Belowground Interactions Impact the Soil Bacterial Community, Soil Fertility, and Crop Yield in Maize/Peanut Intercropping Systems
Abstract
:1. Introduction
2. Results
2.1. Yields in Field Experiments and Plant Properties in Pot Experiments
2.2. Soil Nutrition and Soil Enzyme Activities
2.3. Shifts of the Soil Microbial Community
2.4. qPCR of Specific Bacterial Groups
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. Field Yields, Experimental Plant Property Determination and Soil Sampling
4.3. Measurement of Soil Nutrient and Enzymatic Activities
4.4. DNA Extraction and Terminal Restriction Fragment Length Polymorphism (T-RFLP) Analysis
4.5. Quantification PCR of Bacterial Communities
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NS | Non-separation intercropping |
| SS | Semi-separation intercropping |
| CS | Complete separation intercropping |
| MS | Monoculture |
| NM | Non-separation intercropping maize |
| SM | Semi-separation intercropping maize |
| CM | Complete separation intercropping maize |
| NP | Non-separation intercropping peanut |
| SP | Semi-separation intercropping peanut |
| CP | Complete separation intercropping peanut |
| LER | Land equivalent ratio |
References
- Hauggaard-Nielsen, H.; Jørnsgaard, B.; Kinane, J.; Jensen, E.S. Grain legume–cereal intercropping: The practical application of diversity, competition and facilitation in arable and organic cropping systems. Renew. Agric. Food Syst. 2008, 23, 3–12. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, G.; Bian, X.; Jiang, X.; Zhao, Q. Review of researches on advantages of intercropping. Asian Agric. Res. 2012, 114, 126–132. [Google Scholar]
- Li, L.; Sun, J.; Zhang, F.; Li, X.; Rengel, Z.; Yang, S. Wheat/maize or wheat/soybean strip intercropping: Ii. Recovery or compensation of maize and soybean after wheat harvesting. Field Crop. Res. 2001, 71, 173–181. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, X.; Wu, P.; He, J.; Chen, X.; Gao, Y.; Cao, X. Radiation interception and utilization by wheat/maize strip intercropping systems. Agric. For. Meteorol. 2015, 204, 58–66. [Google Scholar] [CrossRef]
- Morris, R.; Garrity, D.P. Resource capture and utilization in intercropping: Water. Field Crop. Res. 1993, 34, 303–317. [Google Scholar] [CrossRef]
- Li, L.; Tilman, D.; Lambers, H.; Zhang, F.S. Plant diversity and overyielding: Insights from belowground facilitation of intercropping in agriculture. New Phytol. 2014, 203, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, S.M.; Sun, J.H.; Zhou, L.L.; Bao, X.G.; Zhang, H.G.; Zhang, F.S. Diversity enhances agricultural productivity via rhizosphere phosphorus facilitation on phosphorus-deficient soils. Proc. Nat. Acad. Sci. USA 2007, 104, 11192–11196. [Google Scholar] [CrossRef] [PubMed]
- Betencourt, E.; Duputel, M.; Colomb, B.; Desclaux, D.; Hinsinger, P. Intercropping promotes the ability of durum wheat and chickpea to increase rhizosphere phosphorus availability in a low p soil. Soil Biol. Biochem. 2012, 46, 181–190. [Google Scholar] [CrossRef]
- He, Y.; Ding, N.; Shi, J.; Wu, M.; Liao, H.; Xu, J. Profiling of microbial plfas: Implications for interspecific interactions due to intercropping which increase phosphorus uptake in phosphorus limited acidic soils. Soil Biol. Biochem. 2013, 57, 625–634. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Marschner, P. Plant-microbe interactions in the rhizosphere and nutrient cycling. In Nutrient Cycling in Terrestrial Ecosystems; Springer: Berlin/Heidelberg, Germany, 2007; pp. 159–182. [Google Scholar]
- Van de Mortel, J.E.; de Vos, R.C.; Dekkers, E.; Pineda, A.; Guillod, L.; Bouwmeester, K.; van Loon, J.J.; Dicke, M.; Raaijmakers, J.M. Metabolic and transcriptomic changes induced in arabidopsis by the rhizobacterium pseudomonas fluorescens ss101. Plant Physiol. 2012, 160, 2173–2188. [Google Scholar] [CrossRef] [PubMed]
- Sanguin, H.; Sarniguer, A.; Gazengel, K.; Moënne-Loccoz, Y.; Grundmann, G.L. Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytol. 2009, 184, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Weidner, S.; Koller, R.; Latz, E.; Kowalchuk, G.; Bonkowski, M.; Scheu, S.; Jousset, A. Bacterial diversity amplifies nutrient-based plant–soil feedbacks. Funct. Ecol. 2015, 29, 1341–1349. [Google Scholar] [CrossRef]
- Kourtev, P.; Ehrenfeld, J.; Häggblom, M. Experimental analysis of the effect of exotic and native plant species on the structure and function of soil microbial communities. Soil Biol. Biochem. 2003, 35, 895–905. [Google Scholar] [CrossRef]
- Baudoin, E.; Benizri, E.; Guckert, A. Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol. Biochem. 2003, 35, 1183–1192. [Google Scholar] [CrossRef]
- Sinsabaugh, R. Enzymic analysis of microbial pattern and process. Biol. Fertil. Soils 1994, 17, 69–74. [Google Scholar] [CrossRef]
- Awal, M.; Koshi, H.; Ikeda, T. Radiation interception and use by maize/peanut intercrop canopy. Agric. For. Meteorol. 2006, 139, 74–83. [Google Scholar] [CrossRef]
- Zuo, Y.; Zhan, F. Iron and zinc biofortification strategies in dicot plants by intercropping with gramineous species. A review. Agron. Sustain. Dev. 2009, 29, 63–71. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2010, 54, 655–670. [Google Scholar] [CrossRef]
- Wang, M.; Ahrné, S.; Antonsson, M.; Molin, G. T-rflp combined with principal component analysis and 16s rrna gene sequencing: An effective strategy for comparison of fecal microbiota in infants of different ages. J. Microbiol. Methods 2004, 59, 53. [Google Scholar] [CrossRef] [PubMed]
- Courtney, K.C.; Bainard, L.D.; Sikes, B.A.; Koch, A.M.; Maherali, H.; Klironomos, J.N.; Hart, M.M. Determining a minimum detection threshold in terminal restriction fragment length polymorphism analysis. J. Microbiol. Methods 2012, 88, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Xia, H.; Christie, P.; Zhang, Z.; Li, L.; Tang, C. Crop acquisition of phosphorus, iron and zinc from soil in cereal/legume intercropping systems: A critical review. Ann. Bot. 2016, 117, 363. [Google Scholar] [CrossRef] [PubMed]
- Strobel, B.W. Influence of vegetation on low-molecular-weight carboxylic acids in soil solution—A review. Geoderma 2001, 99, 169–198. [Google Scholar] [CrossRef]
- Ström, L.; Owen, A.; Godbold, D.; Jones, D. Organic acid behaviour in a calcareous soil: Sorption reactions and biodegradation rates. Soil Biol. Biochem. 2001, 33, 2125–2133. [Google Scholar] [CrossRef]
- Dick, R.P. Soil enzyme activities as indicators of soil quality. Soil Sci. Soc. Am. J. 1994, 58, 107–124. [Google Scholar]
- Zhang, F.; Shen, J.; Li, L.; Liu, X. An overview of rhizosphere processes related with plant nutrition in major cropping systems in china. Plant Soil 2004, 260, 89–99. [Google Scholar] [CrossRef]
- Pang, P.; Kolenko, H. Phosphomonoesterase activity in forest soils. Soil Biol. Biochem. 1986, 18, 35–39. [Google Scholar] [CrossRef]
- Guan, S.; Zhang, D.; Zhang, Z. Soil Enzyme and Its Research Methods; China Agriculture Press: Beijing, China, 1986. [Google Scholar]
- Gao, Y.; Duan, A.; Sun, J.; Li, F.; Liu, Z.; Liu, H.; Liu, Z. Crop coefficient and water-use efficiency of winter wheat/spring maize strip intercropping. Field Crop. Res. 2009, 111, 65–73. [Google Scholar] [CrossRef]
- Li, L.; Zhang, F.; Li, X.; Christie, P.; Sun, J.; Yang, S.; Tang, C. Interspecific facilitation of nutrient uptake by intercropped maize and faba bean. Nutr. Cycl. Agroecosyst. 2003, 65, 61–71. [Google Scholar] [CrossRef]
- Xiao, X.; Cheng, Z.; Meng, H.; Liu, L.; Li, H.; Dong, Y. Intercropping of green garlic (allium sativum L.) induces nutrient concentration changes in the soil and plants in continuously cropped cucumber (cucumis sativus L.) in a plastic tunnel. PLoS ONE 2013, 8, e62173. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Cheng, Z.; Meng, H.; Khan, M.A.; Li, H. Intercropping with garlic alleviated continuous cropping obstacle of cucumber in plastic tunnel. Acta Agric. Scand. Sect. B Soil Plant Sci. 2012, 62, 696–705. [Google Scholar] [CrossRef]
- Khan, M.A.; Zhihui, C.; Khan, A.R.; Rana, S.J.; Ghazanfar, B. Pepper-garlic intercropping system improves soil biology and nutrient status in plastic tunnel. Int. J. Agric. Biol. 2015, 17, 869–880. [Google Scholar] [CrossRef]
- Dai, C.C.; Chen, Y.; Wang, X.X.; Li, P.D. Effects of intercropping of peanut with the medicinal plant atractylodes lancea on soil microecology and peanut yield in subtropical china. Agrofor. Syst. 2013, 87, 417–426. [Google Scholar] [CrossRef]
- Sharma, S.K.; Ramesh, A.; Sharma, M.P.; Joshi, O.P.; Govaerts, B.; Steenwerth, K.L.; Karlen, D.L. Microbial community structure and diversity as indicators for evaluating soil quality. In Biodiversity, Biofuels, Agroforestry and Conservation Agriculture; Springer: Berlin/Heidelberg, Germany, 2010; pp. 317–358. [Google Scholar]
- Yang, Z.; Yang, W.; Li, S.; Hao, J.; Su, Z.; Sun, M.; Gao, Z.; Zhang, C. Variation of bacterial community diversity in rhizosphere soil of sole-cropped versus intercropped wheat field after harvest. PLoS ONE 2016, 11, e0150618. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, M.; Zhang, H.; Xu, N.; Sun, G. Use of mulberry–soybean intercropping in salt–alkali soil impacts the diversity of the soil bacterial community. Microb. Biotechnol. 2016, 9, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.S.; Wu, L.K.; Chen, J.; Khan, M.A.; Luo, X.M.; Lin, W.X. Biochemical and microbial properties of rhizospheres under maize/ peanut intercropping. J. Integr. Agric. 2016, 15, 101–110. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (pgpr): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Bagyaraj, D.J.; Selvakumar, G.; Sundaram, S.P. Novel plant growth promoting rhizobacteria—prospects and potential. Appl. Soil Ecol. 2015, 95, 38–53. [Google Scholar] [CrossRef]
- Bossio, D.; Scow, K.; Gunapala, N.; Graham, K. Determinants of soil microbial communities: Effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb. Ecol. 1998, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.E.; Daniell, T.J.; White, P.J. Benefits of breeding crops for yield response to soil organisms. In Molecular Microbial Ecology of the Rhizosphere: Volume 1 & 2; Wiley: Hoboken, NJ, USA, 2013; pp. 17–27. [Google Scholar]
- Qiao, Y.J.; Li, Z.Z.; Wang, X.; Zhu, B.; Hu, Y.G.; Zeng, Z.H. Effect of legume-cereal mixtures on the diversity of bacterial communities in the rhizosphere. Plant Soil Environ. 2012, 58, 174–180. [Google Scholar] [CrossRef]
- Yao, H.; Jiao, X.; Wu, F. Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 2006, 284, 195–203. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Sheng, L.; Chu, L.; Gao, J.T.; Azeem, S.; Lin, W. Insight into structure dynamics of soil microbiota mediated by the richness of replanted pseudostellaria heterophylla. Sci. Rep. 2016, 6, 26175. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; He, Z.; Van Nostrand, J.D.; Albrigo, G.; Zhou, J.; Wang, N. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 2012, 6, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.S. Grain yield, symbiotic n₂ fixation and interspecific competition for inorganic n in pea-barley intercrops. Plant Soil 1996, 182, 25–38. [Google Scholar] [CrossRef]
- Badri, D.V.; Weir, T.L.; Lelie, D.V.D.; Vivanco, J.M. Rhizosphere chemical dialogues: Plant–microbe interactions. Curr. Opin. Biotechnol. 2009, 20, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Bouffaud, M.L.; Kyselková, M.; Gouesnard, B.; Grundmann, G.; Muller, D.; Moënne-Loccoz, Y. Is diversification history of maize influencing selection of soil bacteria by roots? Mol. Ecol. 2012, 21, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Houlden, A.; Timms-Wilson, T.M.; Day, M.J.; Bailey, M.J. Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol. Ecol. 2008, 65, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Gschwendtner, S.; Esperschütz, J.; Buegger, F.; Reichmann, M.; Müller, M.; Munch, J.C.; Schloter, M. Effects of genetically modified starch metabolism in potato plants on photosynthate fluxes into the rhizosphere and on microbial degraders of root exudates. FEMS Microbiol. Ecol. 2011, 76, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Mead, R.; Willey, R. The concept of a ‘land equivalent ratio’ and advantages in yields from intercropping. Exp. Agric. 1980, 16, 217–228. [Google Scholar] [CrossRef]
- Jackson, M.L. Soil Chemical Analysis; Prentice-Hall: Upper Saddle River, NJ, USA, 1958. [Google Scholar]
- Wu, L.; Wang, J.; Huang, W.; Wu, H.; Chen, J.; Yang, Y.; Zhang, Z.; Lin, W. Corrigendum: Plant-microbe rhizosphere interactions mediated by rehmannia glutinosa root exudates under consecutive monoculture. Sci. Rep. 2016, 6, 19101. [Google Scholar] [CrossRef] [PubMed]
- Egert, M.; Marhan, S.; Wagner, B.; Scheu, S.; Friedrich, M.W. Molecular profiling of 16s rrna genes reveals diet-related differences of microbial communities in soil, gut, and casts of lumbricus terrestris l.(oligochaeta: Lumbricidae). FEMS Microbiol. Ecol. 2004, 48, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Kent, A.D.; Smith, D.J.; Benson, B.J.; Triplett, E.W. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 2003, 69, 6768–6776. [Google Scholar] [CrossRef] [PubMed]
- Rees, G.N.; Baldwin, D.S.; Watson, G.O.; Perryman, S.; Nielsen, D.L. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: Application of multivariate statistics. Antonie Van Leeuwenhoek 2004, 86, 339–347. [Google Scholar] [CrossRef] [PubMed]
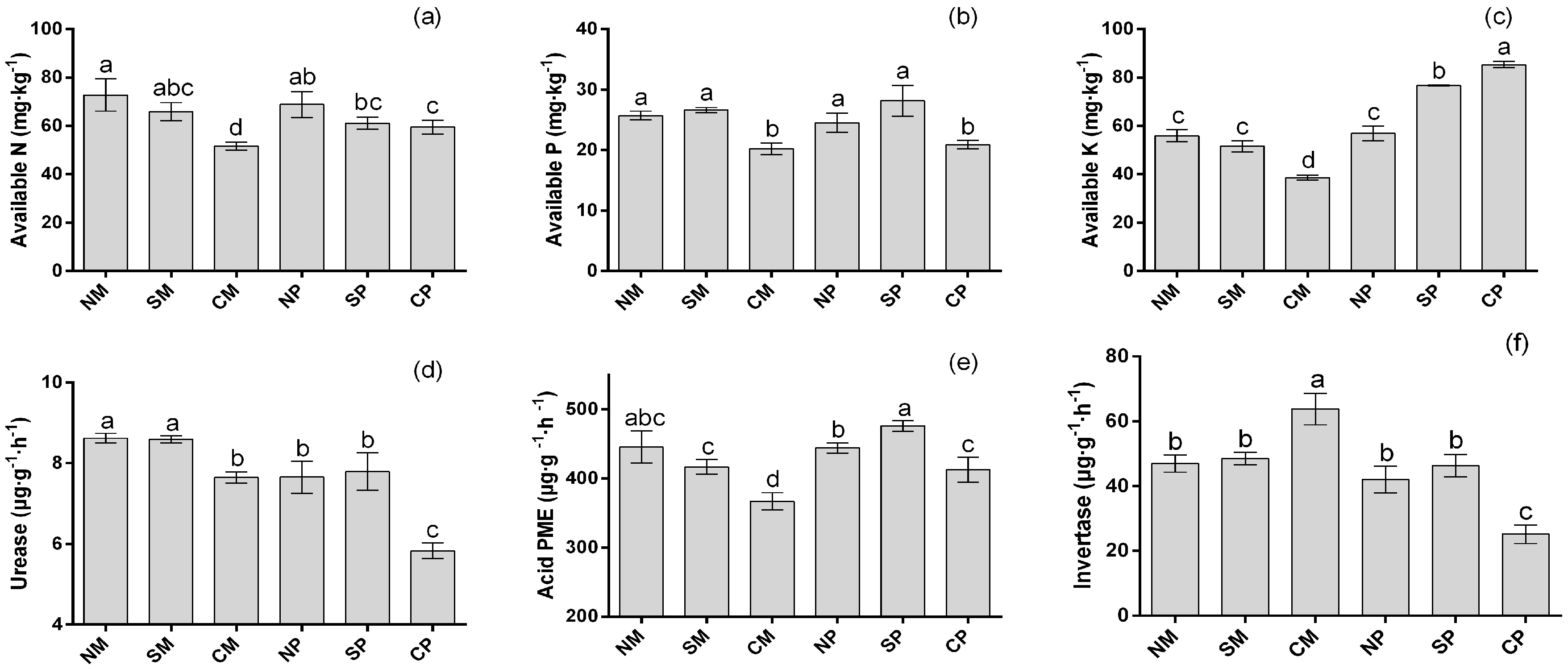
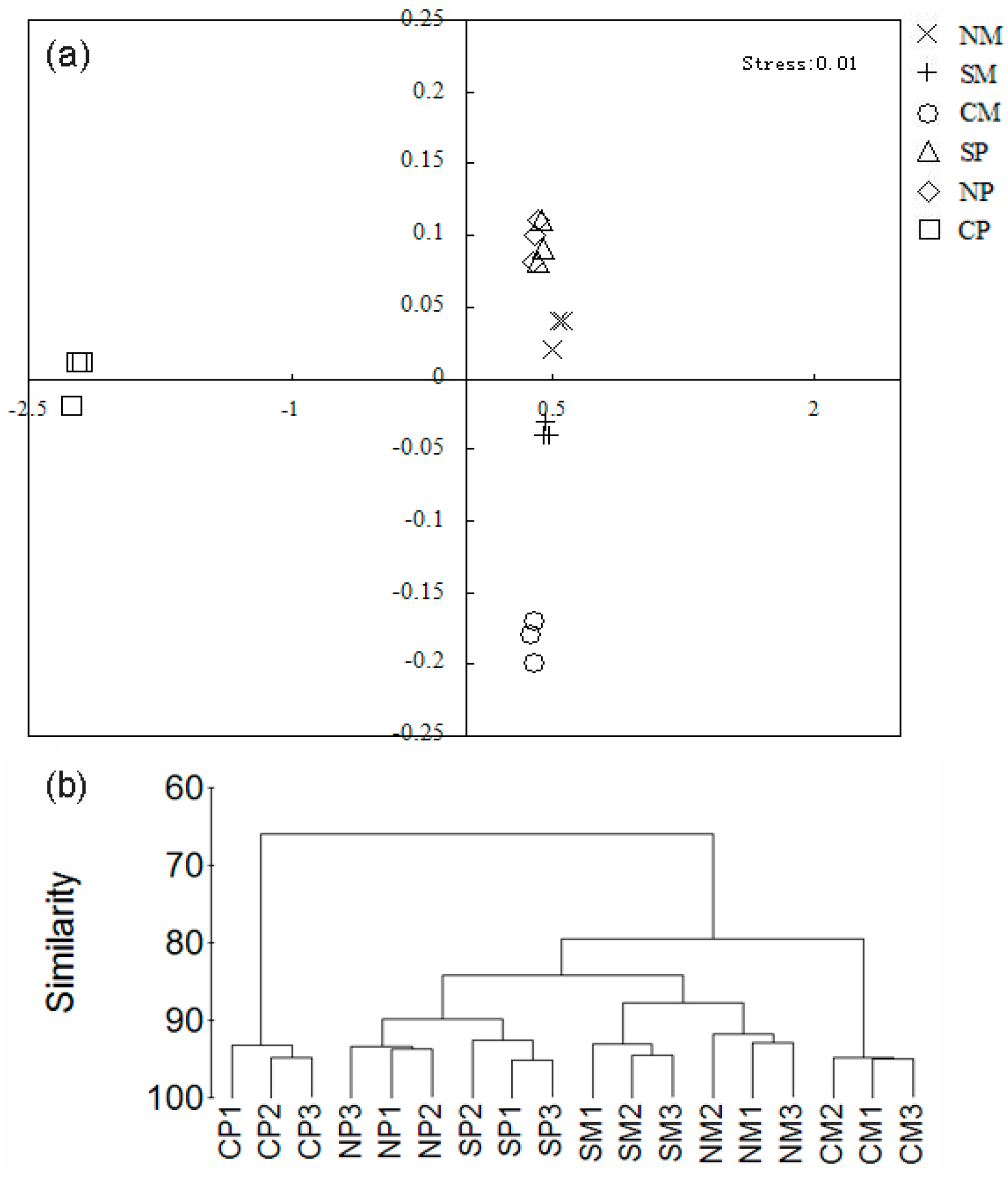
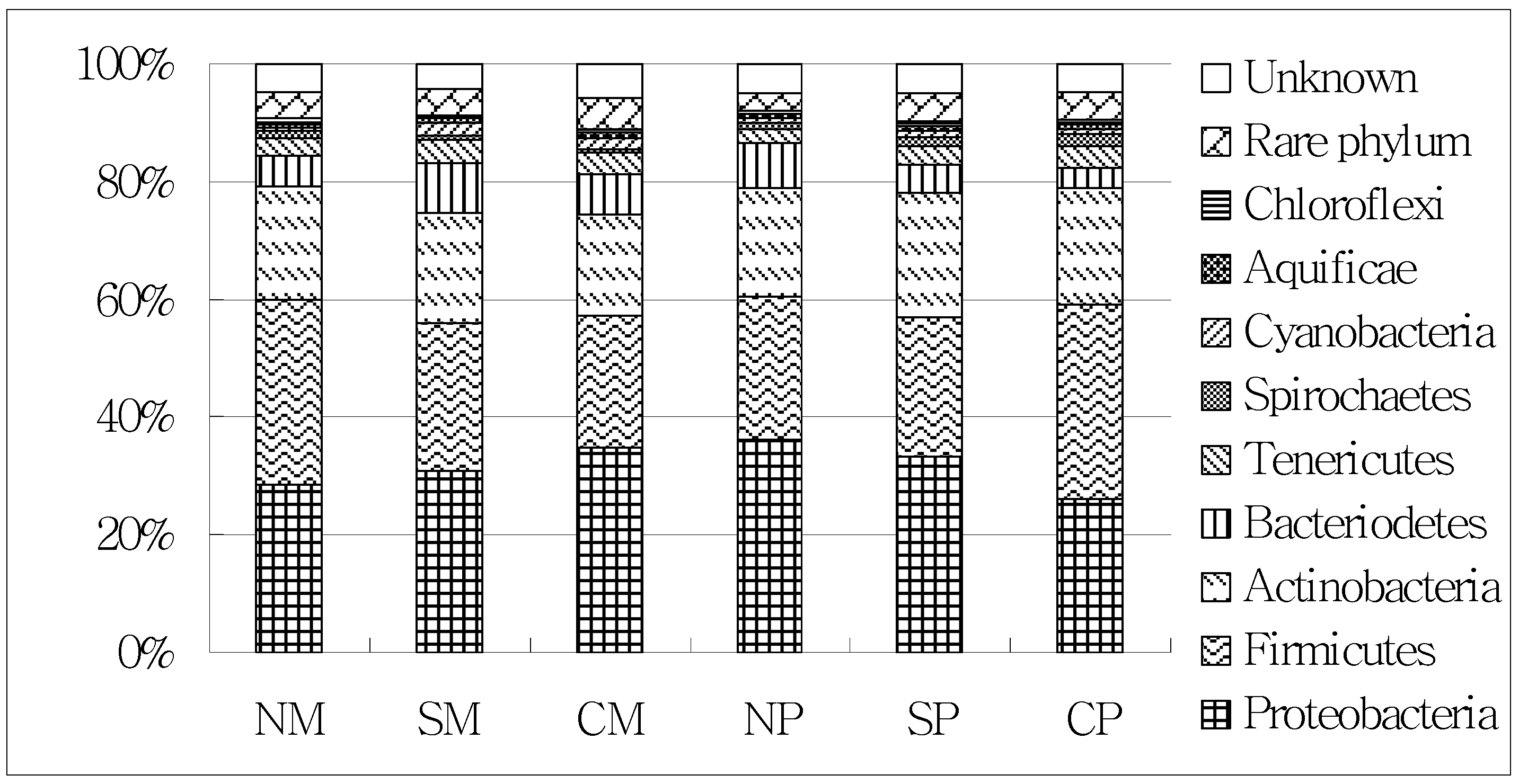
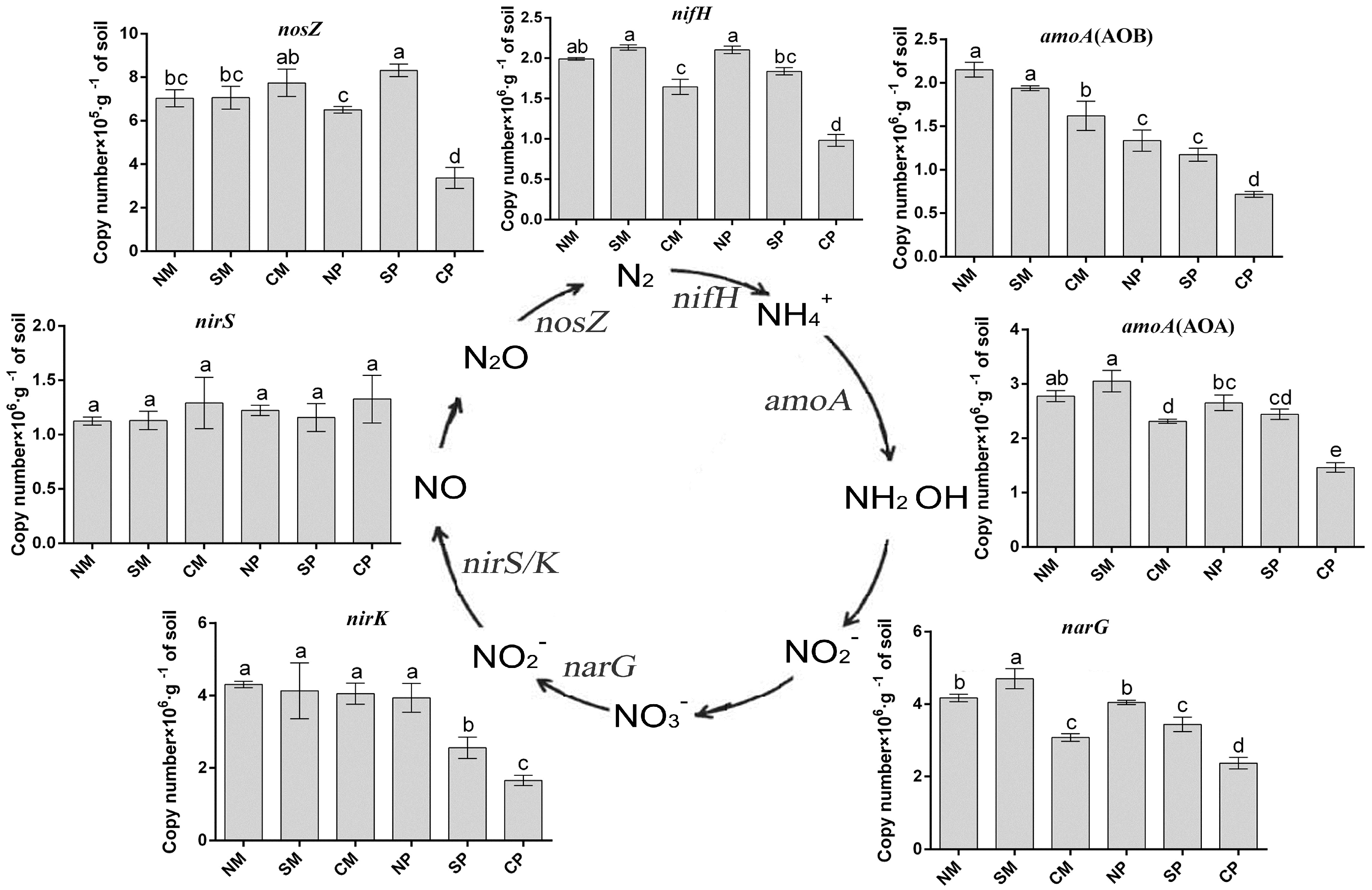

| Treatments | Maize Yield (Mg·ha−1) | Peanut Yield (Mg·ha−1) | LER |
|---|---|---|---|
| 2011 | |||
| NS | 8.62 ± 0.39a | 3.42 ± 0.14a | 1.31 |
| SS | 9.00 ± 0.34a | 3.32 ± 0.14ab | 1.33 |
| CS | 4.69 ± 0.29b | 3.14 ± 0.06b | 0.95 |
| MS | 4.70 ± 0.30b | 3.38 ± 0.07ab | 1 |
| 2012 | |||
| NS | 10.22 ± 0.33a | 3.69 ± 0.35a | 1.11 |
| SS | 9.95 ± 0.22a | 3.61 ± 0.15a | 1.08 |
| CS | 8.15 ± 0.30b | 3.40 ± 0.17a | 0.97 |
| MS | 8.06 ± 0.39b | 3.61 ± 0.27a | 1 |
| Treatments | Biomass (g) | Shoot Biomass (g) | Root Biomass (g) | Pn (μmol CO2·m−2·s−1) | Nodule Number per Plant | Dry Weight per Nodule (mg) |
|---|---|---|---|---|---|---|
| Maize | ||||||
| NS | 118.40 ± 6.26a | 95.13 ± 4.71a | 23.27 ± 1.55a | 40.38 ± 1.81a | / | / |
| SS | 120.22 ± 5.56a | 94.95 ± 4.81a | 25.27 ± 0.75a | 40.23 ± 3.52a | / | / |
| CS | 94.19 ± 2.98b | 76.16 ± 2.58b | 18.03 ± 0.40b | 34.48 ± 1.08b | / | / |
| Peanut | ||||||
| NS | 13.95 ± 0.99a | 12.36 ± 0.76a | 1.59 ± 0.23a | 26.60 ± 1.53a | 528.33 ± 25.42a | 0.51 ± 0.03ab |
| SS | 14.60 ± 1.07a | 12.83 ± 0.70a | 1.77 ± 0.37a | 27.60 ± 1.63a | 626.00 ± 40.00a | 0.57 ± 0.06a |
| CS | 14.30 ± 0.72a | 13.17 ± 0.66a | 1.13 ± 0.06b | 24.98 ± 1.67a | 310.00 ± 50.20b | 0.43 ± 0.04b |
| Treatments | Simpson Index (J) | Shannon-Wiener Index (H) |
|---|---|---|
| Maize | ||
| NS | 0.988a | 5.921a |
| SS | 0.984b | 5.84b |
| CS | 0.982c | 5.71c |
| Peanut | ||
| NS | 0.985a | 5.826a |
| SS | 0.986a | 5.862a |
| CS | 0.982b | 5.596c |
| Contribution (%) | TRFLP-PAT Assignment | Relative Abundance (%) | Functions | ||
|---|---|---|---|---|---|
| NS | SS | CS | |||
| Maize | |||||
| 4.63 | Brevibacillus brevis (D78457) | 3.53 | 3.63 | 0 | Improving root growth, nodulation and pathogen antagonism |
| 3.33 | Paenibacillus sp. | 2.8 | 2.35 | 0 | N-fixation and pathogen antagonism |
| 1.87 | Bacillus sp. | 6 | 5.65 | 4.38 | Improving nodulation, P-solubilization and pathogen antagonism |
| 1.68 | Clone OCS155 (AF001652) | 1.25 | 0 | 3.46 | Unknown |
| 1.58 | No Match | 0.85 | 0.96 | 0.18 | Unknown |
| 1.28 | Polyangium sp. | 0.76 | 0.79 | 1.62 | C cycle |
| 1.21 | Bacillus subtilis (AL009126) | 4.99 | 4.77 | 3.95 | Improving nodulation, P-solubilization and pathogen antagonism |
| 1.19 | Clone T33 (Z93960) | 1.33 | 0.32 | 1.55 | Unknown |
| 1.12 | Pseudomonas sp. | 3.64 | 4.32 | 3.26 | P-solubilizing and pathogen antagonism |
| 1.08 | Acidosphaera (D86512) | 1.4 | 0.8 | 1.02 | Unknown |
| 1.02 | Sphingomonas sp. | 1.52 | 1.24 | 1.89 | C cycle |
| Peanut | |||||
| 2.23 | Bacillus sp. | 8.33 | 8.89 | 5.81 | Improving nodulation, P-solubilization and pathogen antagonism |
| 1.98 | Burkholderia sp. | 5.78 | 5.62 | 3.49 | Legume N-fixing symbiont. Pathogen antagonism and plant growth promotion |
| 1.81 | Clostridium sp. | 1.53 | 1.32 | 3.98 | C cycle |
| 1.6 | Pseudomonas sp. | 1.25 | 1.44 | 0.62 | P-solubilization and pathogen antagonism |
| 1.59 | str. AS2988.(AF060671) | 1.4 | 0.94 | 0 | Unknown |
| 1.5 | Nocardia crassostrae (U92800) | 0.99 | 1.13 | 3.44 | Unknown |
| 1.31 | clone Sva0556. | 0.49 | 0 | 1.8 | Unknown |
| 1.29 | Brevibacillus brevis (D78457) | 3.45 | 3.08 | 2.65 | Improving root growth, nodulation and pathogen antagonism |
| 1.2 | No match | 0 | 0 | 0.87 | Unknown |
| 1.02 | Xylophilus ampelinus (AF078758) | 0.77 | 1.38 | 2.98 | Plant pathology |
| 0.99 | Cytophaga lytica (M62796) | 1.24 | 1.24 | 0 | C cycle |
| 0.95 | Mesorhizobium loti (D14514) | 2.71 | 3.3 | 1.81 | Legume nodulation and N fixation |
| 0.9 | Rhizobium hainanense (U71078) | 1.53 | 1.7 | 0.89 | Legume nodulation and N fixation |
| 0.86 | Afipia clevelandensis (M69186) | 1.9 | 2.3 | 2.5 | Nitrification |
| 0.86 | Sphingomonas sp. (U52146) | 1.35 | 1.59 | 0.66 | C cycle |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Chen, J.; Wu, L.; Luo, X.; Li, N.; Arafat, Y.; Lin, S.; Lin, W. Belowground Interactions Impact the Soil Bacterial Community, Soil Fertility, and Crop Yield in Maize/Peanut Intercropping Systems. Int. J. Mol. Sci. 2018, 19, 622. https://doi.org/10.3390/ijms19020622
Li Q, Chen J, Wu L, Luo X, Li N, Arafat Y, Lin S, Lin W. Belowground Interactions Impact the Soil Bacterial Community, Soil Fertility, and Crop Yield in Maize/Peanut Intercropping Systems. International Journal of Molecular Sciences. 2018; 19(2):622. https://doi.org/10.3390/ijms19020622
Chicago/Turabian StyleLi, Qisong, Jun Chen, Linkun Wu, Xiaomian Luo, Na Li, Yasir Arafat, Sheng Lin, and Wenxiong Lin. 2018. "Belowground Interactions Impact the Soil Bacterial Community, Soil Fertility, and Crop Yield in Maize/Peanut Intercropping Systems" International Journal of Molecular Sciences 19, no. 2: 622. https://doi.org/10.3390/ijms19020622
APA StyleLi, Q., Chen, J., Wu, L., Luo, X., Li, N., Arafat, Y., Lin, S., & Lin, W. (2018). Belowground Interactions Impact the Soil Bacterial Community, Soil Fertility, and Crop Yield in Maize/Peanut Intercropping Systems. International Journal of Molecular Sciences, 19(2), 622. https://doi.org/10.3390/ijms19020622




