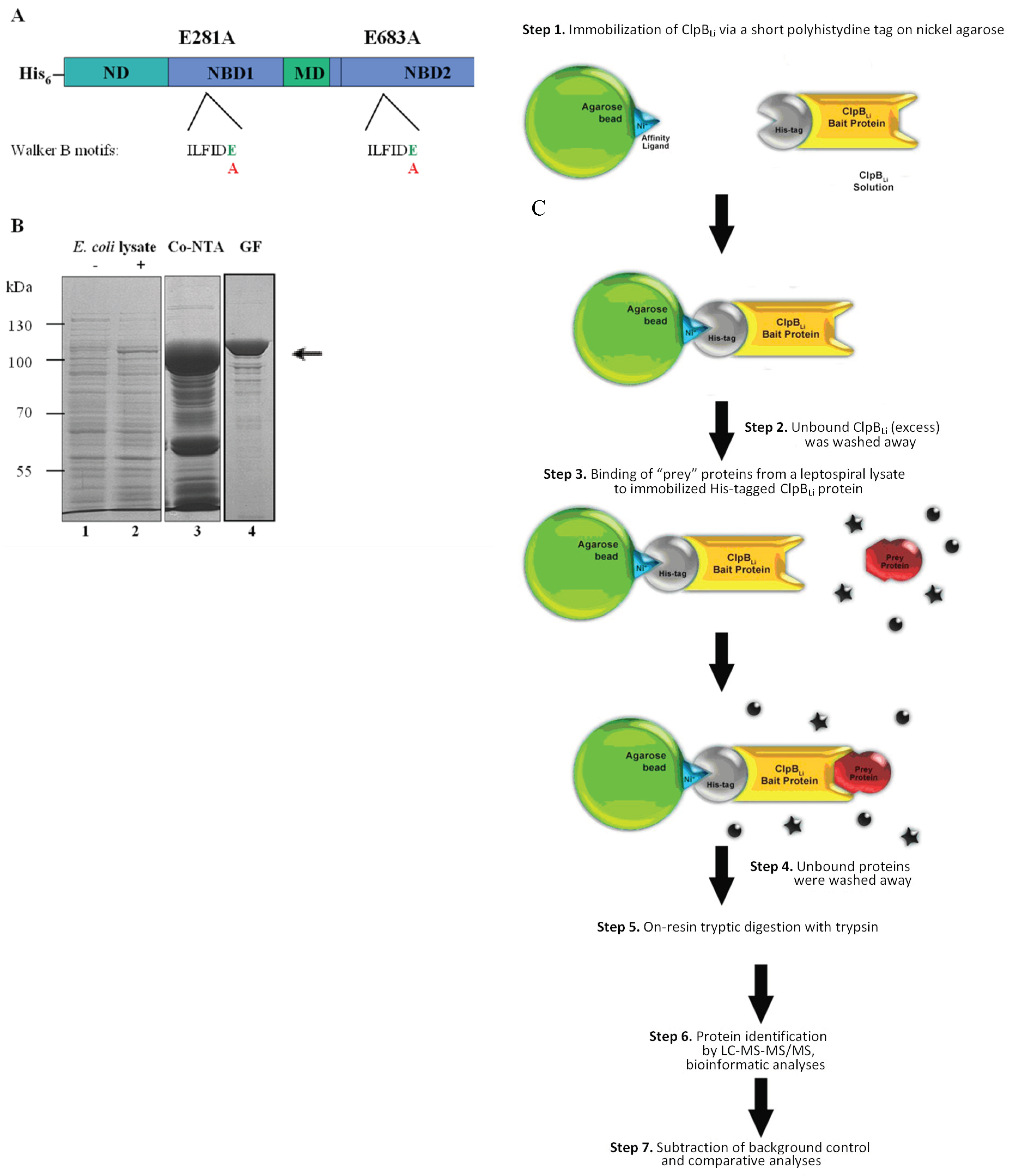
3.2. Construction of ClpBLi-Trap Mutant
3.3. Purification of ClpBLi-Trap (E281A/E683A)
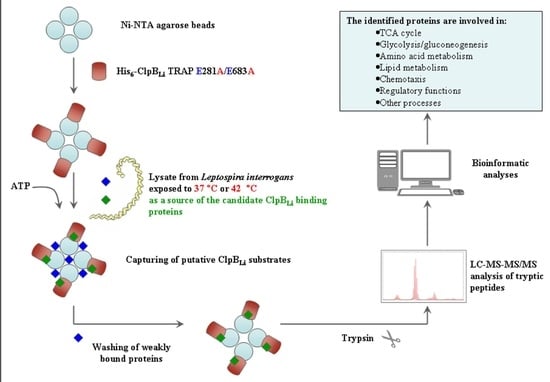
3.4. Affinity Pull-Down Assay and MS Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zolkiewski, M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J. Biol. Chem. 1999, 274, 28083–28086. [Google Scholar] [CrossRef] [PubMed]
- Goloubinoff, P.; Mogk, A.; Ben-Zvi, A.P; Tomoyasu, T.; Bukau, B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 1999, 96, 13732–13737. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Tomoyasu, T.; Goloubinoff, P.; Rűdiger, S.; Röder, D.; Langen, H.; Bukau, B. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999, 18, 6934–6949. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Sowa, M.E.; Watanabe, Y.; Sigler, P.B.; Chiu, W.; Yoshida, M.; Tsai, F.T. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell 2003, 115, 229–240. [Google Scholar] [CrossRef]
- Weibezahn, J.; Tessarz, P.; Schlieker, C.; Zahn, R.; Maglica, Z.; Lee, S.; Zentgraf, H.; Weber-Ban, E.U.; Dougan, D.A; Tsai, F.T.; et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 2004, 119, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Barnett, M.E.; Nagy, M.; Kedzierska, S.; Zolkiewski, M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 2005, 280, 34940–34945. [Google Scholar] [CrossRef] [PubMed]
- Nagy, M.; Guenther, I.; Akoyev, V.; Barnett, M.E.; Zavodszky, M.I.; Kedzierska-Mieszkowska, S.; Zolkiewski, M. Synergistic cooperation between two ClpB isoforms in aggregate reactivation. J. Mol. Biol. 2010, 396, 697–707. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Akoev, V.; Gogol, E.P.; Barnett, M.E.; Zolkiewski, M. Nucleotide-induced switch in oligomerization of the AAA+ ATPase ClpB. Protein Sci. 2004, 13, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Farber, P.; Velvis, A.; Rennella, E.; Latham, M.P.; Kay, L.E. ClpB N-terminal domain plays a regulatory role in protein disaggregation. Proc. Natl. Acad. Sci. USA 2015, 112, E6872–E6881. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, M.E.; Shorter, J. The elusive middle domain of Hsp104 and ClpB: Location and function. Biochim. Biophys. Acta 2012, 1823, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Kedzierska, S.; Akoev, V.; Barnett, M.E.; Zolkiewski, M. Structure and function of the middle domain of ClpB from Escherichia coli. Biochemistry 2003, 42, 14242–14248. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Weaver, C.L.; Lin, J.; Duran, E.C.; Miller, J.M.; Lucius, A.L. Escherichia coli ClpB is a no-n-processive polypeptide translocase. Biochem. J. 2015, 470, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Squires, C.L.; Pedersen, S.; Ross, B.M.; Squires, C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1991, 173, 4254–4262. [Google Scholar] [CrossRef] [PubMed]
- Kannan, T.R.; Musatovova, O.; Gowda, P.; Baseman, J.B. Characterization of a unique ClpB protein of Mycoplasma pneumoniae and its impact on growth. Infect. Immun. 2008, 76, 5082–5092. [Google Scholar] [CrossRef] [PubMed]
- Capestany, C.A.; Tribble, G.D.; Maeda, K.; Demuth, D.R.; Lament, R.J. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 2008, 190, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Chastanet, A.; Derre, I.; Nair, S.; Msadek, T. ClpB, a novel number of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 2004, 186, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Meibom, K.L.; Dubail, I.; Dupuis, M.; Barel, M.; Lenco, J.; Stulik, J.; Golovliov, I.; Sjöstedt, A.; Charbit, A. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 2008, 67, 1384–1401. [Google Scholar] [CrossRef] [PubMed]
- Lourdault, K.; Cerqueira, G.M.; Wunder, E.A., Jr.; Picardeau, M. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 2011, 79, 3711–3717. [Google Scholar] [CrossRef] [PubMed]
- Adler, B.; Lo, M.; Seemann, T.; Murray, G.L. Pathogenesis of leptospirosis: The influence of genomics. Vet. Mirobiol. 2011, 153, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017, 15, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.G.; Nola, L.; O’Grady, L.; More, S.J.; Doherty, L.M. Seroprevalence of Leptospira Hardjo in the Irish suckler cattle population. Ir. Vet. J. 2012, 65, 8. [Google Scholar] [CrossRef] [PubMed]
- Arent, Z.; Kędzierska-Mieszkowska, S. Seroprevalence study of leptospirosis in horses in northern Poland. Vet. Rec. 2013, 172, 269. [Google Scholar] [CrossRef] [PubMed]
- Arent, Z.; Frizzell, C.; Gilmore, C.; Mackie, D.; Ellis, W.A. Isolation of leptospires from genital tract of sheep. Vet. Rec. 2013, 173, 582. [Google Scholar] [CrossRef] [PubMed]
- Arent, Z.J.; Andrews, S.; Adamama-Moraitou, K.; Gilmore, C.; Pardali, D.; Ellis, W.A. Emergence of novel Leptospira serovars: A need for adjusting vaccination policies for dogs? Epidemiol. Infect. 2013, 141, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Ristow, P.; Bourhy, P.; da McBride, F.W.; Figueira, C.P.; Huerre, M.; Ave, P.; Girons, I.S.; Ko, A.I.; Picardeau, M. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 2007, 3, e97. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Sun, A.; Ojcius, D.; Wu, S.; Zhao, J.; Yan, J. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 2009, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.; Picardeau, M.; Haake, D.A.; Sermswan, R.W.; Srikram, A.; Adler, B.; Murray, G.L. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of the flagellum sheath. Infect. Immun. 2012, 80, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Becam, J.; Lambert, A.; Sismeiro, O.; Dillies, M.A.; Jagla, B.; Wunder, E.A., Jr.; Ko, I.; Coppee, J.Y.; Goaran, C.; et al. A putative regulatory genetic locus modulates virulence in the pathogen Leptospira interrogans. Infect. Immun. 2014, 82, 2542–2552. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Srikram, A.; Henry, R.; Puapairoj, A.; Sermswan, R.W.; Adler, B. Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. 2009, 11, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Lourdault, K.; Murray, G.L.; Bartpho, T.; Sermswan, R.W.; Picardeau, M.; Adler, B.; Snarr, B.; Zuerner, R.L.; Cameron, C.E. Leptospira interrogans catalase is required for resistance to H2O2 and for virulence. Infect. Immun. 2012, 80, 3892–3899. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.F.; Chen, H.H.; Ojcius, D.M.; Zhao, X.; Sun, D.; Ge, Y.M.; Zheng, L.L.; Lin, X.; Li, L.J.; Yan, J. Identification of Leptospira interrogans phospholipase C as a novel virulence factor responsible for intracellular free calcium ion elevation during macrophage death. PLoS ONE 2013, 8, e75652. [Google Scholar] [CrossRef] [PubMed][Green Version]
- King, A.M.; Pretre, G.; Bartpho, T.; Sermswan, R.W.; Toma, C.; Suzuki, T.; Eshgh, A.; Picardeau, M.; Adler, B.; Murray, G.L. High-temperature protein G is an essential virulence factor of Leptospira interrogans. Infect. Immun. 2014, 82, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Srikram, A.; Henry, R.; Hartskeerl, R.A.; Sermswan, R.W.; Adler, B. Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol. Microbiol. 2010, 78, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, J.; Arent, Z.; Więckowski, D.; Zolkiewski, M.; Kędzierska-Mieszkowska, S. Immunoreactivity of the AAA+ chaperone ClpB from Leptospira interrogans with sera from Leptospira-infected animals. BMC Microbiol. 2016, 16, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Hu, W.; Me, Y.; Ojcius, D.M.; Lin, X.; Yan, J. A leptospiral AAA+ chaperone-Ntn peptidase complex, HslUV, contributes to the intracellular survival of Leptospira interrogans in hosts and the transmission of leptospirosis. Emerg. Microbes Infect. 2017, 6, e105. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, J.; Modrak-Wójcik, A.; Arent, Z.; Więckowski, D.; Zolkiewski, M.; Bzowska, A.; Kędzierska-Mieszkowska, S. Characterization of the molecular chaperone ClpB from the pathogenic spirochaete Leptospira interrogans. PLoS ONE 2017, 12, e0181118. [Google Scholar] [CrossRef] [PubMed]
- Kool, J.; Jonker, N.; Irth, H.; Niessen, W.M.A. Studing protein-protein affinity and immobilized ligand protein affinity interactions using MS-based methods. Anal. Bioanal. Chem. 2011, 401, 1109–1125. [Google Scholar] [CrossRef] [PubMed]
- Weibezahn, J.; Schlieker, S.; Bukau, B.; Mogk, A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 2003, 278, 32608–32617. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.L.T.O.; Verjovski-Almeida, S.; Van Sluys, M.A.; Monteiro-Vitorello, C.B.; Camargo, L.E.A.; Digiampietri, L.A.; Harstkeerl, R.A.; Ho, P.L.; Marques, M.V.; Oliveira, M.C.; et al. Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 2004, 37, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Matuszewska, E. ATPaza ClpB a usuwanie termicznie zdenaturowanych białek z komórek Escherichia coli. Ph.D. Thesis, University of Gdańsk, Gdańsk, Poland, 2004. [Google Scholar]
- Charon, N.W.; Cockburn, A.; Li, C.; Liu, J.; Miller, K.A.; Miller, M.R.; Motaleb, M.; Wolgemuth, C.W. The unique paradigm of spirochete motility and chemotaxis. Annu. Rev. Microbiol. 2012, 66, 349–370. [Google Scholar] [CrossRef] [PubMed]
- Bender, T.; Lewrenz, I.; Franken, S.; Baitzel, C.; Voos, W. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and Pim1/LON protease. Mol. Biol. Cell 2011, 22, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Fischer, F.; Langer, J.D.; Osiewicz, H.D. Identification of potential mitochondrial ClpXP protease interactions and substrates suggests its central role in energy metabolism. Sci. Rep. 2015, 5, 18375. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.W.; Lei, M.G.; Lee, C.Y. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J. Bacteriol. 2013, 195, 4506–4516. [Google Scholar] [CrossRef] [PubMed]
- Arifuzzaman, M.; Maeda, M.; Itoh, A.; Nishikata, K.; Takita, C.; Saito, R.; Ara, T.; Nakahigashi, K.; Huang, H.C.; Hirai, A.; et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006, 16, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Pinne, M.; Haake, D.A. A comprehensive approach to identification of surface-exposed outer membrane-spanning proteins of Leptospira interrogans. PLoS ONE 2009, 4, e6071. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for quantition of proteins utilizing the principles of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of the structural protein during assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]

| Protein Name | Gene ID a/Gene Name Accession Number | Molecular Mass (kDa) b | Sequence Coverage (%) | Matched Peptides | Score c |
|---|---|---|---|---|---|
| 50S ribosomal protein L19 | LIC11559/rplS gi|446995403 | 15.5 | 13 | 2 | 56 |
| Succinate dehydrogenase flavoprotein subunit | LIC12002/sdhA gi|45657855 | 70.9 | 5 | 4 | 287 |
| Hypothetical protein | gi|446175654 | 15.3 | 9 | 2 | 65 |
| LipL45 | LIC10123 gi|45600754 | 42.3 | 6 | 2 | 173 |
| LipL46 | LIC11885 gi|447001777 | 34.7 | 30 | 8 | 380 |
| Conserved hypothetical protein | LIC11848 gi|45600951 | 32.1 | 9 | 2 | 102 |
| Function and Protein Name | Gene ID a/Gene Name Accession Number | Molecular Mass (kDa) b | Sequence Coverage (%) 37/42 °C | Matched Peptides 37/42 °C | Score c 37/42 °C |
|---|---|---|---|---|---|
| Energy metabolism (16) * | |||||
| Fructose-bisphosphate aldolase | LIC12233 gi|45658082 | 37.8 | 14/- | 3/- | 180/- |
| Triosephosphate isomerase | LIC12094/tpiA gi|45601183 | 27.4 | 11/- | 3/- | 128/- |
| Alcohol dehydrogenase | LIC10253/adh gi|45599391 | 45.9 | 25/4 | 10/2 | 351/114 |
| Putative citrate lyase | LIC11194 gi|45600315 | 38.4 | 9/- | 3/- | 146/- |
| Aconitate hydratase | LIC20249/acnA GI:45655824 | 82.3 | 12/5 | 9/4 | 375/212 |
| Type II citrate synthase | LIC12925/gltA gi|45601997 | 48.6 | -/4 | -/2 | -/118 |
| Matate dehydrogenase | LIC11781/mdh gi|45600887 | 35.1 | 37/19 | 8/4 | 830/273 |
| Succinate dehydrogenase flavoprotein subunit | LIC12002/sdhA gi|45657855 | 71.0 | 11/2 | 5/2 | 229/95 |
| Acyl-CoA hydrolase, thioesterase family protein | LIC11758 gi|45600864 | 16.0 | 17/- | 2/- | 119/- |
| 2,4-dienoyl-CoA reductase | LIC11729/fadH gi|45600834 | 73.8 | 9/- | 4/- | 283/- |
| Electron transfer flavoprotein subunit alpha | LIC10360/etfA gi|45656263 | 28.2 | 43/13 | 9/2 | 611/142 |
| Acyl-CoA dehydrogenase | LIC10583/acd gi|45599716 | 48.7 | 5/- | 3/- | 185/- |
| Acyl-CoA dehydrogenase | LIC13009/acd gi|447196883 | 55.7 | -/8 | -/3 | -/106 |
| Acetyl CoA C-acetyltransferase | LIC12795/phbA gi|446701619 | 48.0 | 10/- | 3/- | 127/- |
| Succinyl-CoA ligase/synthetase subunit β | LIC12573/sucC gi|446613340 | 40.4 | 14/17 | 5/3 | 247/165 |
| 2-oxoglutarate dehydrogenase E1 component | LIC12474/odhA | 103.6 | 5/7 | 7/9 | 275/360 |
| Amino acid metabolism (10) * | |||||
| Acetolactate synthase small subunit | LIC11410/ilvH gi|45600525 | 18.0 | 15/- | 2/- | 65/- |
| N-acetyl-gamma-glutamyl-phosphate reductase | LIC11746/argC gi|45600852 | 37.8 | 5/- | 2/- | 55/- |
| S-adenosyl-l-homocysteine hydrolase | LIC20083/ahcY gi|45655666 | 48.7 | 4/5 | 2/2 | 88/73 |
| Putative branched-chain amino acid aminotransferase | LIC13496/ilvE gi|45602553 | 35.1 | 17/- | 4/- | 207/- |
| Pyridoxal phosphate-dependent aspartate aminotransferase superfamily (ABHA synthase) | LIC12168/aspC gi|45601258 | 44.6 | -/6 | -/2 | -/95 |
| B12-dependent methionine synthase | LIC20085/metH gi|45655668 | 142.5 | 1/1 | 2/2 | 48/79 |
| 3-isopropylmalate dehydrogenase | LIC11768/leuB gi|45657634 | 39.0 | 19/- | 6/- | 355/- |
| d-3-phosphoglycerate dehydrogenase/4-phosphoerythronate dehydrogenase | LIC11992/serA gi|45601085 | 42.3 | 13/- | 3/- | 152/- |
| Cysteine desulfurase | LIC20204/csdB gi|45655784 | 43.8 | 15/- | 3/- | 130/- |
| Ketol-acid reductoisomerase | LIC13393/ilvC gi|45602456 | 35.4 | 16/13 | 5/4 | 271/189 |
| Cysteine synthase | LIC12082/cysK gi|446567601 | 33.2 | 14/- | 2/- | 104/- |
| Nucleotide biosynthesis (1) * | |||||
| Inosine-5′-monophosphate dehydrogenase | LIC11919/guaB gi|45601019 | 56.0 | 6/- | 2/- | 67/- |
| Fatty acid biosynthesis (2) * | |||||
| FAD dependent oxidoreductase/glycerol-3-phosphate dehydrogenase | LIC11699/glpD gi|45600804 | 62.0 | 7/3 | 3/2 | 168/95 |
| Biotin carboxylase | LIC11518/accC gi|446487065 | 102.4 | -/2 | -/2 | -/68 |
| Inorganic ion transport, homeostasis (1) * | |||||
| Potassium transporter TrkA | LIC13175/trkA gi|45602242 | 26.7 | 8/- | 2/- | 107/- |
| Protein degradation (4) | |||||
| Cysteine protease (papain family cysteine protease) | LIC20197 gi|45602748 | 87.8 | 6/5 | 3/2 | 192/118 |
| Aminopeptidase N | LIC12591/pepN gi|45601672 | 102.2 | 6/- | 3/- | 68/- |
| PDZ domain protein, trypsin-like peptidase domain protein/Serine protease MucD precursor | LIC12812/mucD gi|45601887 | 41.2 | 23/12 | 5/2 | 257/176 |
| ATP-dependent ClpP protease ATP-binding subunit ClpX | LIC11418/clpX gi|456986981 | 46.7 | 3/- | 2/- | 168/- |
| Oxidation-reduction processes (3) | |||||
| Molybdopterin oxidoreductase (4Fe-4S-cluster domain protein) | LIC10874 gi|45656765 | 113.6 | 21/- | 14/- | 713/- |
| GMC family oxidoreductase | LIC10037 gi|45655951 | 58.6 | 3/- | 2/- | 69/- |
| Rubrerythrin domain protein | LIC20205 gi|446945174 | 30.6 | 11/21 | 6/12 | 323/535 |
| DNA metabolism (2) * | |||||
| DNA-binding ferritin-like protein | LIC10606/dps gi|45599739 | 18.2 | 47/23 | 5/3 | 397/228 |
| Recombinase RecA | LIC11745/recA gi|446426865 | 39.8 | 6/3 | 5/6 | 240/371 |
| Transcription (4) * | |||||
| ArsR family transcriptional regulator | LIC11617 gi|45600728 | 11.2 | 32/35 | 2/2 | 75/206 |
| DNA-directed RNA polymerase subunit alpha | LIC12846/rpoA gi|45601920 | 36.7 | 36/8 | 13/2 | 474/165 |
| Polyribonucleotide nucleotidyltransferase/polynucleotide phosphorylase | LIC12701/pnpA gi|45601779 | 76.6 | 21/4 | 14/2 | 748/130 |
| Transcription termination factor Rho | LIC12636/rho gi|45601716 | 53.8 | 12/2 | 6/2 | 375/129 |
| Ribosome structure/biogenesis, translation and protein folding (4)* | |||||
| 30S ribosomal protein S15 | LIC12702/rpsO gi|45601780 | 10.3 | -/27 | -/2 | -/78 |
| 30S ribosomal protein S4 | LIC12847/rpsD gi|446057405 | 24.1 | 7/- | 4/- | 133/- |
| 30S ribosomal S3 | LIC12867/rpsC gi|446452098 | 25.7 | 14/- | 2/- | 86/- |
| Elongation factor 4/LepA | LIC12010/lepA gi|45601104 | 67.3 | 11/- | 6/- | 339/- |
| Regulatory function (1) * | |||||
| SAM-dependent methyltransferase | LIC12190/smtA gi|45601280 | 26.1 | 17/- | 2/- | 109/- |
| Cell wall/membrane biogenesis (3) * | |||||
| 2-dehydro-3-deoxyphosphooctonate aldolase/3-deoxy-8-phosphooctulonate synthase | LIC11541/kdsA gi|45600653 | 32.2 | 16/- | 3/- | 190/- |
| LipL71/LruA | LIC11003/lipL71 gi|45600127 | 62.1 | 16/6 | 6/3 | 237/199 |
| Rod shape-determining protein/cell shape determining protein, MreB/Mrl family | LIC11258/mreB gi|456985405 | 37.0 | 9/- | 4/- | 275/- |
| Chemotaxis, motility (2) * | |||||
| Chemotaxis protein | LIC12456/cheA gi|476492777 | 120.0 | 6/- | 6/- | 275/- |
| Methyl-accepting chemotaxis protein | LIC12921/mcpA gi|45601994 | 76.8 | 15/- | 10/- | 605/- |
| Hypothetical proteins (15) | |||||
| Conserved hypothetical protein (with the CBS domain) | LIC12236 gi|45601326 | 16.6 | 29/- | 3/- | 142/- |
| Conserved hypothetical protein (metallo-beta-lactamase superfamily hydrolase) | LIC12478 gi|4560156 | 35.4 | 10/- | 2/- | 76/- |
| Conserved hypothetical protein (with PIN domain) | LIC10215 gi|45656120 | 37.0 | 10/14 | 3/2 | 185/270 |
| Hypothetical protein (TPR protein) | LIC10125 gi|45656035 | 135.3 | 4/3 | 4/3 | 186/212 |
| Conserved hypothetical protein (helicase C-terminal domain protein) | LIC11405 gi|45600520 | 76.9 | 5/3 | 3/2 | 162/124 |
| Conserved hypothetical protein | LIC11274 gi|45600394 | 43.2 | 5/- | 2/- | 94/- |
| Conserved hypothetical protein (region ClpX-like) | LIC10558 gi|45599691 | 17.3 | 15/- | 2/- | 115/- |
| Conserved hypothetical protein (carbohydrate-binding protein, F5/8 type C domain protein) | LIC20001 gi|45602557 | 91.0 | 9/- | 5/- | 205/- |
| Hypothetical protein | LIC13428 gi|45659245 | 54.9 | 15/10 | 6/5 | 438/284 |
| Conserved hypothetical protein | LIC10017 gi|45599164 | 34.3 | -/3 | -/2 | -/88 |
| Conserved hypothetical protein (PaaI family thioesterase) | LIC11209 gi|446570840 | 15.1 | -/23 | -/2 | -/127 |
| Hypothetical protein (aminotransferase) | LIC12198 gi|45601288 | 41.6 | 9/- | 2/- | 144/- |
| Hypothetical protein | LIC10235 gi|45656140 | 10.9 | 22/- | 2/- | 126/204 |
| Hypothetical protein (HEAT repeat domain-containing protein) | LIC10411 gi|446594877 | 17.2 | 14/- | 2/- | 140/- |
| Conserved hypothetical protein (ATPase) | LIC12581 gi|45601662 | 18.3 | 30/- | 2/- | 151/- |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krajewska, J.; Arent, Z.; Zolkiewski, M.; Kędzierska-Mieszkowska, S. Isolation and Identification of Putative Protein Substrates of the AAA+ Molecular Chaperone ClpB from the Pathogenic Spirochaete Leptospira interrogans. Int. J. Mol. Sci. 2018, 19, 1234. https://doi.org/10.3390/ijms19041234
Krajewska J, Arent Z, Zolkiewski M, Kędzierska-Mieszkowska S. Isolation and Identification of Putative Protein Substrates of the AAA+ Molecular Chaperone ClpB from the Pathogenic Spirochaete Leptospira interrogans. International Journal of Molecular Sciences. 2018; 19(4):1234. https://doi.org/10.3390/ijms19041234
Chicago/Turabian StyleKrajewska, Joanna, Zbigniew Arent, Michal Zolkiewski, and Sabina Kędzierska-Mieszkowska. 2018. "Isolation and Identification of Putative Protein Substrates of the AAA+ Molecular Chaperone ClpB from the Pathogenic Spirochaete Leptospira interrogans" International Journal of Molecular Sciences 19, no. 4: 1234. https://doi.org/10.3390/ijms19041234
APA StyleKrajewska, J., Arent, Z., Zolkiewski, M., & Kędzierska-Mieszkowska, S. (2018). Isolation and Identification of Putative Protein Substrates of the AAA+ Molecular Chaperone ClpB from the Pathogenic Spirochaete Leptospira interrogans. International Journal of Molecular Sciences, 19(4), 1234. https://doi.org/10.3390/ijms19041234