Pathobiologic Roles of Epstein–Barr Virus-Encoded MicroRNAs in Human Lymphomas
Abstract
1. Epstein–Barr Virus as an Oncovirus in Human Lymphomas
EBV Latency Programs
2. MicroRNAs
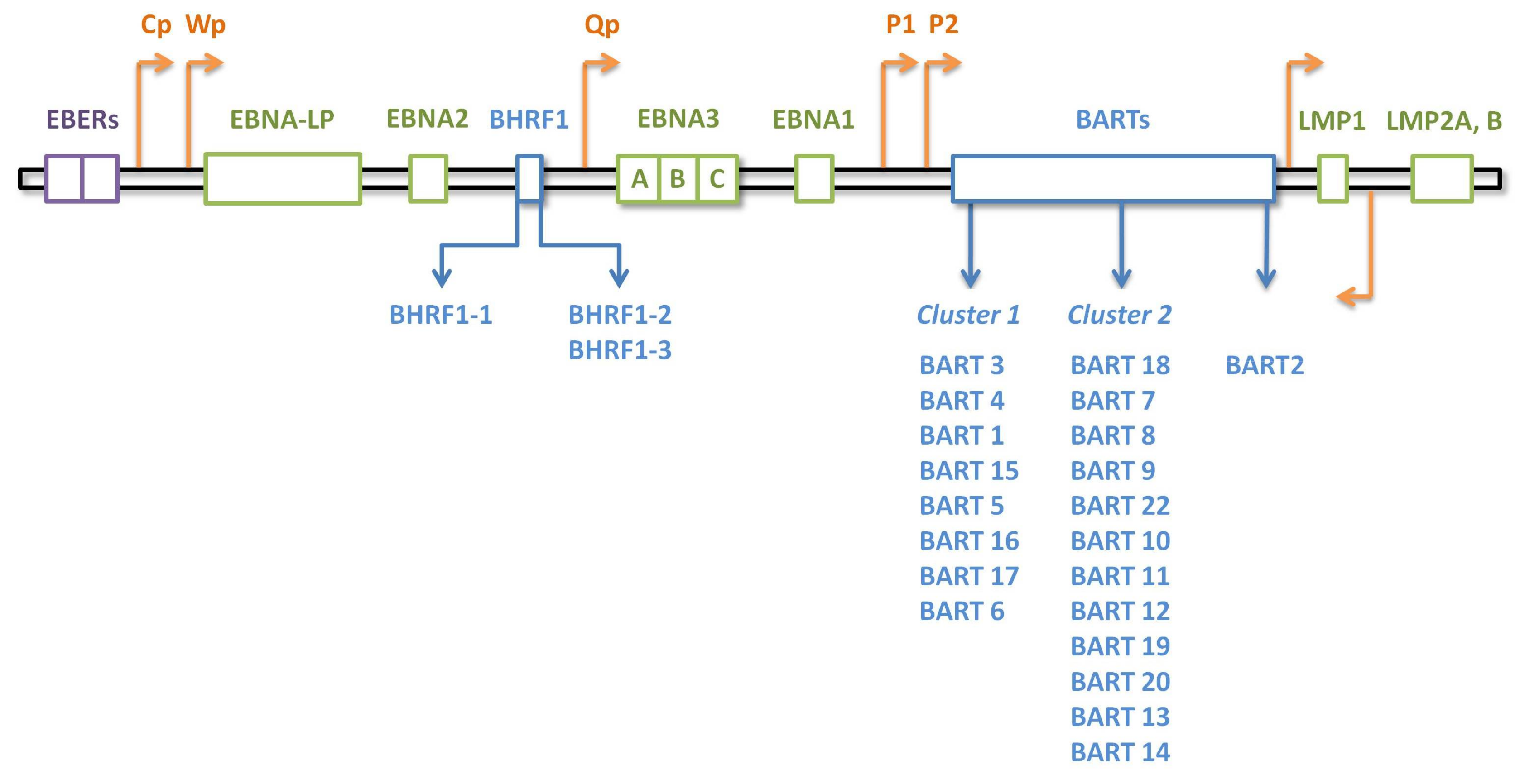
3. EBV-Encoded MicroRNAs in Human Lymphomas
3.1. EBV miRNA Expression Patterns
3.2. Pathologic and Biologic Role of EBV miRNAs
3.2.1. EBV miRNAs Contribute to Evasion from Immunosurveillance
3.2.2. EBV miRNAs Interfere with Several Other Cancer-Related Mechanisms
3.2.3. Molecular Networks and Circuits Define More Complex Roles for EBV-Encoded MicroRNAs
3.2.4. The Pathogenetic Effect of Viral miRNAs Could Be Observed at Global Gene Expression Profile Level
3.2.5. Exosomal Shuttle MicroRNAs Help EBV to Manipulate Its Surrounding Microenvironment
3.3. EBV miRNAs as Molecular Markers for Classification, Diagnosis, and Prognosis
4. Conclusion Remarks
Acknowledgments
Conflicts of Interest
Abbreviations
| BART | BamHI-A rightward transcript |
| BAX | BCL2 Associated X |
| BHRF1 | BamHI fragment H rightward open reading frame 1 |
| BL | Burkitt Lymphoma |
| CASP3 | Caspase 3 |
| CDKN1B | Cyclin Dependent Kinase Inhibitor 1b |
| C/EBP | CCAAT-Enhancer-Binding Protein |
| CLL | Chronic Lymphocytic Leukemia |
| CREBBP | CREB-Binding Protein |
| CTSB | cathepsin B |
| Cp | C Promoter |
| DLBCL | Diffuse Large B Cell Lymphoma |
| EBERs | EBV-Encoded small RNAs |
| eBL | endemic Burkitt Lymphoma |
| EBNA | EBV-Encoded Nuclear Antigen |
| EBNA-LP | EBV-Encoded Nuclear Antigen Leader Protein |
| EBV | Epstein–Barr Virus |
| HHV-4 | Human Herpesvirus 4 |
| HIV-BL | HIV-related Burkitt Lymphoma |
| HIV-DLBCL | HIV-related Diffuse Large B Cell Lymphoma |
| HL | Hodgkin Lymphoma |
| IFI30 | Interferon Gamma Inducible Protein 30 |
| IFN | Interferon |
| IgG | Immunoglobulin G |
| IHC | Immunohistochemistry |
| IL-1 | Interleukin-1 |
| IL-1β | Interleukin-1β |
| IL-12B | Interleukin-12 Subunit Beta |
| IL-18 | Interleukin 18 |
| IL-6 | Interleukin-6 |
| IL-6RB | Interleukin-6 Receptor B |
| IPO7 | Importin 7 |
| IRF | Interferon Regulatory Factor |
| ISH | In Situ Hybridization |
| LCL | Lymphoblastoid Cell Line |
| LGMN | Legumain |
| LMP | Latent Membrane Protein |
| MHC | Major Histocompatibility Complex |
| miRNA | microRNA |
| MTX-LPD | Methotrexate-Associated Lymphoproliferative Disorder |
| NKTL | Natural Killer/T Cell Lymphoma |
| NLRP3 | NLR Family Pyrin Domain Containing 3 |
| p27 | protein 27 |
| P1 | Promoter 1 |
| P2 | Promoter 2 |
| PAL | Pyothorax-Associated Lymphoma |
| PBL | Plasmablastic Lymphoma |
| PRDM1 | pr/set domain 1 |
| PTEN | Phosphatase and Tensin Homolog |
| PTLD | Post-Transplant Lymphoproliferative Disorder |
| Qp | Q Promoter |
| RICTOR | Rapamycin-Insensitive Companion of Mammalian Target of Rapamycin |
| RIG-I | Retinoic-Acid-Inducible Protein 1 |
| S1PR1 | Sphingosin-1-Phosphate Receptor 1 |
| sBL | sporadic Burkitt Lymphoma |
| SNP | Single Nucleotide Polymorphism |
| SUMO | Small Ubiquitin-Like Modifier |
| TAP | Transporter Associated With Antigen Processing |
| T-bet | T-box Protein Expressed in T cells |
| TOMM22 | Translocase of Outer Mitochondrial Membrane 22 |
| VEGFA | Vascular Endothelial Growth Factor A |
| Wp | W Promoter |
References
- McLaughlin-Drubin, M.E.; Munger, K. Viruses associated with human cancer. Biochim. Biophys. Acta 2008, 1782, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Esau, D. Viral causes of lymphoma: The history of epstein–barr virus and human T-lymphotropic virus 1. Virology 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Navari, M.; Fuligni, F.; Laginestra, M.A.; Etebari, M.; Ambrosio, M.R.; Sapienza, M.R.; Rossi, M.; de Falco, G.; Gibellini, D.; Tripodo, C.; et al. Molecular signature of epstein–barr virus-positive burkitt lymphoma and post-transplant lymphoproliferative disorder suggest different roles for epstein–barr virus. Front. Microbiol. 2014, 5, 728. [Google Scholar] [CrossRef] [PubMed]
- Epstein, M.; Achong, B.; Barr, Y. Virus particles in cultured lymphoblasts from burkitt’s lymphoma. Lancet 1964, 283, 702–703. [Google Scholar] [CrossRef]
- Mesri, E.A.; Feitelson, M.A.; Munger, K. Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe 2014, 15, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, M.R.; Mundo, L.; Gazaneo, S.; Picciolini, M.; Vara, P.S.; Sayed, S.; Ginori, A.; lo Bello, G.; del Porro, L.; Navari, M.; et al. MicroRNAs sequencing unveils distinct molecular subgroups of plasmablastic lymphoma. Oncotarget 2017, 8, 107356–107373. [Google Scholar] [CrossRef] [PubMed]
- Mundo, L.; Ambrosio, M.R.; Picciolini, M.; lo Bello, G.; Gazaneo, S.; del Porro, L.; Lazzi, S.; Navari, M.; Onyango, N.; Granai, M.; et al. Unveiling another missing piece in EBV-driven lymphomagenesis: EBV-encoded micrornas expression in eber-negative burkitt lymphoma cases. Front. Microbiol. 2017, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Sekizuka, T.; Uehara, T.; Hishima, T.; Mine, S.; Fukumoto, H.; Sato, Y.; Hasegawa, H.; Kuroda, M.; Katano, H. Next-generation sequencing of mirnas in clinical samples of epstein–barr virus-associated B-cell lymphomas. Cancer Med. 2017, 6, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Navari, M.; Etebari, M.; de Falco, G.; Ambrosio, M.R.; Gibellini, D.; Leoncini, L.; Piccaluga, P.P. The presence of epstein–barr virus significantly impacts the transcriptional profile in immunodeficiency-associated burkitt lymphoma. Front. Microbiol. 2015, 6, 556. [Google Scholar] [CrossRef] [PubMed]
- Ferrajoli, A.; Ivan, C.; Ciccone, M.; Shimizu, M.; Kita, Y.; Ohtsuka, M.; D’Abundo, L.; Qiang, J.; Lerner, S.; Nouraee, N.; et al. Epstein–barr virus microRNAs are expressed in patients with chronic lymphocytic leukemia and correlate with overall survival. eBioMedicine 2015, 2, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Komabayashi, Y.; Kishibe, K.; Nagato, T.; Ueda, S.; Takahara, M.; Harabuchi, Y. Circulating epstein–barr virus-encoded microRNAs as potential biomarkers for nasal natural killer/T-cell lymphoma. Hematol. Oncol. 2017, 35, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Grinde, B. Herpesviruses: Latency and reactivation—Viral strategies and host response. J. Oral Microbiol. 2013, 5, 22766. [Google Scholar] [CrossRef] [PubMed]
- Speck, S.H.; Ganem, D. Viral latency and its regulation: Lessons from the γ-herpesviruses. Cell Host Microbe 2010, 8, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Munz, C. Epstein–barr virus nuclear antigen 1: From immunologically invisible to a promising T cell target. J. Exp. Med. 2004, 199, 1301–1304. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.S.; Lu, F.; Lieberman, P.M. Epigenetic regulation of EBV and KSHV latency. Curr. Opin. Virol. 2013, 3, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Schelcher, C.; Valencia, S.; Delecluse, H.J.; Hicks, M.; Sinclair, A.J. Mutation of a single amino acid residue in the basic region of the epstein–barr virus (EBV) lytic cycle switch protein zta (bzlf1) prevents reactivation of ebv from latency. J. Virol. 2005, 79, 13822–13828. [Google Scholar] [CrossRef] [PubMed]
- Kenney, S.C. Reactivation and Lytic Replication of EBV; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Grywalska, E.; Rolinski, J. Epstein–barr virus-associated lymphomas. Semin. Oncol. 2015, 42, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Li, L.; Young, K.H. EBV-driven B-cell lymphoproliferative disorders: From biology, classification and differential diagnosis to clinical management. Exp. Mol. Med. 2015, 47, e132. [Google Scholar] [CrossRef] [PubMed]
- Weiss, L.M.; Chen, Y.-Y. EBER in situ hybridization for epstein–barr virus. In Hematological Malignancies; Springer: Berlin, Germany, 2013; pp. 223–230. [Google Scholar]
- Kelly, G.L.; Stylianou, J.; Rasaiyaah, J.; Wei, W.; Thomas, W.; Croom-Carter, D.; Kohler, C.; Spang, R.; Woodman, C.; Kellam, P.; et al. Different patterns of epstein–barr virus latency in endemic burkitt lymphoma (BL) lead to distinct variants within the BL-associated gene expression signature. J. Virol. 2013, 87, 2882–2894. [Google Scholar] [CrossRef] [PubMed]
- Kelly, G.L.; Milner, A.E.; Tierney, R.J.; Croom-Carter, D.S.; Altmann, M.; Hammerschmidt, W.; Bell, A.I.; Rickinson, A.B. Epstein–barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A,-3b, and-3c expression in burkitt’s lymphoma cells and with increased resistance to apoptosis. J. Virol. 2005, 79, 10709–10717. [Google Scholar] [CrossRef] [PubMed]
- Gion, Y.; Iwaki, N.; Takata, K.; Takeuchi, M.; Nishida, K.; Orita, Y.; Tachibana, T.; Yoshino, T.; Sato, Y. Clinicopathological analysis of methotrexate-associated lymphoproliferative disorders: Comparison of diffuse large B-cell lymphoma and classical hodgkin lymphoma types. Cancer Sci. 2017, 108, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein–barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789. [Google Scholar] [CrossRef] [PubMed]
- Kis, L.L.; Gerasimčik, N.; Salamon, D.; Persson, E.K.; Nagy, N.; Klein, G.; Severinson, E.; Klein, E. Stat6 signaling pathway activated by the cytokines IL-4 and IL-13 induces expression of the epstein–barr virus—Encoded protein LMP-1 in absence of EBNA-2: Implications for the type II EBV latent gene expression in hodgkin lymphoma. Blood 2011, 117, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.K.; Tao, Q.; Srivastava, G.; Ho, F. Nasal NK-and T-cell lymphomas share the same type of epstein-barr virus latency as nasopharyngeal carcinoma and hodgkin’s disease. Int. J. Cancer 1996, 68, 285–290. [Google Scholar] [CrossRef]
- Laytragoon-Lewin, N.; Chen, F.; Avila-Cariño, J.; Zou, J.Z.; Mellstedt, H.; Ernberg, I.; Klein, G. Epstein–barr virus (EBV)-carrying cells of a chronic lymphocytic leukemia (CLL) subpopulation express EBNA1 and lmps but not EBNA2 in vivo. Int. J. Cancer 1995, 63, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.J.; Beltran, B.E.; Miranda, R.N.; Paydas, S.; Winer, E.S.; Butera, J.N. Epstein–barr virus–positive diffuse large B-cell lymphoma of the elderly: What we know so far. Oncologist 2011, 16, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene LIN-4 encodes small RNAs with antisense complementarity to LIN-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. Micrornas: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. Microrna dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2017, 9, 852. [Google Scholar] [CrossRef] [PubMed]
- Onnis, A.; Navari, M.; Antonicelli, G.; Morettini, F.; Mannucci, S.; de Falco, G.; Vigorito, E.; Leoncini, L. Epstein–barr nuclear antigen 1 induces expression of the cellular microrna HSA-MIR-127 and impairing b-cell differentiation in EBV-infected memory B cells. New insights into the pathogenesis of burkitt lymphoma. Blood Cancer J. 2012, 31, 29. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of micrornas in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Anastasiadou, E.; Esteller, M.; He, L.; Slack, F.J. The inescapable influence of noncoding RNAs in cancer. Cancer Res. 2015, 75, 5206–5210. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. Mirbase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Cosmopoulos, K.; Pegtel, M.; Hopmans, E.; Murray, P.; Middeldorp, J.; Shapiro, M.; Thorley-Lawson, D.A. A novel persistence associated ebv mirna expression profile is disrupted in neoplasia. PLoS Pathog. 2011, 7, e1002193. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Cullen, B.R. Ebv noncoding rnas. In Epstein–Barr Virus; Springer: Berlin, Germany, 2015; Volume 2, pp. 181–217. [Google Scholar]
- Edwards, R.H.; Marquitz, A.R.; Raab-Traub, N. Epstein–barr virus bart microRNAs are produced from a large intron prior to splicing. J. Virol. 2008, 82, 9094–9106. [Google Scholar] [CrossRef] [PubMed]
- Al-Mozaini, M.; Bodelon, G.; Karstegl, C.E.; Jin, B.; Al-Ahdal, M.; Farrell, P.J. Epstein–barr virus bart gene expression. J. Gen. Virol. 2009, 90, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Marquitz, A.R.; Mathur, A.; Edwards, R.H.; Raab-Traub, N. Host Gene Expression Is Regulated by Two Types of Noncoding RNAs Transcribed from the Epstein-Barr Virus BamHI A Rightward Transcript Region. J. Virol. 2015, 89, 11256–11268. [Google Scholar] [CrossRef] [PubMed]
- Poling, B.C.; Price, A.M.; Luftig, M.A.; Cullen, B.R. The epstein–barr virus MIR-BHRF1 microRNAs regulate viral gene expression in CIS. Virology 2017, 512, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Piccaluga, P.P.; Navari, M.; de Falco, G.; Ambrosio, M.R.; Lazzi, S.; Fuligni, F.; Bellan, C.; Rossi, M.; Sapienza, M.R.; Laginestra, M.A.; et al. Virus-encoded microrna contributes to the molecular profile of EBV-positive burkitt lymphomas. Oncotarget 2016, 7, 224–240. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, M.R.; Navari, M.; Di Lisio, L.; Leon, E.A.; Onnis, A.; Gazaneo, S.; Mundo, L.; Ulivieri, C.; Gomez, G.; Lazzi, S.; et al. The epstein–barr-encoded BART-6-3p microrna affects regulation of cell growth and immuno response in burkitt lymphoma. Infect. Agent Cancer 2014, 9, 1750–9378. [Google Scholar] [CrossRef] [PubMed]
- Oduor, C.I.; Movassagh, M.; Kaymaz, Y.; Chelimo, K.; Otieno, J.; Ong’echa, J.M.; Moormann, A.M.; Bailey, J.A. Human and epstein–barr virus mirna profiling as predictive biomarkers for endemic burkitt lymphoma. Front. Microbiol. 2017, 8, 501. [Google Scholar] [CrossRef] [PubMed]
- Hooykaas, M.J.; Kruse, E.; Wiertz, E.J.; Lebbink, R.J. Comprehensive profiling of functional epstein–barr virus mirna expression in human cell lines. BMC Genom. 2016, 17, 644. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Menegatti, J.; Motsch, N.; Hart, M.; Eichner, N.; Reinhardt, R.; Meister, G.; Grasser, F.A. miRNA expression profiling of epstein–barr virus-associated NKTL cell lines by illumina deep sequencing. FEBS Open Bio 2016, 6, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Fink, S.E.; Gandhi, M.K.; Nourse, J.P.; Keane, C.; Jones, K.; Crooks, P.; Johrens, K.; Korfel, A.; Schmidt, H.; Neumann, S.; et al. A comprehensive analysis of the cellular and EBV-specific micrornaome in primary CNS PTLD identifies different patterns among EBV-associated tumors. Am. J. Transplant. 2014, 14, 2577–2587. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Vella, S.; Miele, M.; Timoneri, F.; di Bella, M.; Bosi, S.; Sciveres, M.; Conaldi, P.G. Global profiling of viral and cellular non-coding RNAs in epstein–barr virus-induced lymphoblastoid cell lines and released exosome cargos. Cancer Lett. 2017, 388, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Hoshina, S.; Sekizuka, T.; Kataoka, M.; Hasegawa, H.; Hamada, H.; Kuroda, M.; Katano, H. Profile of exosomal and intracellular microrna in γ-herpesvirus-infected lymphoma cell lines. PLoS ONE 2016, 11, e0162574. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, R.; Fitzsimmons, L.; Thomas, W.A.; Kelly, G.L.; Rowe, M.; Bell, A.I. Quantitative studies of epstein–barr virus-encoded microRNAs provide novel insights into their regulation. J. Virol. 2011, 85, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Huang, J.; Wu, F.Y.; Liao, G.; Hutt-Fletcher, L.; Hayward, S.D. Regulation of expression of the epstein–barr virus bamhi-A rightward transcripts. J. Virol. 2005, 79, 1724–1733. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, R.J.A.; Tong, S.; Zhang, G.; Zong, J.; Chen, Y.; Jin, D.-Y.; Chen, M.-R.; Pan, J.; Chen, H. NF-κB signaling regulates expression of epstein–barr virus bart microRNAs and long noncoding RNAs in nasopharyngeal carcinoma. J. Virol. 2016, 90, 6475–6488. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.N.; Song, Y.-J.; Lee, S.K. The role of promoter methylation in epstein–barr virus (EBV) microrna expression in EBV-infected B cell lines. Exp. Mol. Med. 2011, 43, 401. [Google Scholar] [CrossRef] [PubMed]
- Haar, J.; Contrant, M.; Bernhardt, K.; Feederle, R.; Diederichs, S.; Pfeffer, S.; Delecluse, H.-J. The expression of a viral microrna is regulated by clustering to allow optimal B cell transformation. Nucleic Acids Res. 2015, 44, 1326–1341. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.H.; Lin, X.; Shumilov, A.; Bernhardt, K.; Feederle, R.; Poirey, R.; Kopp-Schneider, A.; Pereira, B.; Almeida, R.; Delecluse, H.J. The biological properties of different epstein–barr virus strains explain their association with various types of cancers. Oncotarget 2017, 8, 10238–10254. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Palser, A.; Elgueta Karstegl, C.; Middeldorp, J.M.; Ramayanti, O.; Cohen, J.I.; Hildesheim, A.; Fellner, M.D.; Wiels, J.; White, R.E.; et al. Natural variation of epstein–barr virus genes, proteins, and primary microRNA. J. Virol. 2017, 91, e00375-17. [Google Scholar] [CrossRef] [PubMed]
- De Paschale, M.; Clerici, P. Serological diagnosis of epstein–barr virus infection: Problems and solutions. World J. Virol. 2012, 1, 31. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Grogan, E.; Rowe, D.; Rooney, C.; Heston, L.; Eastman, R.; Andiman, W.; Niederman, J.; Lenoir, G.; Henle, W.; et al. Selective lack of antibody to a component of EB nuclear antigen in patients with chronic active epstein–barr virus infection. J. Infect. Dis. 1987, 156, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Vetter, V.; Kreutzer, L.; Bauer, G. Differentiation of primary from secondary anti-EBNA-1-negative cases by determination of avidity of VCA-IGG. Clin. Diagn. Virol. 1994, 2, 29–39. [Google Scholar] [CrossRef]
- Hjalgrim, H.; Friborg, J.; Melbye, M. The Epidemiology of EBV and Its Association with Malignant Disease; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Martin, J.N. The Epidemiology of Kshv and Its Association with Malignant Disease; University of California: San Francisco, CA, USA, 2007. [Google Scholar]
- Sunagawa, K.; Hishima, T.; Fukumoto, H.; Hasegawa, H.; Katano, H. Conserved sequences of bart and bhrf regions encoding viral microRNAs in epstein–barr virus-associated lymphoma. BMC Res. Notes 2017, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Ressing, M.E.; van Gent, M.; Gram, A.M.; Hooykaas, M.J.; Piersma, S.J.; Wiertz, E.J. Immune evasion by epstein–barr virus. In Epstein–Barr Virus; Springer: Berlin, Germany, 2015; Volume 2, pp. 355–381. [Google Scholar]
- Apcher, S.; Daskalogianni, C.; Manoury, B.; Fåhraeus, R. Epstein–barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mrna translation initiation and antigen presentation. PLoS Pathog. 2010, 6, e1001151. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Qin, Z.; Wang, J.; Zheng, X.; Lu, J.; Zhang, X.; Wei, L.; Peng, Q.; Zheng, Y.; Ou, C.; et al. Epstein–barr virus miR-BART6-3p inhibits the RIG-I pathway. J. Innate Immun. 2017, 9, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.Y.; Ichinohe, T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015, 23, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Haneklaus, M.; Gerlic, M.; Kurowska-Stolarska, M.; Rainey, A.A.; Pich, D.; McInnes, I.B.; Hammerschmidt, W.; O’Neill, L.A.; Masters, S.L. Cutting edge: MiR-223 and EBV miR-bart15 regulate the NLRP3 inflammasome and IL-1β production. J. Immunol. 2012, 189, 3795–3799. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Yu, Y.; Zhao, H.P. EBVbart63p and cellular microRNA197 compromise the immune defense of host cells in ebvpositive burkitt lymphoma. Mol. Med. Rep. 2017, 15, 1877–1883. [Google Scholar] [CrossRef] [PubMed]
- Vereide, D.T.; Seto, E.; Chiu, Y.F.; Hayes, M.; Tagawa, T.; Grundhoff, A.; Hammerschmidt, W.; Sugden, B. Epstein–barr virus maintains lymphomas via its mirnas. Oncogene 2014, 33, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.V.; Wade, C.M.; Kang, H.M.; Alper, S.; Rutledge, H.; Lackford, B.; Eskin, E.; Daly, M.J.; Schwartz, D.A. Identification of novel genes that mediate innate immunity using inbred mice. Genetics 2009, 183, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Dolken, L.; Malterer, G.; Erhard, F.; Kothe, S.; Friedel, C.C.; Suffert, G.; Marcinowski, L.; Motsch, N.; Barth, S.; Beitzinger, M.; et al. Systematic analysis of viral and cellular microrna targets in cells latently infected with human gamma-herpesviruses by risc immunoprecipitation assay. Cell Host Microbe 2010, 7, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Skinner, C.M.; Ivanov, N.S.; Barr, S.A.; Chen, Y.; Skalsky, R.L. An epstein–barr virus microRNA blocks interleukin-1 (IL-1) signaling by targeting IL-1 receptor 1. J. Virol. 2017, 91, e00530-17. [Google Scholar] [CrossRef] [PubMed]
- Hooykaas, M.J.G.; van Gent, M.; Soppe, J.A.; Kruse, E.; Boer, I.G.J.; van Leenen, D.; Groot Koerkamp, M.J.A.; Holstege, F.C.P.; Ressing, M.E.; Wiertz, E.; et al. EBV microrna BART16 suppresses type I ifn signaling. J. Immunol. 2017, 198, 4062–4073. [Google Scholar] [CrossRef] [PubMed]
- Albanese, M.; Tagawa, T.; Bouvet, M.; Maliqi, L.; Lutter, D.; Hoser, J.; Hastreiter, M.; Hayes, M.; Sugden, B.; Martin, L.; et al. Epstein–barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA 2016, 113, E6467–E6475. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, T.; Albanese, M.; Bouvet, M.; Moosmann, A.; Mautner, J.; Heissmeyer, V.; Zielinski, C.; Lutter, D.; Hoser, J.; Hastreiter, M.; et al. Epstein–barr viral mirnas inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J. Exp. Med. 2016, 213, 2065–2080. [Google Scholar] [CrossRef] [PubMed]
- Feederle, R.; Linnstaedt, S.D.; Bannert, H.; Lips, H.; Bencun, M.; Cullen, B.R.; Delecluse, H.-J. A viral microrna cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011, 7, e1001294. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.; Pfuhl, T.; Mamiani, A.; Ehses, C.; Roemer, K.; Kremmer, E.; Jäker, C.; Höck, J.; Meister, G.; Grässer, F.A. Epstein–barr virus-encoded microrna miR-bart2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2007, 36, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Moosmann, A.; Grömminger, S.; Walz, N.; Grundhoff, A.; Hammerschmidt, W. MicroRNAs of epstein–barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010, 6, e1001063. [Google Scholar] [CrossRef] [PubMed]
- Ning, S. Innate immune modulation in EBV infection. Herpesviridae 2011, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Everett, H.; McFadden, G. Viruses and apoptosis: Meddling with mitochondria. Virology 2001, 288, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Harold, C.; Cox, D.; Riley, K.J. Epstein–barr viral microRNAs target caspase 3. Virol. J. 2016, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Nie, K.; Redmond, D.; Liu, Y.; Elemento, O.; Knowles, D.M.; Tam, W. EBV-miR-BHRF1-2 targets PRDM1/BLIMP1: Potential role in EBV lymphomagenesis. Leukemia 2016, 30, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Bu, Y.; Liang, Y.; Zhang, F.; Zhang, H.; Li, S. Epstein–barr virus (EBV)-bamhi-a rightward transcript (bart)-6 and cellular microRNA-142 synergistically compromise immune defense of host cells in EBV-positive burkitt lymphoma. Med. Sci. Monit. 2016, 22, 4114–4120. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, K.; Haar, J.; Tsai, M.H.; Poirey, R.; Feederle, R.; Delecluse, H.J. A viral microRNA cluster regulates the expression of PTEN, p27 and of a BCL-2 homolog. PLoS Pathog. 2016, 12, e1005405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Duy Le, T.; Liu, L.; He, J.; Li, J. Identifying mirna synergistic regulatory networks in heterogeneous human data via network motifs. Mol. Biosyst. 2016, 12, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Garg, N.; Bigi, R.; Yadav, S.; Campese, A.F.; Lapenta, C.; Spada, M.; Cuomo, L.; Botta, A.; Belardelli, F.; et al. Epstein–barr virus infection induces miR-21 in terminally differentiated malignant B cells. Int. J. Cancer 2015, 137, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Riley, K.J.; Rabinowitz, G.S.; Yario, T.A.; Luna, J.M.; Darnell, R.B.; Steitz, J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012, 31, 2207–2221. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Huang, W.T.; Yang, L.W.; Lin, C.W. The PTEN-AKT-MTOR/RICTOR pathway in nasal natural killer cell lymphoma is activated by miR-494-3p via PTEN but inhibited by miR-142-3p via rictor. Am. J. Pathol. 2015, 185, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Callegari, S.; Gastaldello, S.; Faridani, O.R.; Masucci, M.G. Epstein–barr virus encoded microRNAs target sumo-regulated cellular functions. FEBS J. 2014, 281, 4935–4950. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.; Linnstaedt, S.D.; Esoda, C.; Krisko, J.F.; Martinez-Torres, F.; Delecluse, H.J.; Cullen, B.R.; Garcia, J.V. A cluster of virus-encoded microRNAs accelerates acute systemic epstein–barr virus infection but does not significantly enhance virus-induced oncogenesis in vivo. J. Virol. 2013, 87, 5437–5446. [Google Scholar] [CrossRef] [PubMed]
- Teow, S.Y.; Liew, K.; Khoo, A.S.; Peh, S.C. Pathogenic role of exosomes in epstein–barr virus (EBV)-associated cancers. Int. J. Biol. Sci. 2017, 13, 1276–1286. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, W.; Xiao, J.; Cao, B. The role of exosomes and “exosomal shuttle microrna” in tumorigenesis and drug resistance. Cancer Lett. 2015, 356, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H. Clinical relevance of circulating, cell-free and exosomal microRNAs in plasma and serum of breast cancer patients. Oncol. Res. Treat. 2017, 40, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Rechavi, O.; Erlich, Y.; Amram, H.; Flomenblit, L.; Karginov, F.V.; Goldstein, I.; Hannon, G.J.; Kloog, Y. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small rnas. Genes Dev. 2009, 23, 1971–1979. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Wurdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Kawanishi, E.; Yoshida, R.; Yoshiyama, H. Exosomes derived from epstein–barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J. Virol. 2013, 87, 10334–10347. [Google Scholar] [CrossRef] [PubMed]
- Yogev, O.; Henderson, S.; Hayes, M.J.; Marelli, S.S.; Ofir-Birin, Y.; Regev-Rudzki, N.; Herrero, J.; Enver, T. Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. PLoS Pathog. 2017, 13, e1006524. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Faruq, O.; Vecchione, A. MicroRNA: Diagnostic perspective. Front. Med. 2015, 2, 51. [Google Scholar] [CrossRef] [PubMed]
- De Souza, M.F.; Kuasne, H.; de Camargo Barros-Filho, M.; Cilião, H.L.; Marchi, F.A.; Fuganti, P.E.; Paschoal, A.R.; Rogatto, S.R.; de Syllos Cólus, I.M. Circulating mrnas and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef] [PubMed]
- Palser, A.L.; Grayson, N.E.; White, R.E.; Corton, C.; Correia, S.; Ba Abdullah, M.M.; Watson, S.J.; Cotten, M.; Arrand, J.R.; Murray, P.G.; et al. Genome diversity of epstein–barr virus from multiple tumor types and normal infection. J. Virol. 2015, 89, 5222–5237. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E.R.; Kara, M.; Coleman, C.B.; Grau, K.R.; Oko, L.M.; Krueger, B.J.; Renne, R.; van Dyk, L.F.; Tibbetts, S.A. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. MBio 2014, 5, e00981-14. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.K.; Currie, M.J.; Robinson, B.A.; Morrin, H.; Phung, Y.; Pearson, J.F.; Anderson, T.P.; Potter, J.D.; Walker, L.C. Cytomegalovirus and epstein–barr virus in breast cancer. PLoS ONE 2015, 10, e0118989. [Google Scholar] [CrossRef] [PubMed]
- Pai, T.; Gupta, S.; Gurav, M.; Nag, S.; Shet, T.; Patil, A.; Desai, S. Evidence for the association of epstein–barr virus in breast cancer in indian patients using in-situ hybridization technique. Breast J. 2018, 24, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M. Smooth muscle tumors of soft tissue and non-uterine viscera: Biology and prognosis. Mod. Pathol. 2014, 27, S17–S29. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.W.; Choi, Y.; Kwon, O.K.; Lee, S.S.; Chung, H.Y.; Yu, W.; Bae, H.I.; Seo, A.N.; Kang, H.; Lee, S.K.; et al. High level of viral microRNA-BART20-5p expression is associated with worse survival of patients with epstein–barr virus-associated gastric cancer. Oncotarget 2017, 8, 14988–14994. [Google Scholar] [CrossRef] [PubMed]
- Treece, A.L.; Duncan, D.L.; Tang, W.; Elmore, S.; Morgan, D.R.; Dominguez, R.L.; Speck, O.; Meyers, M.O.; Gulley, M.L. Gastric adenocarcinoma microrna profiles in fixed tissue and in plasma reveal cancer-associated and epstein–barr virus-related expression patterns. Lab. Investig. 2016, 96, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Li, Z.H.; Chen, S.; Chan, J.Y.; Yin, M.; Zhang, M.J.; Wong, T.S. Epstein–barr virus encoded microRNA BART7 regulates radiation sensitivity of nasopharyngeal carcinoma. Oncotarget 2017, 8, 20297–20308. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chan, J.Y.; Wong, S.T.; Wei, W.I. The role of epstein–barr virus-encoded microRNA BART7 status of resection margins in the prediction of local recurrence after salvage nasopharyngectomy for recurrent nasopharyngeal carcinoma. Cancer 2015, 121, 2358–2366. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.Y.; Yi, Y.H.; Chang, K.P.; Chang, Y.S.; Chen, S.J.; Chen, H.C. The epstein–barr virus-encoded microrna miR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PLoS Pathog. 2014, 10, e1003974. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; He, D.D.; Liang, H.W.; Yang, D.; Yue, H.; Zhang, X.M.; Wang, R.; Li, B.; Yang, H.X.; Liu, Y.; et al. The identification of up-regulated EBV-miR-BHRF1-2-5p targeting MALT1 and EBV-MIR-BHRF1-3 in the circulation of patients with multiple sclerosis. Clin. Exp. Immunol. 2017, 189, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Menendez, S.; Fernandez-Moran, M.; Fernandez-Vega, I.; Perez-Alvarez, A.; Villafani-Echazu, J. Epstein–barr virus and multiple sclerosis. From evidence to therapeutic strategies. J. Neurol. Sci. 2016, 361, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Jang, S.-I.; Ong, H.L.; Perez, P.; Tandon, M.; Ambudkar, I.; Illei, G.; Alevizos, I. Targeting the Ca2+ sensor STIM1 by exosomal transfer of EBV-miR-BART13-3p is associated with sjögren’s syndrome. EBioMedicine 2016, 10, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Dasari, V.; Bhatt, K.H.; Smith, C.; Khanna, R. Designing an effective vaccine to prevent epstein–barr virus-associated diseases: Challenges and opportunities. Expert Rev. Vac. 2017, 16, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Tensen, C.P.; Vermeer, M.H. Microrna-155 potentiates tumour development in mycosis fungoides. Br. J. Dermatol. 2017, 177, 618–620. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Li, J.; Zhang, X.; Lu, Y.; Wang, J.; Lyu, X.; Chen, Y.; Liu, J.; Cai, H.; Wang, Y.; et al. Gold nano-particles (AUNPS) carrying anti-EBV-miR-BART7-3p inhibit growth of EBV-positive nasopharyngeal carcinoma. Oncotarget 2015, 6, 7838–7850. [Google Scholar] [CrossRef] [PubMed]
| Latency Type | Expressed Product | Example | Reference |
|---|---|---|---|
| Type I | EBNA1, EBERs, BART miRNAs | BL, MTX-LPD, PBL | [9,23,24] |
| Type II | EBNA1, LMP1, LMP2s, EBERs, BART miRNAs | HL, PBL, MTX-LPD, NKTL, CLL, PTLD, DLBCL of the elderly | [6,23,24,25,26,27,28] |
| Type III | EBNA1, EBNA2, EBNA3s, LMP1, LMP2s, EBERs, BART miRNAs (non-to-low), BHRF1 | PTLD, DLBCL of elderly, PAL, HIV-DLBCL, LCL | [3,8,28] |
| MicroRNA | Target | Related Process/Application | Reference |
|---|---|---|---|
| BART and BHRF1 Families | EBNA1 | Latency Regulation | [77] |
| BHRF1 Family | PTEN | Cell Proliferation, Apoptosis | [87] |
| p27 | Cell Cycle Progression | [87] | |
| Unidentified | Transformation Capacity, Cell Cycle Progression | [79] | |
| Unidentified | Acute Systemic EBV Infection | [94] | |
| Latent Proteins | Latency Regulation | [79] | |
| miR-BHRF1-1 | Not Applicable | Survival Marker in CLL | [10] |
| miR-BHRF1-2 | CTSB | CD4+ T Cell Response | [77,78] |
| PRDM1 | Cell Cycle Progression, Apoptosis | [85] | |
| IL-12B | CD4+ T Cell Response | [77,78] | |
| miR-BHRF1-2-5p | IL-1 receptor | Innate Immunity | [75] |
| miR-BHRF1-3 | TAP2 | CD8+ T Cell Response | [77,78] |
| miR-BART1-3p | IL-12B | CD4+ T Cell Response | [77,78] |
| IFI30 | CD4+ T Cell Response | [77,78] | |
| CASP3 | Apoptosis | [72] | |
| miR-BART1-5p | IFI30 | CD4+ T-Cell Response | [77,78] |
| CASP3 | Apoptosis | [84] | |
| Not Applicable | Diagnostic Marker for NKTL | [11] | |
| miR-BART2-5p | CTSB | CD4+ T-Cell Response | [77,78] |
| LGMN | CD4+ T-Cell Response | [77,78] | |
| IL-12B | CD4+ T-Cell Response | [77,78] | |
| CASP3 | Apoptosis | [84] | |
| BALF5 | Latency Regulation | [80] | |
| Not Applicable | Diagnostic Marker for NKTL | ||
| miR-BART3-3p | IPO7 | Innate Immunity | [72,74] |
| CASP3 | Apoptosis | [84] | |
| miR-BART4-5p | CASP3 | Apoptosis | [84] |
| miR-BART6-3p | PTEN * | Cell Proliferation, Apoptosis | [86] |
| IL-6RB ** | Innate Immunity | [71] | |
| RIG-I | Innate Immunity | [68] | |
| miR-BART7-3p | CASP3 | Apoptosis | [84] |
| Not Applicable | Diagnostic Marker for NKTL | [11] | |
| miR-BART8-5p | CASP3 | Apoptosis | [84] |
| miR-BART10-3p | IL-12B | CD4+ T Cell Response | [77,78] |
| miR-BART13-3p | CASP3 | Apoptosis | [84] |
| Not Applicable | Diagnostic Marker for NKTL | [11] | |
| miR-BART15 | NLRP3 | Innate Immunity | [70] |
| miR-BART16 | S1PR1 | Cell Growth/Mobility *** | [48] |
| CREBBP | Innate Immunity | [76] | |
| IPO7 | Innate Immunity | [72] | |
| CASP3 | Apoptosis | [72] | |
| TOMM22 | Apoptosis | [74] | |
| miR-BART17 | TAP2 | CD8+ T-Cell Response | [77] |
| miR-BART20-5p | T-bet **** | Transcription Regulation of Cytotoxic NK Cells | [92] |
| miR-BART22 | CASP3 | Apoptosis | [84] |
| IL-12B | CD4+ T Cell Response | [77,78] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navari, M.; Etebari, M.; Ibrahimi, M.; Leoncini, L.; Piccaluga, P.P. Pathobiologic Roles of Epstein–Barr Virus-Encoded MicroRNAs in Human Lymphomas. Int. J. Mol. Sci. 2018, 19, 1168. https://doi.org/10.3390/ijms19041168
Navari M, Etebari M, Ibrahimi M, Leoncini L, Piccaluga PP. Pathobiologic Roles of Epstein–Barr Virus-Encoded MicroRNAs in Human Lymphomas. International Journal of Molecular Sciences. 2018; 19(4):1168. https://doi.org/10.3390/ijms19041168
Chicago/Turabian StyleNavari, Mohsen, Maryam Etebari, Mostafa Ibrahimi, Lorenzo Leoncini, and Pier Paolo Piccaluga. 2018. "Pathobiologic Roles of Epstein–Barr Virus-Encoded MicroRNAs in Human Lymphomas" International Journal of Molecular Sciences 19, no. 4: 1168. https://doi.org/10.3390/ijms19041168
APA StyleNavari, M., Etebari, M., Ibrahimi, M., Leoncini, L., & Piccaluga, P. P. (2018). Pathobiologic Roles of Epstein–Barr Virus-Encoded MicroRNAs in Human Lymphomas. International Journal of Molecular Sciences, 19(4), 1168. https://doi.org/10.3390/ijms19041168






