Atomic Force Microscopy for Protein Detection and Their Physicoсhemical Characterization
Abstract
1. Introduction
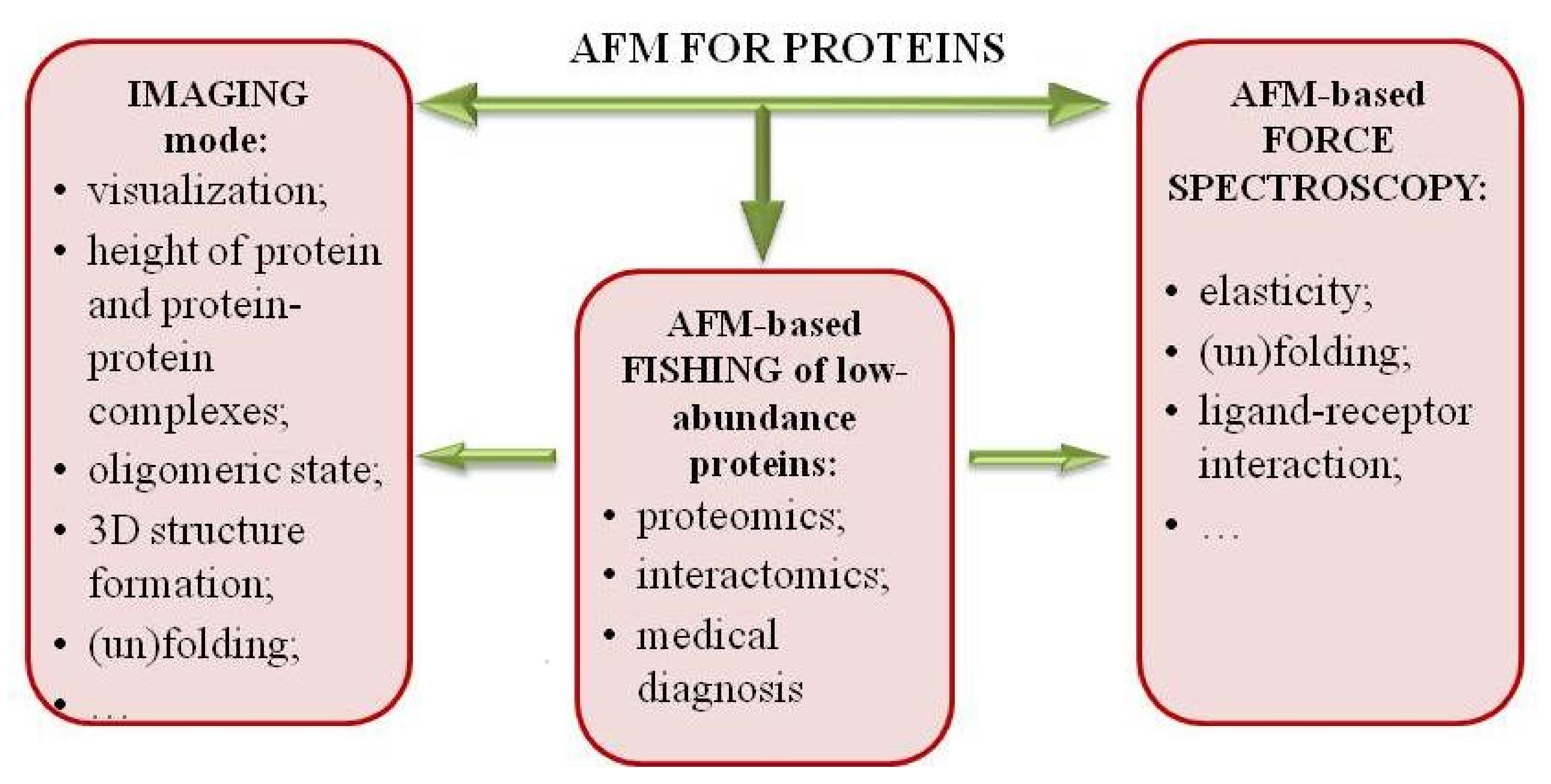
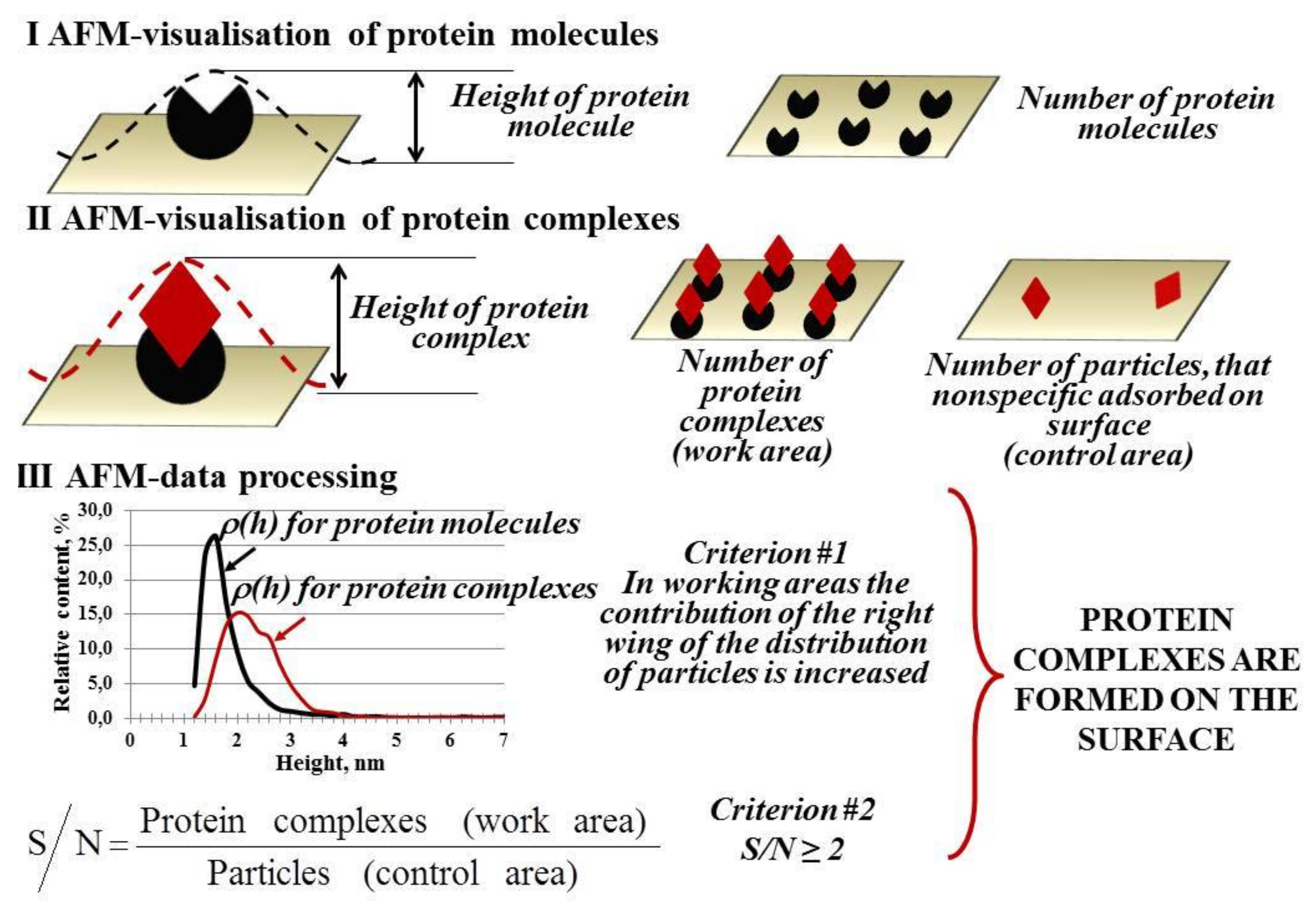
2. Atomic Force Microscopy (AFM) Visualization of Proteins
3. Force Spectroscopy Mode
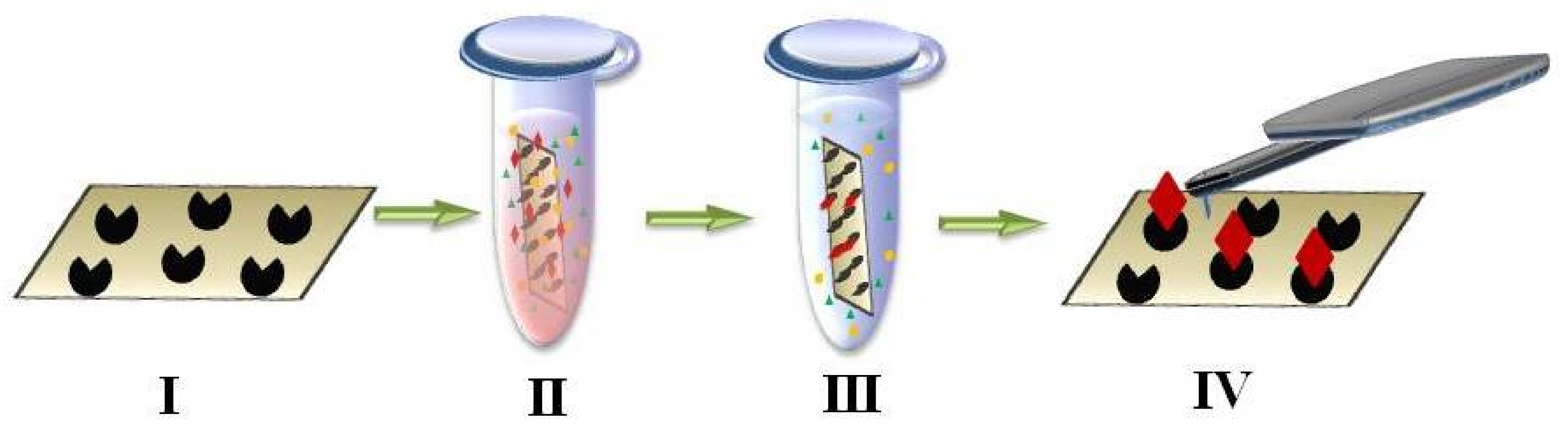
4. AFM-Based Molecular Detector of Low-Abundance Proteins
- (1)
- fishing of biomolecules from a big volume of biological fluid onto a small surface (concentrating instead of conventional removal of high-abundance proteins by chromatography and electrophoresis used for protein matrix separation);
- (2)
- high-sensitivity detection of caught molecules using AFM-based molecular detector (registering and counting the single molecules and molecular complexes).
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hinterdorfer, P.; Oijen, A. Handbook of Single-Molecule Biophysics. CERN Doc. Serv. 2009. [Google Scholar] [CrossRef]
- Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Application of atomic force microscopy for characteristics of single intermolecular interactions. Biochem. Mosc. 2012, 77, 1536–1552. [Google Scholar] [CrossRef] [PubMed]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.L. Force spectroscopy unveils hidden protein-folding states. Phys. Today 2017, 70, 16–18. [Google Scholar] [CrossRef]
- Hoh, J.H.; Schoenenberger, C.A. Surface morphology and mechanical properties of MDCK monolayers by atomic force microscopy. J. Cell Sci. 1994, 107, 1105–1114. [Google Scholar] [PubMed]
- Henderson, E.; Haydon, P.G.; Sakaguchi, D.S. Actin filament dynamics in living glial cells imaged by atomic force microscopy. Science 1992, 257, 1944–1946. [Google Scholar] [CrossRef] [PubMed]
- Hoh, J.H.; Lal, R.; John, S.A.; Revel, J.P.; Arnsdorf, M.F. Atomic force microscopy and dissection of gap junctions. Science 1991, 253, 1405–1408. [Google Scholar] [CrossRef] [PubMed]
- Mou, J.; Yang, J.; Shao, Z. Atomic Force Microscopy of Cholera Toxin B-oligomers Bound to Bilayers of Biologically Relevant Lipids. J. Mol. Biol. 1995, 248, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Hansma, H.G.; Vesenka, J.; Siegerist, C.; Kelderman, G.; Morrett, H.; Sinsheimer, R.L.; Elings, V.; Bustamante, C.; Hansma, P.K. Reproducible imaging and dissection of plasmid DNA under liquid with the atomic force microscope. Science 1992, 256, 1180–1184. [Google Scholar] [CrossRef] [PubMed]
- Limanskaya, O.Y.; Limanskii, A.P. Imaging compaction of single supercoiled DNA molecules by atomic force microscopy. Gen. Physiol. Biophys. 2008, 27, 322–337. [Google Scholar] [PubMed]
- Kwak, K.J.; Kudo, H.; Fujihira, M. Imaging stretched single DNA molecules by pulsed-force-mode atomic force microscopy. Ultramicroscopy 2003, 97, 249–255. [Google Scholar] [CrossRef]
- Egger, M.; Ohnesorge, F.; Weisenhorn, A.L.; Heyn, S.P.; Drake, B.; Prater, C.B.; Gould, S.A.C.; Hansma, P.K.; Gaub, H.E. Wet lipid-protein membranes imaged at submolecular resolution by atomic force microscopy. J. Struct. Biol. 1990, 103, 89–94. [Google Scholar] [CrossRef]
- Hansma, H.G.; Gould, S.A.C.; Hansma, P.K.; Gaub, H.E.; Longo, M.L.; Zasadzinski, J.A.N. Imaging nanometer scale defects in Langmuir-Blodgett films with the atomic force microscope. Langmuir 1991, 7, 1051–1054. [Google Scholar] [CrossRef]
- Chi, L.F.; Anders, M.; Fuchs, H.; Johnston, R.R.; Ringsdorf, H. Domain Structures in Langmuir-Blodgett Films Investigated by Atomic Force Microscopy. Science 1993, 259, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Mou, J.; Shao, Z. Structure and stability of pertussis toxin studied by in situ atomic force microscopy. FEBS Lett. 1994, 338, 89–92. [Google Scholar] [CrossRef]
- Müller, D.J.; Fotiadis, D.; Scheuring, S.; Müller, S.A.; Engel, A. Electrostatically Balanced Subnanometer Imaging of Biological Specimens by Atomic Force Microscope. Biophys. J. 1999, 76, 1101–1111. [Google Scholar] [CrossRef]
- Schabert, F.A.; Henn, C.; Engel, A. Native Escherichia coli OmpF porin surfaces probed by atomic force microscopy. Science 1995, 268, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.P. Imaging proteins with atomic force microscopy: An overview. Curr. Protein Pept. Sci. 2005, 6, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Kada, G.; Kienberger, F.; Hinterdorfer, P. Atomic force microscopy in bionanotechnology. Nano Today 2008, 3, 12–19. [Google Scholar] [CrossRef]
- Müller, D.J.; Engel, A.; Carrascosa, J.L.; Vélez, M. The bacteriophage φ29 head–tail connector imaged at high resolution with the atomic force microscope in buffer solution. EMBO J. 1997, 16, 2547–2553. [Google Scholar] [CrossRef] [PubMed]
- Bykov, I.V.; Bykov, V.A. Attraction and repulsion regime of semicontact mode atomic force microscopy. Automatized methods for optimization of operation in attraction regime. Izvestiya Vysshikh Uchebnykh Zavedenii. Materialy Elektronnoi Tekhniki 2008, 1, 75–77. [Google Scholar]
- Engel, A.; Müller, D.J. Observing single biomolecules at work with the atomic force microscope. Nat. Struct. Mol. Biol. 2000, 7, 715. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T.R.; Grütter, P.; Horne, D.; Rugar, D. Frequency modulation detection using high-Q cantilevers for enhanced force microscope sensitivity. J. Appl. Phys. 1991, 69, 668–673. [Google Scholar] [CrossRef]
- Putman, C.A.J.; van der Werf, K.O.; de Grooth, B.G.; Van Hulst, N.F.; Greve, J. Tapping mode atomic force microscopy in liquid. Appl. Phys. Lett. 1994, 64, 2454–2456. [Google Scholar] [CrossRef]
- Garcia, R.; Herruzo, E.T. The emergence of multifrequency force microscopy. Nat. Nanotechnol. 2012, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Hansma, P.K.; Cleveland, J.P.; Radmacher, M.; Walters, D.A.; Hillner, P.E.; Bezanilla, M.; Fritz, M.; Vie, D.; Hansma, H.G.; Prater, C.B.; et al. Tapping mode atomic force microscopy in liquids. Appl. Phys. Lett. 1994, 64, 1738–1740. [Google Scholar] [CrossRef]
- Ido, S.; Kimura, K.; Oyabu, N.; Kobayashi, K.; Tsukada, M.; Matsushige, K.; Yamada, H. Beyond the Helix Pitch: Direct Visualization of Native DNA in Aqueous Solution. ACS Nano 2013, 7, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Ido, S.; Kimiya, H.; Kobayashi, K.; Kominami, H.; Matsushige, K.; Yamada, H. Immunoactive two-dimensional self-assembly of monoclonal antibodies in aqueous solution revealed by atomic force microscopy. Nat. Mater. 2014, 13, 264. [Google Scholar] [CrossRef] [PubMed]
- Kienberger, F.; Stroh, C.; Kada, G.; Moser, R.; Baumgartner, W.; Pastushenko, V.; Rankl, C.; Schmidt, U.; Müller, H.; Orlova, E.; LeGrimellec, C. Dynamic force microscopy imaging of native membranes. Ultramicroscopy 2003, 97, 229–237. [Google Scholar] [CrossRef]
- Scheuring, S.; Seguin, J.; Marco, S.; Levy, D.; Breyton, C.; Robert, B.; Rigaud, J.L. AFM characterization of tilt and intrinsic flexibility of rhodobacter sphaeroides light harvesting complex 2 (LH2). J. Mol. Biol. 2003, 325, 569–580. [Google Scholar] [CrossRef]
- Radmacher, M.; Fritz, M.; Cleveland, J.P.; Walters, D.A.; Hansma, P.K. Imaging adhesion forces and elasticity of lysozyme adsorbed on mica with the atomic force microscope. Langmuir 1994, 10, 3809–3814. [Google Scholar] [CrossRef]
- Alonso, J.L.; Goldmann, W.H. Feeling the forces: Atomic force microscopy in cell biology. Life Sci. 2003, 72, 2553–2560. [Google Scholar] [CrossRef]
- Fotiadis, D.; Scheuring, S.; Müller, S.A.; Engel, A.; Müller, D.J. Imaging and manipulation of biological structures with the AFM. Micron 2002, 33, 385–397. [Google Scholar] [CrossRef]
- Zlatanova, J.; Lindsay, S.M.; Leuba, S.H. Single molecule force spectroscopy in biology using the atomic force microscope. Prog. Biophys. Mol. Biol. 2000, 74, 37–61. [Google Scholar] [CrossRef]
- Stolz, M.; Stoffler, D.; Aebi, U.; Goldsbury, C. Monitoring biomolecular interactions by time-lapse atomic force microscopy. J. Struct. Biol. 2000, 131, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Smith, S.B.; Liphardt, J.; Smith, D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000, 10, 279–285. [Google Scholar] [CrossRef]
- Rief, M.; Oesterhelt, F.; Heymann, B.; Gaub, H.E. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science 1997, 275, 1295–1297. [Google Scholar] [CrossRef] [PubMed]
- Vengasandra, S.; Sethumadhavan, G.; Yan, F.; Wang, R. Studies on the protein−receptor interaction by atomic force microscopy. Langmuir 2003, 19, 10940–10946. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.Y. Effect of protein–protein interactions on protein aggregation kinetics. J. Chem. Phys. 2003, 119, 10972–10976. [Google Scholar] [CrossRef]
- Thomson, N.H.; Fritz, M.; Radmacher, M.; Cleveland, J.P.; Schmidt, C.F.; Hansma, P.K. Protein tracking and detection of protein motion using atomic force microscopy. Biophys. J. 1996, 70, 2421–2431. [Google Scholar] [CrossRef]
- Kiselyova, O.I.; Yaminsky, I.V. Atomic force microscopy of protein complexes. In Atomic Force Microscopy; Humana Press: New York, NY, USA, 2004; pp. 217–230. [Google Scholar]
- Silva, L.P. Atomic force microscopy investigation of ribonuclease A. Protein Pept. Lett. 2001, 8, 343–347. [Google Scholar] [CrossRef]
- Wagner, P. Immobilization strategies for biological scanning probe microscopy 1. FEBS Lett. 1998, 430, 112–115. [Google Scholar] [CrossRef]
- Müller, D.J.; Amrein, M.; Engel, A. Adsorption of biological molecules to a solid support for scanning probe microscopy. J. Struct. Biol. 1997, 119, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Hafner, J.H.; Cheung, C.-L.; Woolley, A.T.; Lieber, C.M. Structural and functional imaging with carbon nanotube AFM probes. Prog. Biophys. Mol. Biol. 2001, 77, 73–110. [Google Scholar] [CrossRef]
- Alessandrini, A.; Facci, P. AFM: A versatile tool in biophysics. Meas. Sci. Technol. 2005, 16, R65. [Google Scholar] [CrossRef]
- Schneider, S.W.; Lärmer, J.; Henderson, R.M.; Oberleithner, H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflüg. Arch. 1998, 435, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Thomson, N.H. Imaging the substructure of antibodies with tapping-mode AFM in air: The importance of a water layer on mica. J. Microsc. 2005, 217, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Ramos, J.; Perrino, A.P.; Garcia, R. Dependence of the volume of an antibody on the force applied in a force microscopy experiment in liquid. Ultramicroscopy 2016, 171, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Barinov, N.; Ivanov, N.; Kopylov, A.; Klinov, D.; Zavyalova, E. Direct visualization of the oligomeric state of hemagglutinins of influenza virus by high-resolution atomic force microscopy. Biochimie 2018, 146, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, Y.D.; Frantsuzov, P.A.; Pleshakova, T.O.; Ziborov, V.S.; Svetlov, S.K.; Krokhin, N.V.; Konev, V.A.; Kovalev, O.B.; Uchaĭkin, V.F.; Iastrebova, O.N.; et al. Atomic force microscopy detection of serological markers of viral hepatites B and C. Biochem. Mosc. Suppl. Ser. B Biomed. Chem. 2010, 4, 117–122. [Google Scholar] [CrossRef]
- Archakov, A.I.; Bachmanova, G.I. Cytochrome P-450 and active oxygen. Trends Biochem. Sci. 1990. [Google Scholar] [CrossRef]
- Neeli, R.; Girvan, H.M.; Lawrence, A.; Warren, M.J.; Leys, D.; Scrutton, N.S.; Munro, A.W. The dimeric form of flavocytochrome P450 BM3 is catalytically functional as a fatty acid hydroxylase. FEBS Lett. 2005, 579, 5582–5588. [Google Scholar] [CrossRef] [PubMed]
- Narhi, L.O.; Fulco, A.J. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 1986, 261, 7160–7169. [Google Scholar] [PubMed]
- Bernhardt, R. Cytochrome P450: Structure, function, and generation of reactive oxygen species. In Reviews of Physiology Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 1995; Volume 127, pp. 137–221. [Google Scholar]
- De Montellano, P.R.O. Cytochrome P450: Structure, Mechanism, and Biochemistry; Springer Science & Business Media: Berlin, Germany, 2005. [Google Scholar]
- Lewis, D.F.V. Guide to Cytochromes: Structure and Function; CRC Press: Boca Raton, FL, USA, 1996. [Google Scholar]
- Archakov, A.I.; Ivanov, Y.D. Application of AFM and optical biosensor for investigation of complexes formed in P450-containing monooxygenase systems. Biochim. Biophys. Acta Proteins Proteom. 2011, 1814, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, Y.D.; Bukharina, N.S.; Frantsuzov, P.A.; Pleshakova, T.O.; Kanashenko, S.L.; Medvedeva, N.V.; Argentova, V.V.; Zgoda, V.G.; Munro, A.W.; Archakov, A.I. AFM study of cytochrome CYP102A1 oligomeric state. Soft Matter 2012, 8, 4602–4608. [Google Scholar] [CrossRef]
- Bukharina, N.S.; Pleshakova, T.O.; Ziborov, V.S.; Fokin, D.A.; Ivanova, N.D.; Ivanov, Y.D. Atomic force microscopy analysis of deformation of single cytochrome CYP102A1 molecules. Sci. Adv. Mater. 2017, 9, 135–143. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Bukharina, N.S.; Frantsuzov, P.A.; Pleshakova, T.O.; Krohin, N.V.; Kanashenko, S.L.; Archakov, A.I. Oligomeric state investigation of flavocytochrome CYP102A1 using AFM with standard and supersharp probes. Biochem. Mosc. Suppl. Ser. B Biomed. Chem. 2012, 6, 218–224. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Bukharina, N.S.; Pleshakova, T.O.; Frantsuzov, P.A.; Krokhin, N.V.; Ziborov, V.S.; Archakov, A.I. Atomic force microscopy visualization and measurement of the activity and physicochemical properties of single monomeric and oligomeric enzymes. Biophysics 2011, 56, 892–896. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Frantsuzov, P.A.; Bykov, V.A.; Besedin, S.P.; Hoa, G.H.B.; Archakov, A.I. Comparative investigation of PdR by usual and ultrafine atomic force microscopy. Anal. Methods 2010, 2, 688–693. [Google Scholar] [CrossRef]
- Kuznetsov, V.Y.; Ivanov, Y.D.; Archakov, A.I. Atomic force microscopy revelation of molecular complexes in the multiprotein cytochrome P450 2B4-containing system. Proteomics 2004, 4, 2390–2396. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, D.A.; Massino, Y.S.; Segal, O.L.; Smirnova, M.B.; Pavlova, E.V.; Gurevich, K.G.; Gnedenko, O.V.; Ivanov, Y.D.; Kolyaskina, G.I.; Archakov, A.I.; et al. Analysis of the binding of bispecific monoclonal antibodies with immobilized antigens (human IgG and horseradish peroxidase) using a resonant mirror biosensor. J. Immunol. Methods 2002, 261, 103–118. [Google Scholar] [CrossRef]
- Kuznetsov, V.Y.; Ivanov, Y.D.; Bykov, V.A.; Saunin, S.A.; Fedorov, I.A.; Lemeshko, S.V.; Hoa, H.B.; Archakov, A.I. Atomic force microscopy detection of molecular complexes in multiprotein P450cam containing monooxygenase system. Proteomics 2002, 2, 1699–1705. [Google Scholar] [CrossRef]
- Kiselyova, O.I.; Yaminsky, I.V.; Ivanov, Y.D.; Kanaeva, I.P.; Kuznetsov, V.Y.; Archakov, A.I. AFM study of membrane proteins, cytochrome P450 2B4, and NADPH- cytochrome P450 reductase and their complex formation. Arch. Biochem. Biophys. 1999, 371, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, Y.D.; Frantsuzov, P.A.; Zöllner, A.; Medvedeva, N.V.; Archakov, A.I.; Reinle, W.; Bernhardt, R. Atomic Force Microscopy Study of Protein-Protein Interactions in the Cytochrome CYP11A1 (P450scc)-Containing Steroid Hydroxylase System. Nanoscale Res. Lett. 2011, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Hashemi, M.; Lv, Z.; Maity, S.; Rochet, J.C.; Lyubchenko, Y.L. A novel pathway for amyloids self-assembly in aggregates at nanomolar concentration mediated by the interaction with surfaces. Sci. Rep. 2017, 7, 45592. [Google Scholar] [CrossRef] [PubMed]
- Uchihashi, T.; Scheuring, S. Applications of high-speed atomic force microscopy to real-time visualization of dynamic biomolecular processes. Biochim. Biophys. Acta BBA Gen. Subj. 2018, 1862, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Ando, T. High-speed AFM imaging. Curr. Opin. Struct. Biol. 2014, 28, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hashemi, M.; Lv, Z.; Williams, B.; Popov, K.I.; Dokholyan, N.V.; Lyubchenko, Y.L. High-speed atomic force microscopy reveals structural dynamics of α-synuclein monomers and dimers. J. Chem. Phys. 2018, 148, 123322. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Oh, B.K.; Bae, Y.M.; Paek, S.H.; Lee, W.H.; Choi, J.W. Fabrication of self-assembled protein A monolayer and its application as an immunosensor. Biosens. Bioelectron. 2003, 19, 185–192. [Google Scholar] [CrossRef]
- Lacava, B.M.; Azevedo, R.B.; Silva, L.P.; Lacava, Z.G.M.; Neto, K.S.; Buske, N.; Bakuzis, A.F.; Morais, P.C. Particle sizing of magnetite-based magnetic fluid using atomic force microscopy: A comparative study with electron microscopy and birefringence. Appl. Phys. Lett. 2000, 77, 1876–1878. [Google Scholar] [CrossRef]
- Silva, L.P.; Lacava, Z.G.M.; Buske, N.; Morais, P.C.; Azevedo, R.B. Atomic force microscopy and transmission electron microscopy of biocompatible magnetic fluids: A comparative analysis. J. Nanopart. Res. 2004, 6, 209–213. [Google Scholar] [CrossRef]
- Muller, D.J. Out and In: Simplifying Membrane Protein Studies by AFM. Biophys. J. 2006, 91, 3133–3134. [Google Scholar] [CrossRef] [PubMed]
- Worcester, D.L.; Miller, R.G.; Bryant, P.J. Atomic force microscopy of purple membranes. J. Microsc. 1988, 152, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Worcester, D.L.; Kim, H.S.; Miller, R.G.; Bryant, P.J. Imaging bacteriorhodopsin lattices in purple membranes with atomic force microscopy. J. Vac. Sci. Technol. Vac. Surf. Films 1990, 8, 403–405. [Google Scholar] [CrossRef]
- Müller, D.J.; Kessler, M.; Oesterhelt, F.; Möller, C.; Oesterhelt, D.; Gaub, H. Stability of Bacteriorhodopsin α-Helices and Loops Analyzed by Single-Molecule Force Spectroscopy. Biophys. J. 2002, 83, 3578–3588. [Google Scholar] [CrossRef]
- Müller, D.J.; Schabert, F.A.; Büldt, G.; Engel, A. Imaging purple membranes in aqueous solutions at sub-nanometer resolution by atomic force microscopy. Biophys. J. 1995, 68, 1681–1686. [Google Scholar] [CrossRef]
- Seelert, H.; Poetsch, A.; Dencher, N.A.; Engel, A.; Stahlberg, H.; Müller, D.J. Structural biology: Proton-powered turbine of a plant motor. Nature 2000, 405, 418. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Jung, G.B.; Kim, K.S.; Lee, G.-J.; Park, H.-K. Medical Applications of Atomic Force Microscopy and Raman Spectroscopy. J. Nanosci. Nanotechnol. 2014, 14, 71–97. [Google Scholar] [CrossRef] [PubMed]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491. [Google Scholar] [CrossRef] [PubMed]
- Bykov, I.V. Point measurements of topography, interaction forces and local properties: New approach to the complex analysis in atomic force microscopy. Nauchnoe Priborostr. 2009, 19, 38–43. [Google Scholar]
- Berquand, A.; Hella-Monika, K.; Andreas, H.; Jan, M.; Petra, K. Expression of tumor suppressors PTEN and TP53 in isogenic glioblastoma U-251MG Cells affects cellular mechanical properties—An AFM-based quantitative investigation. JJAP Conf. Proc. 2013. [Google Scholar] [CrossRef]
- Rehana, A.; Alam Mohammad, T.; Atsushi, I. Pretransition and progressive softening of bovine carbonic anhydrase II as probed by single molecule atomic force microscopy. Protein Sci. 2009, 14, 1447–1457. [Google Scholar]
- Rosa-Zeiser, A.; Weilandt, E.; Hild, S.; Marti, O. The simultaneous measurement of elastic, electrostatic and adhesive properties by scanning force microscopy: Pulsed-force mode operation. Meas. Sci. Technol. 1997, 8, 1333. [Google Scholar] [CrossRef]
- Maivald, P.; Butt, H.J.; Gould, S.A.C.; Prater, C.B.; Drake, B.; Gurley, J.A.; Elings, V.B.; Hansma, P.K. Using force modulation to image surface elasticities with the atomic force microscope. Nanotechnology 1991, 2, 103. [Google Scholar] [CrossRef]
- Lee, G.U.; Kidwell, D.A.; Colton, R.J. Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir 1994, 10, 354–357. [Google Scholar] [CrossRef]
- Moy, V.T.; Florin, E.-L.; Gaub, H.E. Adhesive forces between ligand and receptor measured by AFM. Colloids Surf. Physicochem. Eng. Asp. 1994, 93, 343–348. [Google Scholar] [CrossRef]
- Lo, Y.-S.; Simons, J.; Beebe, T.P. Temperature dependence of the biotin−avidin bond-rupture force studied by atomic force microscopy. J. Phys. Chem. B 2002, 106, 9847–9852. [Google Scholar] [CrossRef]
- Harada, Y.; Kuroda, M.; Ishida, A. Specific and quantized antigen−antibody interaction measured by atomic force microscopy. Langmuir 2000, 16, 708–715. [Google Scholar] [CrossRef]
- Allen, S.; Chen, X.; Davies, J.; Davies, M.C.; Dawkes, A.C.; Edwards, J.C.; Roberts, C.J.; Sefton, J.; Tendler, S.J.; Williams, P.M. Detection of antigen−antibody binding events with the atomic force microscope. Biochemistry 1997, 36, 7457–7463. [Google Scholar] [CrossRef] [PubMed]
- Horton, M.; Charras, G.; Lehenkari, P. Analysis of ligand–receptor interactions in cells by atomic force microscopy. J. Recept. Signal Transduct. 2002, 22, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, O.H.; Snel, M.M.; Cambi, A.; Greve, J.; De Grooth, B.G.; Figdor, C.G. Biomolecular interactions measured by atomic force microscopy. Biophys. J. 2000, 79, 3267–3281. [Google Scholar] [CrossRef]
- Chtcheglova, L.A.; Shubeita, G.T.; Sekatskii, S.K.; Dietler, G. Force spectroscopy with a small dithering of AFM tip: A method of direct and continuous measurement of the spring constant of single molecules and molecular complexes. Biophys. J. 2004, 86, 1177–1184. [Google Scholar] [CrossRef]
- Zapotoczny, S.; Auletta, T.; de Jong, M.R.; Schönherr, H.; Huskens, J.; van Veggel, F.C.; Reinhoudt, D.N.; Vancso, G.J. Chain length and concentration dependence of β-cyclodextrin−ferrocene host−guest complex rupture forces probed by dynamic force spectroscopy. Langmuir 2002, 18, 6988–6994. [Google Scholar] [CrossRef]
- Baumgartner, W.; Hinterdorfer, P.; Schindler, H. Data analysis of interaction forces measured with the atomic force microscope. Ultramicroscopy 2000, 82, 85–95. [Google Scholar] [CrossRef]
- Allison, D.P.; Hinterdorfer, P.; Han, W. Biomolecular force measurements and the atomic force microscope. Curr. Opin. Biotechnol. 2002, 13, 47–51. [Google Scholar] [CrossRef]
- Li, P.-C.; Makarov, D.E. Ubiquitin-like protein domains show high resistance to mechanical unfolding similar to that of the I27 domain in titin: Evidence from simulations. J. Phys. Chem. B 2004, 108, 745–749. [Google Scholar] [CrossRef]
- Janovjak, H.; Kessler, M.; Oesterhelt, D.; Gaub, H.; Müller, D.J. Unfolding pathways of native bacteriorhodopsin depend on temperature. EMBO J. 2003, 22, 5220–5229. [Google Scholar] [CrossRef] [PubMed]
- Carrion-Vazquez, M.; Oberhauser, A.F.; Fowler, S.B.; Marszalek, P.E.; Broedel, S.E.; Clarke, J.; Fernandez, J.M. Mechanical and chemical unfolding of a single protein: A comparison. Proc. Natl. Acad. Sci. USA 1999, 96, 3694–3699. [Google Scholar] [CrossRef] [PubMed]
- Brockwell, D.J.; Paci, E.; Zinober, R.C.; Beddard, G.S.; Olmsted, P.D.; Smith, D.A.; Perham, R.N.; Radford, S.E. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat. Struct. Mol. Biol. 2003, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F.; Martínez-Martín, D.; Medalsy, I.; Alsteens, D.; Müller, D.J. Multiparametric imaging of biological systems by force-distance curve–based AFM. Nat. Methods 2013, 10, 847. [Google Scholar] [CrossRef] [PubMed]
- Introduction to Brukers ScanAsyst and PeakForce Tapping Atomic Force Microscopy Technology. Available online: https://www.bruker.com/Introduction_to_Brukers_ScanAsyst_and_PeakForce_Tapping_Atomic_Force_Microscopy_Technology_AFM_AN133.pdf (accessed on 22 February 2018).
- Viani, M.B.; Schäffer, T.E.; Paloczi, G.T.; Pietrasanta, L.I.; Smith, B.L.; Thompson, J.B.; Richter, M.; Rief, M.; Gaub, H.E.; Plaxco, K.W.; Cleland, A.N. Fast imaging and fast force spectroscopy of single biopolymers with a new atomic force microscope designed for small cantilevers. Rev. Sci. Instrum. 1999, 70, 4300–4303. [Google Scholar] [CrossRef]
- Kienberger, F.; Ebner, A.; Gruber, H.J.; Hinterdorfer, P. Molecular recognition imaging and force spectroscopy of single biomolecules. Acc. Chem. Res. 2006, 39, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Dettmann, W.; Gaub, H.E. Atomic force microscope imaging contrast based on molecular recognition. Biophys. J. 1997, 72, 445–448. [Google Scholar] [CrossRef]
- Senapati, S.; Lindsay, S. Recent progress in molecular recognition imaging using atomic force microscopy. Acc. Chem. Res. 2016, 49, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Raab, A.; Han, W.; Badt, D.; Smith-Gill, S.J.; Lindsay, S.M.; Schindler, H.; Hinterdorfer, P. Antibody recognition imaging by force microscopy. Nat. Biotechnol. 1999, 17, 901. [Google Scholar] [CrossRef] [PubMed]
- Stroh, C.; Wang, H.; Bash, R.; Ashcroft, B.; Nelson, J.; Gruber, H.; Lohr, D.; Lindsay, S.M.; Hinterdorfer, P. Single-molecule recognition imaging microscopy. Proc. Natl. Acad. Sci. USA 2004, 101, 12503–12507. [Google Scholar] [CrossRef] [PubMed]
- Creasey, R.; Sharma, S.; Gibson, C.T.; Craig, J.E.; Ebner, A.; Becker, T.; Hinterdorfer, P.; Voelcker, N.H. Atomic force microscopy-based antibody recognition imaging of proteins in the pathological deposits in Pseudoexfoliation Syndrome. Ultramicroscopy 2011, 111, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Stroh, C.M.; Ebner, A.; Geretschläger, M.; Freudenthaler, G.; Kienberger, F.; Kamruzzahan, A.S.M.; Smith-Gill, S.J.; Gruber, H.J.; Hinterdorfer, P. Simultaneous topography and recognition imaging using force Microscopy. Biophys. J. 2004, 87, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Fuhrmann, A.; Ros, R.; Kutner, L.O.; Schneeweis, L.A.; Navoa, R.; Steger, K.; Xie, L.; Yonan, C.; Abraham, R.; et al. Antibody-unfolding and metastable-state binding in force spectroscopy and recognition imaging. Biophys. J. 2011, 100, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Chtcheglova, L.A.; Wildling, L.; Waschke, J.; Drenckhahn, D.; Hinterdorfer, P. AFM functional imaging on vascular endothelial cells. J. Mol. Recognit. 2010, 23, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Chen, Q.; Wu, Y.; Qi, X.; Zhou, A. Simultaneous topographic and recognition imaging of epidermal growth factor receptor (EGFR) on single human breast cancer cells. Biochim. Biophys. Acta BBA Biomembr. 2015, 1848, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, Y.D.; Bukharina, N.S.; Pleshakova, T.O.; Frantsuzov, P.A.; Andreeva, E.Y.; Kaysheva, A.L.; Zgoda, V.G.; Izotov, A.A.; Pavlova, T.I.; Ziborov, V.S.; et al. Atomic force microscopy fishing and mass spectrometry identification of gp120 on immobilized aptamers. Int. J. Nanomed. 2014, 9, 4659–4670. [Google Scholar]
- Creasey, R.; Sharma, S.; Craig, J.E.; Gibson, C.T.; Ebner, A.; Hinterdorfer, P.; Voelcker, N.H. Detecting protein aggregates on untreated human tissue samples by atomic force microscopy recognition imaging. Biophys. J. 2010, 99, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Barattin, R.; Voyer, N. Chemical modifications of AFM tips for the study of molecular recognition events. Chem. Commun. 2008, 0, 1513–1532. [Google Scholar] [CrossRef] [PubMed]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Archakov, A.; Ivanov, Y.; Lisitsa, A.; Zgoda, V. Biospecific irreversible fishing coupled with atomic force microscopy for detection of extremely low-abundant proteins. Proteomics 2009, 9, 1326–1343. [Google Scholar] [CrossRef] [PubMed]
- Archakov, A.I.; Ivanov, Y.D.; Lisitsa, A.V.; Zgoda, V.G. AFM fishing nanotechnology is the way to reverse the Avogadro number in proteomics. Proteomics 2007, 7, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Archakov, A.I.; Ivanov, Y.D. Analytical nanobiotechnology for medicine diagnostics. Mol. Biosyst. 2007, 3, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L. The clinical plasma proteome: A survey of clinical assays for proteins in plasma and serum. Clin. Chem. 2010, 56, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.N.; Zgoda, V.G.; Maynskova, M.A.; Ronzhina, N.L.; Belyakova, N.V.; Legina, O.K.; Archakov, A.I. Experimental estimation of proteome size for cells and human plasma. Biochem. Mosc. Suppl. Ser. B Biomed. Chem. 2015, 9, 305–311. [Google Scholar] [CrossRef]
- Pleshakova, T.O.; Shumov, I.D.; Ivanov, Y.D.; Malsagova, K.A.; Kaysheva, A.L.; Archakov, A.I. AFM-based technologies as the way towards the reverse Avogadro number. Biochem. Mosc. Suppl. Ser. B Biomed. Chem. 2015, 9, 244–257. [Google Scholar] [CrossRef]
- Ivanov, A.S.; Medvedev, A.; Ershov, P.; Molnar, A.; Mezentsev, Y.; Yablokov, E.; Kaluzhsky, L.; Gnedenko, O.; Buneeva, O.; Haidukevich, I.; et al. Protein interactomics based on direct molecular fishing on paramagnetic particles: Practical realization and further SPR validation. Proteomics 2014, 14, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Kaysheva, A.L.; Bayzyanova, J.M.; Anashkina, A.S.; Uchaikin, V.F.; Shumov, I.D.; Ziborov, V.S.; Konev, V.A.; Archakov, A.I.; Ivanov, Y.D. Advantages of aptamers as ligands upon protein detection by AFM-based fishing. Anal. Methods 2017, 9, 6049–6060. [Google Scholar] [CrossRef]
- Kaysheva, A.L.; Ivanov, Y.D.; Zgoda, V.G.; Frantsuzov, P.A.; Pleshakova, T.O.; Krokhin, N.V.; Ziborov, V.S.; Archakov, A.I. Visualization and identification of hepatitis C viral particles by atomic force microscopy combined with MS/MS analysis. Biochem. Mosc. Suppl. Ser. B Biomed. Chem. 2010, 4, 15–24. [Google Scholar] [CrossRef]
- Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. Combined use of irreversible binding and MRM technology for low- and ultralow copy-number protein detection and quantitation. Proteomics 2013, 13, 727–742. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, Y.D.; Kaysheva, A.L.; Frantsuzov, P.A.; Pleshakova, T.O.; Krohin, N.V.; Izotov, A.A.; Shumov, I.D.; Uchaikin, V.F.; Konev, V.A.; Ziborov, V.S.; et al. Detection of hepatitis C virus core protein in serum by atomic force microscopy combined with mass spectrometry. Int. J. Nanomed. 2015, 10, 1597–1608. [Google Scholar]
- Kaysheva, A.L.; Ivanov, Y.D.; Frantsuzov, P.A.; Krohin, N.V.; Pavlova, T.I.; Uchaikin, V.F.; Konev, V.А.; Kovalev, O.B.; Ziborov, V.S.; Archakov, A.I. Mass spectrometric detection of the amino acid sequence polymorphism of the hepatitis C virus antigen. J. Virol. Methods 2016, 229, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Kaysheva, A.L.; Bayzyanova, J.М.; Anashkina, А.S.; Uchaikin, V.F.; Ziborov, V.S.; Konev, V.A.; Archakov, A.I.; Ivanov, Y.D. The detection of hepatitis c virus core antigen using AFM chips with immobolized aptamers. J. Virol. Methods 2018, 251, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Nettikadan, S.R.; Johnson, J.C.; Vengasandra, S.G.; Muys, J.; Henderson, E. ViriChip: A solid phase assay for detection and identification of viruses by atomic force microscopy. Nanotechnology 2004, 15, 383. [Google Scholar] [CrossRef]
- Huff, J.L.; Lynch, M.P.; Nettikadan, S.; Johnson, J.C.; Vengasandra, S.; Henderson, E. Label-Free Protein and Pathogen Detection Using the Atomic Force Microscope. J. Biomol. Screen. 2004, 9, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Shumov, I.D.; Kanashenko, S.L.; Ziborov, V.S.; Ivanov, Y.D.; Archakov, A.I.; Pleshakova, T.O. Formation of sensor array on the AFM chip surface by magnetron sputtering. J. Phys. Conf. Ser. 2017, 789, 012053. [Google Scholar] [CrossRef]
- BioScope Resolve Overview|BioAFM Atomic Force Microscope|Bruker. Available online: https://www.bruker.com/ru/products/surface-and-dimensional-analysis/atomic-force-microscopes/bioscope-resolve/overview.html (accessed on 29 March 2018).
- Wu, J.; Ding, G.; Chen, X.; Han, T.; Li, Y.; Wei, J. Development of a multiprobe instrument for measuring microstructure surface topography. Sens. Actuators Phys. 2017, 263, 363–368. [Google Scholar] [CrossRef]
- Khanal, D.; Kondyurin, A.; Hau, H.; Knowles, J.C.; Levinson, O.; Ramzan, I.; Fu, D.; Marcott, C.; Chrzanowski, W. Biospectroscopy of nanodiamond-induced alterations in conformation of intra- and extracellular proteins: A nanoscale IR study. Anal. Chem. 2016, 88, 7530–7538. [Google Scholar] [CrossRef] [PubMed]
- Kassies, R.; van der Werf, K.O.; Lenferink, A.; Hunter, C.N.; Olsen, J.D.; Subramaniam, V.; Otto, C. Combined AFM and confocal fluorescence microscope for applications in bio-nanotechnology. J. Microsc. 2005, 217, 109–116. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pleshakova, T.O.; Bukharina, N.S.; Archakov, A.I.; Ivanov, Y.D. Atomic Force Microscopy for Protein Detection and Their Physicoсhemical Characterization. Int. J. Mol. Sci. 2018, 19, 1142. https://doi.org/10.3390/ijms19041142
Pleshakova TO, Bukharina NS, Archakov AI, Ivanov YD. Atomic Force Microscopy for Protein Detection and Their Physicoсhemical Characterization. International Journal of Molecular Sciences. 2018; 19(4):1142. https://doi.org/10.3390/ijms19041142
Chicago/Turabian StylePleshakova, Tatyana O., Natalia S. Bukharina, Alexander I. Archakov, and Yuri D. Ivanov. 2018. "Atomic Force Microscopy for Protein Detection and Their Physicoсhemical Characterization" International Journal of Molecular Sciences 19, no. 4: 1142. https://doi.org/10.3390/ijms19041142
APA StylePleshakova, T. O., Bukharina, N. S., Archakov, A. I., & Ivanov, Y. D. (2018). Atomic Force Microscopy for Protein Detection and Their Physicoсhemical Characterization. International Journal of Molecular Sciences, 19(4), 1142. https://doi.org/10.3390/ijms19041142





