Abstract
Bacterial ClpB is an ATP-dependent Hsp100 chaperone that reactivates aggregated proteins in cooperation with the DnaK chaperone system and promotes survival of bacteria under stress conditions. A large number of publications also indicate that ClpB supports the virulence of bacteria, including a pathogenic spirochaete Leptospira interrogans responsible for leptospirosis in both animals and humans. However, the exact role of ClpB in bacterial pathogenicity remains poorly characterized. It can be assumed that ClpB, due to its role as the molecular chaperone, mediates refolding of essential bacterial proteins, including the known virulence factors, which may become prone to aggregation under infection-induced stresses. In this study, we identified putative substrates of ClpB from L. interrogans (ClpBLi). For this purpose, we used a proteomic approach combining the ClpB-Trap affinity pull-down assays, Liquid chromatography-tandem mass spectrometry (LC-MS-MS/MS), and bioinformatics analyses. Most of the identified proteins were enzymes predominantly associated with major metabolic pathways like the tricarboxylic acid (TCA) cycle, glycolysis–gluconeogenesis and amino acid and fatty acid metabolism. Based on our proteomic study, we suggest that ClpB can support the virulence of L. interrogans by protecting the conformational integrity and catalytic activity of multiple metabolic enzymes, thus maintaining energy homeostasis in pathogen cells.
1. Introduction
Bacterial ClpB is a molecular chaperone belonging to the Hsp100 subfamily of the AAA+ ATPases that cooperates with the DnaK chaperone system in solubilization and reactivation of aggregated proteins [1,2,3,4,5,6,7]. Like other Hsp100s, ClpB assembles into barrel-shaped hexamers in the presence of ATP [8]. Each ClpB monomer contains an N-terminal domain (ND) and two ATP-binding domains (NBD1, NBD2) with all characteristic and conserved sequence motifs of AAA+ ATPases, including Walker A and Walker B, and a coiled-coil middle domain (MD) inserted at the end of NBD1 (Figure 1A). ND of ClpB binds and recognizes protein substrates [9], whereas MD determines functional interactions with the DnaK chaperone system required for protein disaggregation both in vivo and in vitro [10,11]. It has been demonstrated that the mechanism of protein disaggregation mediated by ClpB involves the translocation of substrates through the central channel of the hexameric ring driven by the hydrolysis of ATP [5]. However, a recent study found that protein disaggregation might occur through one or two translocation steps, followed by rapid dissociation and rebinding of ClpB to a protein aggregate [12].
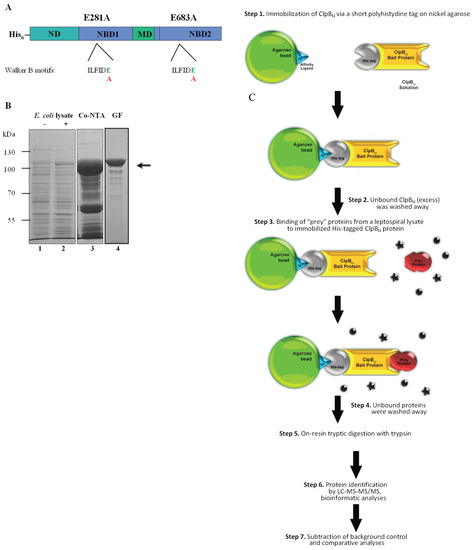
Figure 1.
Schematic representation of the ClpBLi-Trap protein and the experimental strategy used in this study. (A) Domain structure of His6-tagged ClpBLi-Trap used for affinity pull-down experiments: N-terminal domain (ND) involved in recognition and binding of protein substrates, nucleotide binding domain 1 (NBD1), middle coiled-coil domain (MD) and nucleotide binding domain 2 (NBD2). The conserved sequences of the Walker B motifs are shown. The positions of residues within the Walker B motifs changed in this study are indicated. (B) The Coomassie blue-stained SDS-PAGE gel showing the lysates from E. coli cells carrying the recombinant plasmid expressing ClpBLi-Trap without induction (−) (lane 1) and induced with IPTG (+) (lane 2), and the representative fractions obtained following the cobalt affinity column (Co-NTA, lane 3) and gel filtration (GF, lane 4). The arrow indicates the position of His6-tagged ClpBLi-Trap (~98.5 kDa). The positions of protein markers (in kDa), PageRuler Prestained Protein Ladder (Thermo Scientific, Rockford, IL, USA), are shown on the left. (C) Overview of the experimental strategy used for trapping the putative protein substrates of ClpBLi. In the ClpB-Trap affinity pull-down experiments, Ni-NTA agarose was used instead of Co-NTA resin. The ● and * symbols indicate unbound proteins.
ClpB plays a crucial role not only in the survival of bacteria under stressful conditions [7,13], but also in supporting the virulence of some bacterial pathogens, including a pathogenic spirochete Leptospira interrogans [14,15,16,17,18] responsible for leptospirosis affecting animals and humans worldwide. It is estimated that over 1 million human cases of severe leptospirosis occur worldwide each year, with approximately 60,000 deaths from this disease [19,20]. It is worth noting that leptospirosis is also a serious economic problem in many countries, including the European Union. Each year, there are significant economic losses due to reproductive disorders in cattle, sheep, pigs, and horses that are linked to leptospirosis. Moreover, many serological and microbiological studies indicate a high rate of infections in domestic animals [21,22,23,24]. Despite the severity of leptospirosis and its global importance, the molecular mechanisms of the disease pathogenesis are not well understood, mainly due to a lack of standard genetic tools for use in Leptospira species. Identification of the Leptospira virulence factors and characterization of their activity is particularly important for understanding the mechanisms of the disease. To date, several virulence factors have been described in Leptospira, including the ompA-like surface lipoprotein, Loa22 [25], proteins involved in spirochete motility: FliY [26], FlaA2 [27] and LB139 [28], a heme oxygenase, HemO, which is essential for heme-iron utilization [29]; a catalase, KatE, required for resistance to extracellular oxidative stress [30]; phospholipase C, associated with Leptospira-induced macrophage death [31] and HtpG, the highly conserved molecular chaperone from the Hsp90 family [32]. A key virulence factor of Leptospira (common to all Gram-negative bacteria) is lipopolysaccharide (LPS), an important component of the bacterial outer membrane [33]. The molecular chaperone ClpB is also among the known leptospiral virulence factors because the L. interrogans ClpB mutant is avirulent, as opposed to its parental strain [18]. The deficiency of ClpB in L. interrogans also resulted in bacterial growth defects under oxidative and heat stress. As shown previously, the presence of ClpB in kidney tissues of Leptospira-infected hamsters and its immunogenicity [34] also support ClpB’s role in the pathogenicity of Leptospira. However, further studies are needed to elucidate ClpB’s role in virulence. In previous studies, we have demonstrated that the recombinant ClpB from L. interrogans (ClpBLi) displays the aggregate-reactivation activity that may support the survival of L. interrogans under host-induced stress, which is likely to cause denaturation and aggregation of pathogen proteins [35]. Interestingly, we found that ClpBLi may mediate disaggregation of some aggregated proteins without the assistance of the DnaK system [36]. In this study, we constructed a His6-tagged ClpBLi-Trap variant with mutations of the Walker B motif in both ATP-binding domains to identify the putative substrates for ClpBLi by using the protein–protein-interaction-based pull-down strategy [37] coupled with mass spectrometry (MS) analysis. The majority of ClpB-interacting proteins were associated with fundamental metabolic pathways like the TCA cycle, glycolysis–gluconeogenesis, or amino acid and fatty acid metabolism. Thus, our results suggest a possible role of ClpBLi in controlling the energy metabolism of the Leptospira cell under stress. The remaining ClpB-interacting proteins were associated with other essential cellular processes like transcription, protein synthesis, cell wall and membrane biogenesis, spirochete motility, and chemotaxis.
2. Results and Discussion
To reveal the underlying mechanism by which the ClpB chaperone may influence virulence traits in L. interrogans, we have attempted for the first time to identify the Leptospira proteins that can be recognized and potentially reactivated by ClpBLi in cells under environmental stress, including changes in temperature. First, we produced a “substrate trap” variant of ClpBLi (ClpB-Trap; Figure 1A) with mutations within the Walker B motif of both ATP-binding domains (E281A/E683A) based on the work of Weibezahn et al. [38]. These authors showed that ClpB from E. coli with the same mutations in the Walker B motifs binds ATP, but is deficient in the ATP hydrolysis and therefore forms stable complexes with its protein substrates. Additionally, ClpB-Trap was engineered to contain an N-terminal polyhistidine tag. After two-step purification (Figure 1B), His6-tagged ClpBLi-Trap was immobilized on nickel agarose beads and used to capture its potential substrates from the cellular lysates of Leptospira followed by mass-spectrometry-based proteomics. Our strategy for isolation and screening of the ClpBLi substrates is summarized in Figure 1C. The lysates were prepared from L. interrogans serovar Copenhageni cultures exposed to thermal stress at two temperatures, 37 °C (mild heat shock conditions) and 42 °C (severe heat stress) (see Section 3.1). Equal amounts of the total protein lysates (1 µg) were analyzed by SDS-PAGE with Coomassie blue staining (Figure 2A). No apparent differences between the protein profiles obtained under the two heat shock conditions were observed. In the most prominent band of the gel, at approximately 70–80 kDa, we identified mostly GroEL by using LC-MS-MS/MS analysis, although DnaK was also present. Control samples were prepared in parallel with the primary samples (see Section 3.4) to test the effect of ClpBLi binding to the agarose beads and its possible interactions with the endogenous proteins of Leptospira. In the case of the control samples (Figure 2B,E), the trapped proteins were eluted with 250 mM imidazole buffer and analyzed by SDS-PAGE and Coomassie blue staining. The last wash fractions (Figure 2B; LW) were also analyzed in the same way to confirm that all unbound proteins had been washed away from the agarose beads.
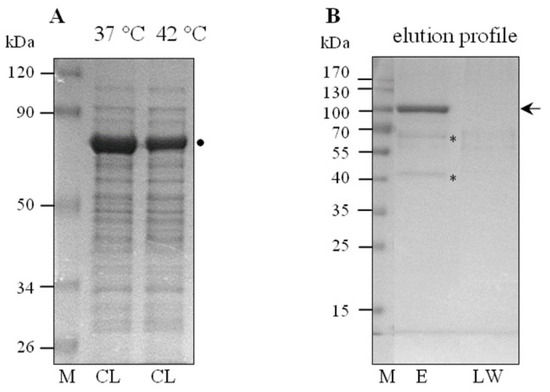
Figure 2.
SDS-PAGE analysis of the lysates of Leptospira cells (CL) cultured at 37 °C or 42 °C (A) and a representative sample of the elution profile of the ClpBLi-Trap binding proteins (B); (M), Prestained Protein Molecular Weight Markers (kDa); Thermo Scientific. (E), the eluted fraction; (LW), the last wash fraction. 12.5% polyacrylamide gels separated using different run-time, were stained with Coomassie Brilliant Blue. The arrow indicates the position of the His6-tagged ClpBLi-Trap protein (~98.5 kDa). The * symbols indicate the putative ClpBLi protein substrates, and the ● symbol indicates GroEL/DnaK identified by LC-MS-MS/MS analysis.
The above experiments were also carried out using His6-tagged HtrA (~51-kDa) as bait (Table 1). The proteins bound to HtrA did not overlap with those captured by ClpBLi, with the exception of succinate dehydrogenase (Table 2). We believe that a comparison with HtrA, which is not related to ClpB and acts as an oligomeric periplasmic serine protease, further validates our strategy for identifying ClpB-specific interactions.

Table 1.
Control protein profile eluted from Ni2+-NTA agarose using His6-tagged HtrA as bait.

Table 2.
Proteins of L. interrogans serovar Copenhageni bound to His6-tagged ClpBLi-Trap.
Table 2 shows a list of all the proteins identified in this study as candidate substrates or partners of ClpBLi after elimination of the background control sample (Table S1, Supplementary Materials) and the pie chart (Figure 3) shows the distribution of these proteins among different functional classes. In total, 68 proteins were identified as ClpB interactors, of which 62 proteins were associated with the lysate prepared from cells submitted to mild heat shock at 37 °C and six additional proteins were obtained when cells were heat-shocked at 42 °C (26 proteins were found in both these fractions). Among the potential ClpB substrates, 15 proteins were annotated as “hypothetical proteins.” The majority of the remaining identified proteins were involved in the central metabolism and energy production. Among them, 10 proteins were assigned to amino acid metabolism and six proteins (i.e., aconitate hydratase, citrate (Si)-synthase, malate dehydrogenase, succinate dehydrogenase flavoprotein subunit, citrate lyase, succinyl-CoA ligase) were involved in the TCA cycle. Another class of the ClpB-interacting proteins was enzymes involved in lipid metabolism (glycerol-3-phosphate dehydrogenase, 2,4-dienoyl-CoA reductase, acyl-CoA hydrolase, acyl-CoA dehydrogenases, acetyl CoA C-acetyltransferase, and biotin carboxylase). Several of the proteins identified in this study were linked to other essential cellular processes, such as ribosome biogenesis (30S ribosomal proteins: S3, S4 and S15), translation (elongation factor 4/LepA), redox homeostasis, or proteolysis. Interestingly, it has been shown that ClpB co-sediments with ribosomes isolated from E. coli cells exposed to heat shock at 45 °C and interacts with some ribosomal proteins [40].
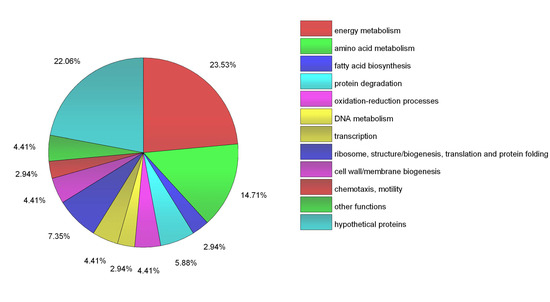
Figure 3.
Functional classification of 68 identified proteins. The pie-chart created using OriginLab software (OriginPro 2016, Northampton, MA, USA) shows the distribution of these proteins into their biological processes in percentage.
Among the proteins identified in this study (see Table 2) was SAM-dependent methyltransferase, which catalyzes the methylation of biomolecules, including amino acids, proteins, and DNA. In addition, two identified proteins were directly associated with chemotaxis and spirochete motility, which support the L. interrogans virulence in the hamster model for leptospirosis [41]. The remaining proteins were associated with the cell wall or membrane biogenesis.
The identified proteins, including the enzymes of major metabolic pathways like the TCA cycle, glycolysis–gluconeogenesis, amino acid and lipid metabolism (Table 2), may require the assistance of ClpBLi during heat shock. In fact, it has been demonstrated that key metabolic enzymes are heat-sensitive and aggregation-prone and therefore are often inactivated by stress [42]. Stress conditions induce structural destabilization, unfolding and, ultimately, aggregation of enzymatic components of the major metabolic pathways.
Interestingly, Fischer and co-workers [43] have recently found that the mitochondrial ClpXP protease in Podospora anserina is mainly associated with enzymes involved in TCA cycle, amino acid and fatty acid metabolism, and subunits of electron transport chain complex. In the ClpXP complex, the ATPase ClpX is responsible for substrate recognition and contains structural domains homologous to those found in ClpB. Many proteins involved in energy metabolism and also in protein translation, transcription, DNA metabolism and fatty acid metabolism, were also reported as substrates of the Staphylococcus aureus ClpC chaperone that is an ATP-dependent Hsp100 chaperone like ClpB [44].
Furthermore, we have previously found that ClpBLi is able to reactivate thermally inactivated fructose-bisphosphate aldolase, one of the identified metabolic enzymes, even in the absence of the DnaK chaperone system from E. coli [36].
We propose that the key metabolic enzymes are the main substrates for the molecular chaperone ClpBLi and preservation of their activity under stress conditions depends on the ClpBLi disaggregase activity. It is likely that the metabolic enzymes have an important impact on the growth of Leptospira cells and the leptospiral pathogenicity. We suggest that ClpBLi influences virulence traits in L. interrogans mainly through preservation of the activity of metabolic enzymes.
The remaining identified ClpBLi substrates, 2-dehydro-3-deoxyphosphooctonate aldolase responsible for biosynthesis of the oligosaccharide core of LPS, an essential virulence factor in all Gram-negative bacteria, the chemotaxis proteins, or a membrane lipoprotein LruA (LipL71), may support the Leptospira virulence. Other potential substrates of ClpBLi include proteins involved in the mRNA metabolism (polyribonucleotide nucleotidyltransferase) or transcription (DNA-directed RNA polymerase α subunit, ArsR family transcriptional regulator). Thus, ClpBLi could indirectly influence gene expression in leptospiral cells. It is noteworthy that Arifuzzaman et al., 2006 [45] observed interactions between a multi-subunit complex of RNA polymerase (RNAP) from E. coli and some chaperones, including ClpB. Those data suggest that not only DnaK, but also ClpB, may assist the assembly of RNAP.
In summary, we performed the first identification of the potential protein substrates of ClpBLi. The majority of these proteins is associated with energy-generating metabolism and may have an important impact on the grown and pathogenicity of Leptospira. Further in vitro studies will determine whether the proteins identified in this work interact directly with ClpB or if they share interacting partners with the chaperone. Our results suggest a possible role of ClpBLi in maintaining the energy-generating metabolism of the Leptospira cell and strongly support the ClpB’s importance in the leptospiral virulence. We believe that our results help explain the previously established role of the molecular chaperone ClpB in supporting bacterial pathogenicity.
3. Materials and Methods
3.1. Leptospira Strain, Growth Conditions, and Cell Lysate Preparation
L. interrogans serovar Copenhageni strain B42 was grown in liquid Ellinghausen McCollough Jonhson and Harris medium (EMJH) at 30 °C until mid-exponential phase (OD420 = ~0.3) then transferred to 37 or 42 °C for 4 or 2 h, respectively (protein aggregate formation). After exposure to thermal stress, a total of 100 mL of cells were harvested by centrifugation at 6000× g for 10 min at room temperature and cell lysates were prepared as previously described [46] with some modifications. Briefly, leptospires were washed twice with phosphate-buffered saline (PBS, pH 7.4), 5 mM MgCl2 and resuspended in lysis buffer (10 mM Tris/HCl, pH 8.0, 2 mM EDTA, 25 mM NaCl, 1 mM PMSF protease inhibitor) containing 1mg/mL of lysozyme. The suspension was incubated for 5 min at 4 °C and then subjected to three cycles of freezing (−80 °C) and thawing (room temperature) with vigorous vortexing. Next, DNase I (to a final concentration of 5 µg/mL) was added and the cell suspensions were incubated on ice for 20 min, sonicated with 20% amplitude in 5 s pulses for 30 s using a microtip Vibra Cell sonicator. The insoluble materials (unbroken cells and cell debris) were removed by centrifugation at 6000× g for 10 min at 4 °C. The soluble supernatants (total cell lysates), including unfolded or aggregated proteins, were used for screening protein substrates/partners of ClpBLi. Protein concentration in each cell lysates was determined by the Bradford method [47] using BSA as a standard. For assessment of the lysis efficiency, SDS-PAGE electrophoresis (Bio-Rad, Hercules, CA, USA) was performed as described previously [48] using 12.5% polyacrylamide gels followed by staining with Coomassie Brillant Blue.
3.2. Construction of ClpBLi-Trap Mutant
To construct a ClpB-Trap variant useful for trapping ClpB substrates, we replaced the conserved glutamic acid in each of the two Walker B motifs (E281 and E683 in the NBD1 and NBD2, respectively) with alanine. The mutations were introduced by the QuickChange II site-directed mutagenesis method (Agilent Technologies, Santa Clara, CA, USA) using primers with the desired mutation and Pfu Turbo DNA polymerase (Agilent Technologies). The pET28ClpBLi construct [34] was used as the template DNA in a mutagenic PCR reaction. The generated construct was confirmed by DNA sequencing (Genomed S.A., Warsaw, Poland).
3.3. Purification of ClpBLi-Trap (E281A/E683A)
The His6-tagged ClpB-Trap protein was overproduced from the recombinant plasmid pET28b in E. coli BL21 (DE3) strain (Novagen/Merck, Darmstadt, Germany) and purified in two steps using HisPur cobalt resin (Thermo Scientific, Rockford, IL, USA) followed by Superdex 200 gel filtration as previously described [34]. Fractions containing ClpBLi were identified by SDS-PAGE and staining with Coomassie blue. ClpB concentration was estimated from absorption at 280 nm using for ClpBLi the extinction coefficient ε0.1% = 0.445 (mg/mL)−1 cm−1 calculated from its amino acid composition by ProtParam [49].
3.4. Affinity Pull-Down Assay and MS Analysis
His6-tagged ClpBLi-Trap (1.5 μM) in buffer A (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 20 mM imidazole) was incubated with 15 μL of Ni-NTA agarose (Macherey-Nagel, Düren, Germany) suspended in the same buffer A for 3 h at 4 °C. Agarose beads were washed twice with buffer A and then the soluble protein fractions (~100 µg proteins) prepared from L. interrogans cultures submitted to thermal stress as described above were added. We used an excess of His-tagged ClpBLi-Trap over potential binding proteins (prey proteins). Thus, competition for binding to Ni-NTA between the prey proteins and ClpB is unlikely. After a 30-min incubation in the presence of 2 mM ATP at room temperature, the agarose beads were washed with buffer A containing 2 mM ATP (15 times with 200 μL), then eluted with buffer A containing 250 mM imidazole to test ClpBLi binding efficiency to the beads or suspended in water and used as a “bead proteome” for identification of proteins interacting with ClpBLi. For this purpose, LC-MS-MS/MS analysis of tryptic peptides obtained after trypsin cleavage of the separated proteins was performed at the MS LAB IBB PAN (Warsaw, Poland). The resulting MS/MS spectra were submitted to the program Mascot and searched against the NCBI-nr database (57,412,064 sequences and 20,591,031,683 residues). The search was restricted to L. interrogans proteins (104,694 sequences). Positive hits were identified with at least two unique peptides present in two independent biological replicates of each sample with a Mascot ion score above 30. Proteins found in the background control sample (the agarose beads incubated with the total cell lysates, described above, in the absence of the His6-tagged ClpBLi-Trap protein; Table S1, Supplementary Materials) were eliminated from the set of candidate ClpB binding proteins.
As another control, we performed an affinity pull-down assay using a protein unrelated to ClpB, His6-tagged HtrA (1.5 µM), as bait (Table 1), and the same experimental conditions as described above, with one exception—namely, no ATP was added to the buffers.
The two control experiments described above were performed to ensure that the detected interactions between His6-tagged ClpBLi and proteins from the leptospiral lysates were linked to the ATP-dependent function of ClpB and not to nonspecific binding.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/4/1234/s1.
Acknowledgments
This work was supported by Preludium Grant 2015/17/N/NZ6/03493 (to Joanna Krajewska) from the National Science Center (Poland) and UG grant for Young Scientists 538-L136-B246-17 (to Joanna Krajewska). The equipment used was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. We are grateful to Janusz Dębski from the Institute of Biochemistry and Biophysics (Polish Academy of Sciences) for his assistance with the data analyses, Daniel Więckowski for preparing Figure 1C, and Urszula Zarzecka for the purified HtrA protein.
Author Contributions
Joanna Krajewska and Zbigniew Arent performed the experiments. Sabina Kędzierska-Mieszkowska designed the experiments, analyzed the data, and wrote the paper. Michal Zolkiewski assisted in data analyses and the preparation of manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zolkiewski, M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J. Biol. Chem. 1999, 274, 28083–28086. [Google Scholar] [CrossRef] [PubMed]
- Goloubinoff, P.; Mogk, A.; Ben-Zvi, A.P; Tomoyasu, T.; Bukau, B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 1999, 96, 13732–13737. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Tomoyasu, T.; Goloubinoff, P.; Rűdiger, S.; Röder, D.; Langen, H.; Bukau, B. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999, 18, 6934–6949. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Sowa, M.E.; Watanabe, Y.; Sigler, P.B.; Chiu, W.; Yoshida, M.; Tsai, F.T. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell 2003, 115, 229–240. [Google Scholar] [CrossRef]
- Weibezahn, J.; Tessarz, P.; Schlieker, C.; Zahn, R.; Maglica, Z.; Lee, S.; Zentgraf, H.; Weber-Ban, E.U.; Dougan, D.A; Tsai, F.T.; et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 2004, 119, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Barnett, M.E.; Nagy, M.; Kedzierska, S.; Zolkiewski, M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 2005, 280, 34940–34945. [Google Scholar] [CrossRef] [PubMed]
- Nagy, M.; Guenther, I.; Akoyev, V.; Barnett, M.E.; Zavodszky, M.I.; Kedzierska-Mieszkowska, S.; Zolkiewski, M. Synergistic cooperation between two ClpB isoforms in aggregate reactivation. J. Mol. Biol. 2010, 396, 697–707. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Akoev, V.; Gogol, E.P.; Barnett, M.E.; Zolkiewski, M. Nucleotide-induced switch in oligomerization of the AAA+ ATPase ClpB. Protein Sci. 2004, 13, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Farber, P.; Velvis, A.; Rennella, E.; Latham, M.P.; Kay, L.E. ClpB N-terminal domain plays a regulatory role in protein disaggregation. Proc. Natl. Acad. Sci. USA 2015, 112, E6872–E6881. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, M.E.; Shorter, J. The elusive middle domain of Hsp104 and ClpB: Location and function. Biochim. Biophys. Acta 2012, 1823, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Kedzierska, S.; Akoev, V.; Barnett, M.E.; Zolkiewski, M. Structure and function of the middle domain of ClpB from Escherichia coli. Biochemistry 2003, 42, 14242–14248. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Weaver, C.L.; Lin, J.; Duran, E.C.; Miller, J.M.; Lucius, A.L. Escherichia coli ClpB is a no-n-processive polypeptide translocase. Biochem. J. 2015, 470, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Squires, C.L.; Pedersen, S.; Ross, B.M.; Squires, C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1991, 173, 4254–4262. [Google Scholar] [CrossRef] [PubMed]
- Kannan, T.R.; Musatovova, O.; Gowda, P.; Baseman, J.B. Characterization of a unique ClpB protein of Mycoplasma pneumoniae and its impact on growth. Infect. Immun. 2008, 76, 5082–5092. [Google Scholar] [CrossRef] [PubMed]
- Capestany, C.A.; Tribble, G.D.; Maeda, K.; Demuth, D.R.; Lament, R.J. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 2008, 190, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Chastanet, A.; Derre, I.; Nair, S.; Msadek, T. ClpB, a novel number of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 2004, 186, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Meibom, K.L.; Dubail, I.; Dupuis, M.; Barel, M.; Lenco, J.; Stulik, J.; Golovliov, I.; Sjöstedt, A.; Charbit, A. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 2008, 67, 1384–1401. [Google Scholar] [CrossRef] [PubMed]
- Lourdault, K.; Cerqueira, G.M.; Wunder, E.A., Jr.; Picardeau, M. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 2011, 79, 3711–3717. [Google Scholar] [CrossRef] [PubMed]
- Adler, B.; Lo, M.; Seemann, T.; Murray, G.L. Pathogenesis of leptospirosis: The influence of genomics. Vet. Mirobiol. 2011, 153, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017, 15, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.G.; Nola, L.; O’Grady, L.; More, S.J.; Doherty, L.M. Seroprevalence of Leptospira Hardjo in the Irish suckler cattle population. Ir. Vet. J. 2012, 65, 8. [Google Scholar] [CrossRef] [PubMed]
- Arent, Z.; Kędzierska-Mieszkowska, S. Seroprevalence study of leptospirosis in horses in northern Poland. Vet. Rec. 2013, 172, 269. [Google Scholar] [CrossRef] [PubMed]
- Arent, Z.; Frizzell, C.; Gilmore, C.; Mackie, D.; Ellis, W.A. Isolation of leptospires from genital tract of sheep. Vet. Rec. 2013, 173, 582. [Google Scholar] [CrossRef] [PubMed]
- Arent, Z.J.; Andrews, S.; Adamama-Moraitou, K.; Gilmore, C.; Pardali, D.; Ellis, W.A. Emergence of novel Leptospira serovars: A need for adjusting vaccination policies for dogs? Epidemiol. Infect. 2013, 141, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Ristow, P.; Bourhy, P.; da McBride, F.W.; Figueira, C.P.; Huerre, M.; Ave, P.; Girons, I.S.; Ko, A.I.; Picardeau, M. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 2007, 3, e97. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Sun, A.; Ojcius, D.; Wu, S.; Zhao, J.; Yan, J. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 2009, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.; Picardeau, M.; Haake, D.A.; Sermswan, R.W.; Srikram, A.; Adler, B.; Murray, G.L. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of the flagellum sheath. Infect. Immun. 2012, 80, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Becam, J.; Lambert, A.; Sismeiro, O.; Dillies, M.A.; Jagla, B.; Wunder, E.A., Jr.; Ko, I.; Coppee, J.Y.; Goaran, C.; et al. A putative regulatory genetic locus modulates virulence in the pathogen Leptospira interrogans. Infect. Immun. 2014, 82, 2542–2552. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Srikram, A.; Henry, R.; Puapairoj, A.; Sermswan, R.W.; Adler, B. Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. 2009, 11, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Eshghi, A.; Lourdault, K.; Murray, G.L.; Bartpho, T.; Sermswan, R.W.; Picardeau, M.; Adler, B.; Snarr, B.; Zuerner, R.L.; Cameron, C.E. Leptospira interrogans catalase is required for resistance to H2O2 and for virulence. Infect. Immun. 2012, 80, 3892–3899. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.F.; Chen, H.H.; Ojcius, D.M.; Zhao, X.; Sun, D.; Ge, Y.M.; Zheng, L.L.; Lin, X.; Li, L.J.; Yan, J. Identification of Leptospira interrogans phospholipase C as a novel virulence factor responsible for intracellular free calcium ion elevation during macrophage death. PLoS ONE 2013, 8, e75652. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; Pretre, G.; Bartpho, T.; Sermswan, R.W.; Toma, C.; Suzuki, T.; Eshgh, A.; Picardeau, M.; Adler, B.; Murray, G.L. High-temperature protein G is an essential virulence factor of Leptospira interrogans. Infect. Immun. 2014, 82, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.L.; Srikram, A.; Henry, R.; Hartskeerl, R.A.; Sermswan, R.W.; Adler, B. Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol. Microbiol. 2010, 78, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, J.; Arent, Z.; Więckowski, D.; Zolkiewski, M.; Kędzierska-Mieszkowska, S. Immunoreactivity of the AAA+ chaperone ClpB from Leptospira interrogans with sera from Leptospira-infected animals. BMC Microbiol. 2016, 16, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Hu, W.; Me, Y.; Ojcius, D.M.; Lin, X.; Yan, J. A leptospiral AAA+ chaperone-Ntn peptidase complex, HslUV, contributes to the intracellular survival of Leptospira interrogans in hosts and the transmission of leptospirosis. Emerg. Microbes Infect. 2017, 6, e105. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, J.; Modrak-Wójcik, A.; Arent, Z.; Więckowski, D.; Zolkiewski, M.; Bzowska, A.; Kędzierska-Mieszkowska, S. Characterization of the molecular chaperone ClpB from the pathogenic spirochaete Leptospira interrogans. PLoS ONE 2017, 12, e0181118. [Google Scholar] [CrossRef] [PubMed]
- Kool, J.; Jonker, N.; Irth, H.; Niessen, W.M.A. Studing protein-protein affinity and immobilized ligand protein affinity interactions using MS-based methods. Anal. Bioanal. Chem. 2011, 401, 1109–1125. [Google Scholar] [CrossRef] [PubMed]
- Weibezahn, J.; Schlieker, S.; Bukau, B.; Mogk, A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 2003, 278, 32608–32617. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.L.T.O.; Verjovski-Almeida, S.; Van Sluys, M.A.; Monteiro-Vitorello, C.B.; Camargo, L.E.A.; Digiampietri, L.A.; Harstkeerl, R.A.; Ho, P.L.; Marques, M.V.; Oliveira, M.C.; et al. Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 2004, 37, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Matuszewska, E. ATPaza ClpB a usuwanie termicznie zdenaturowanych białek z komórek Escherichia coli. Ph.D. Thesis, University of Gdańsk, Gdańsk, Poland, 2004. [Google Scholar]
- Charon, N.W.; Cockburn, A.; Li, C.; Liu, J.; Miller, K.A.; Miller, M.R.; Motaleb, M.; Wolgemuth, C.W. The unique paradigm of spirochete motility and chemotaxis. Annu. Rev. Microbiol. 2012, 66, 349–370. [Google Scholar] [CrossRef] [PubMed]
- Bender, T.; Lewrenz, I.; Franken, S.; Baitzel, C.; Voos, W. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and Pim1/LON protease. Mol. Biol. Cell 2011, 22, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Fischer, F.; Langer, J.D.; Osiewicz, H.D. Identification of potential mitochondrial ClpXP protease interactions and substrates suggests its central role in energy metabolism. Sci. Rep. 2015, 5, 18375. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.W.; Lei, M.G.; Lee, C.Y. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J. Bacteriol. 2013, 195, 4506–4516. [Google Scholar] [CrossRef] [PubMed]
- Arifuzzaman, M.; Maeda, M.; Itoh, A.; Nishikata, K.; Takita, C.; Saito, R.; Ara, T.; Nakahigashi, K.; Huang, H.C.; Hirai, A.; et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006, 16, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Pinne, M.; Haake, D.A. A comprehensive approach to identification of surface-exposed outer membrane-spanning proteins of Leptospira interrogans. PLoS ONE 2009, 4, e6071. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for quantition of proteins utilizing the principles of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of the structural protein during assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).