Prediction of Effective Drug Combinations by an Improved Naïve Bayesian Algorithm
Abstract
:1. Introduction
2. Results and Discussion
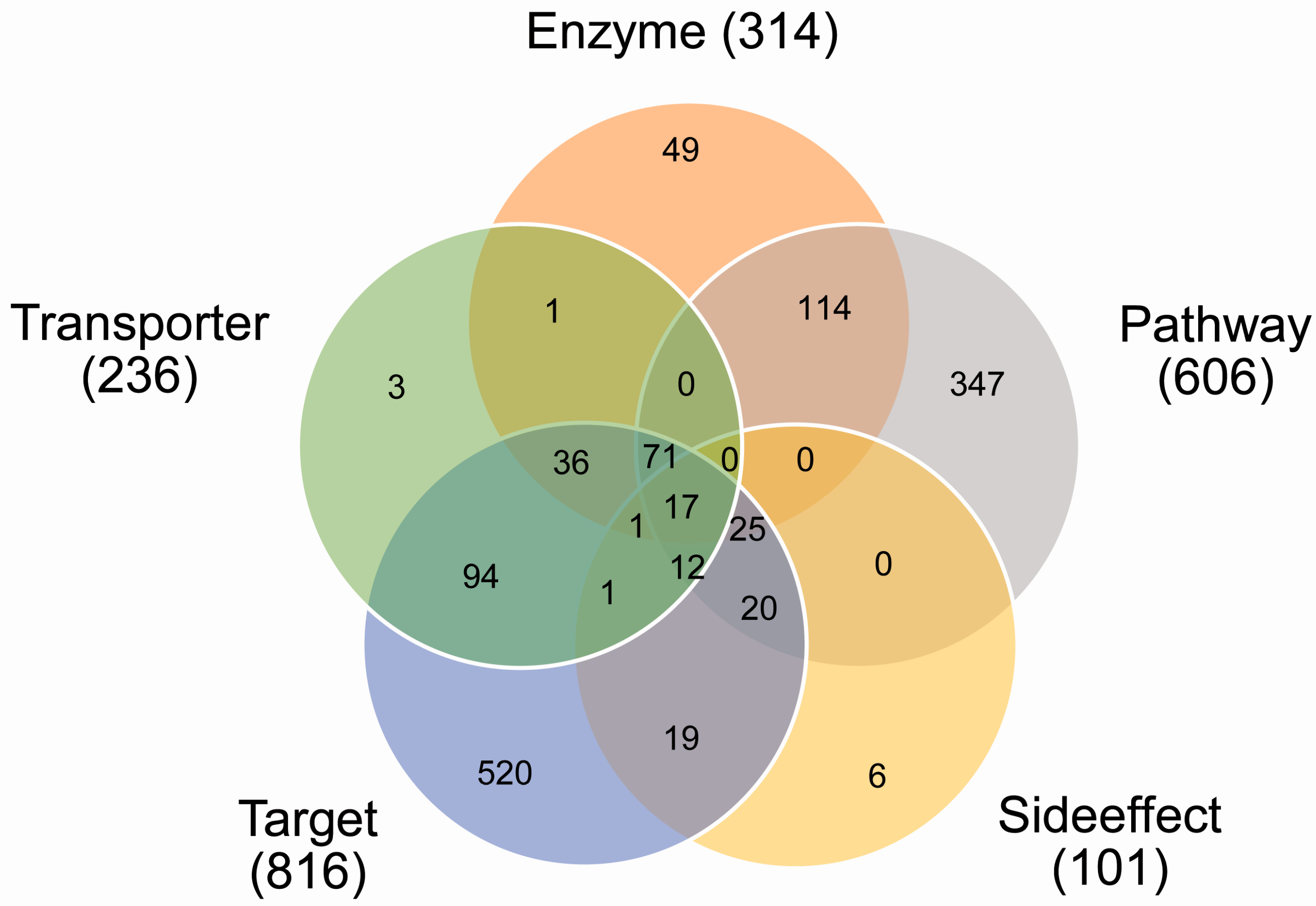
2.1. Coverage of Drug Combinations by Different Features
2.2. The Impact of Different Ratios of Positive-to-Negative Samples on the Classification
2.3. The Impact of Different Compositions of Negative Samples on the Classification
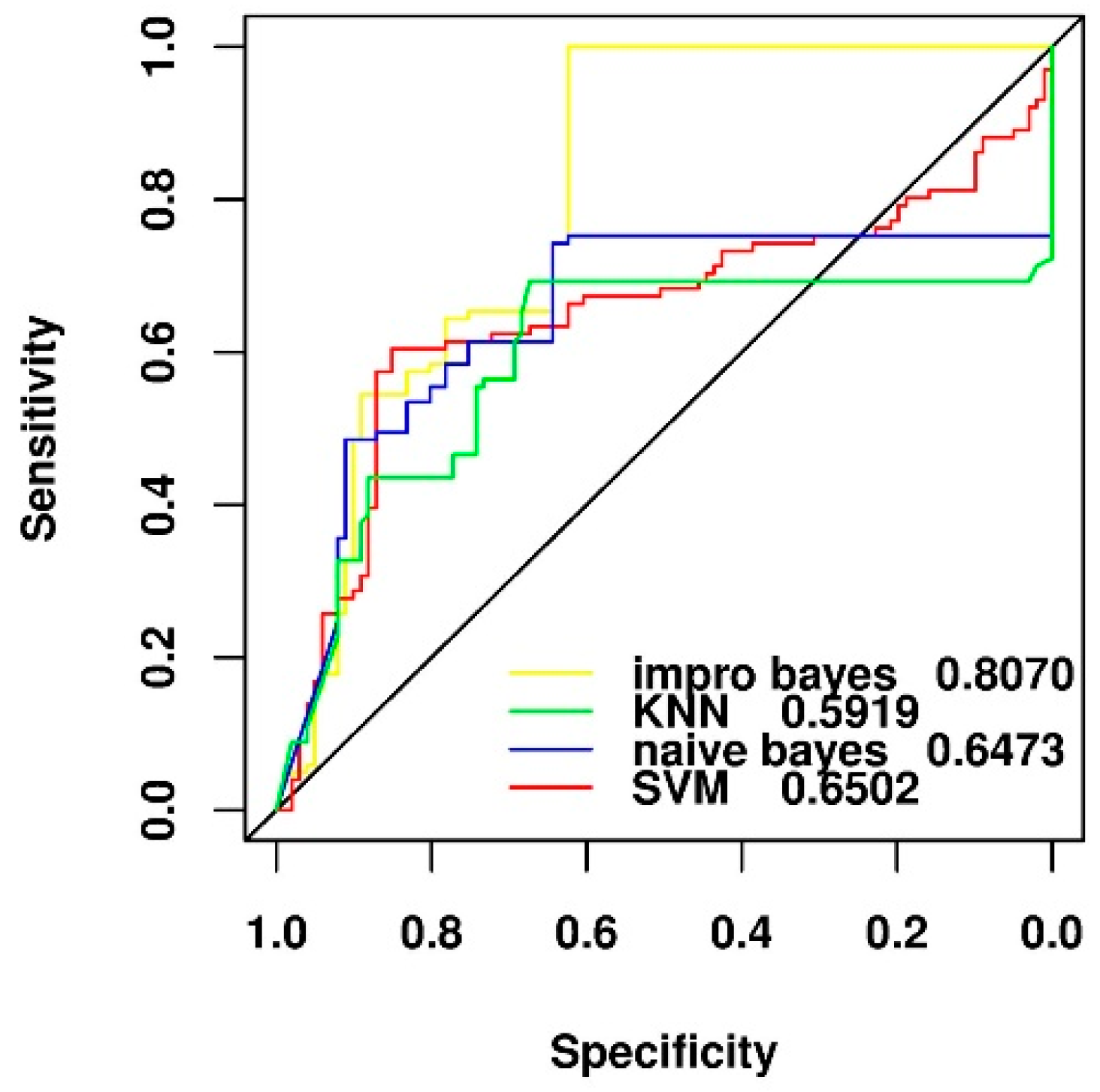
2.4. Comparison of Different Feature Selection and Classification Algorithms
2.5. Comparison of Two Different Feature Selection Methods
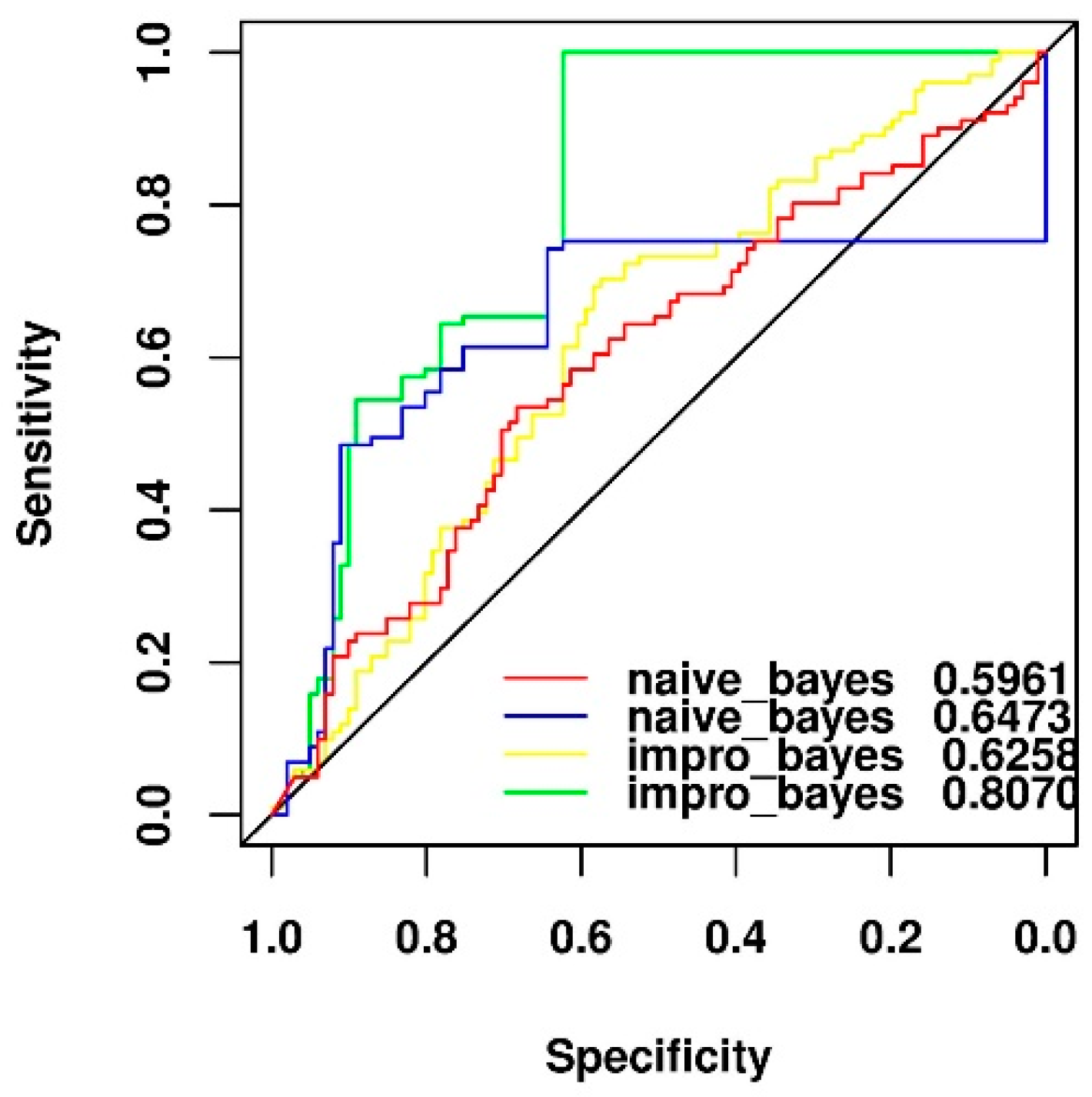
2.6. Performance Comparison of Original and y-Randomization Data Sets
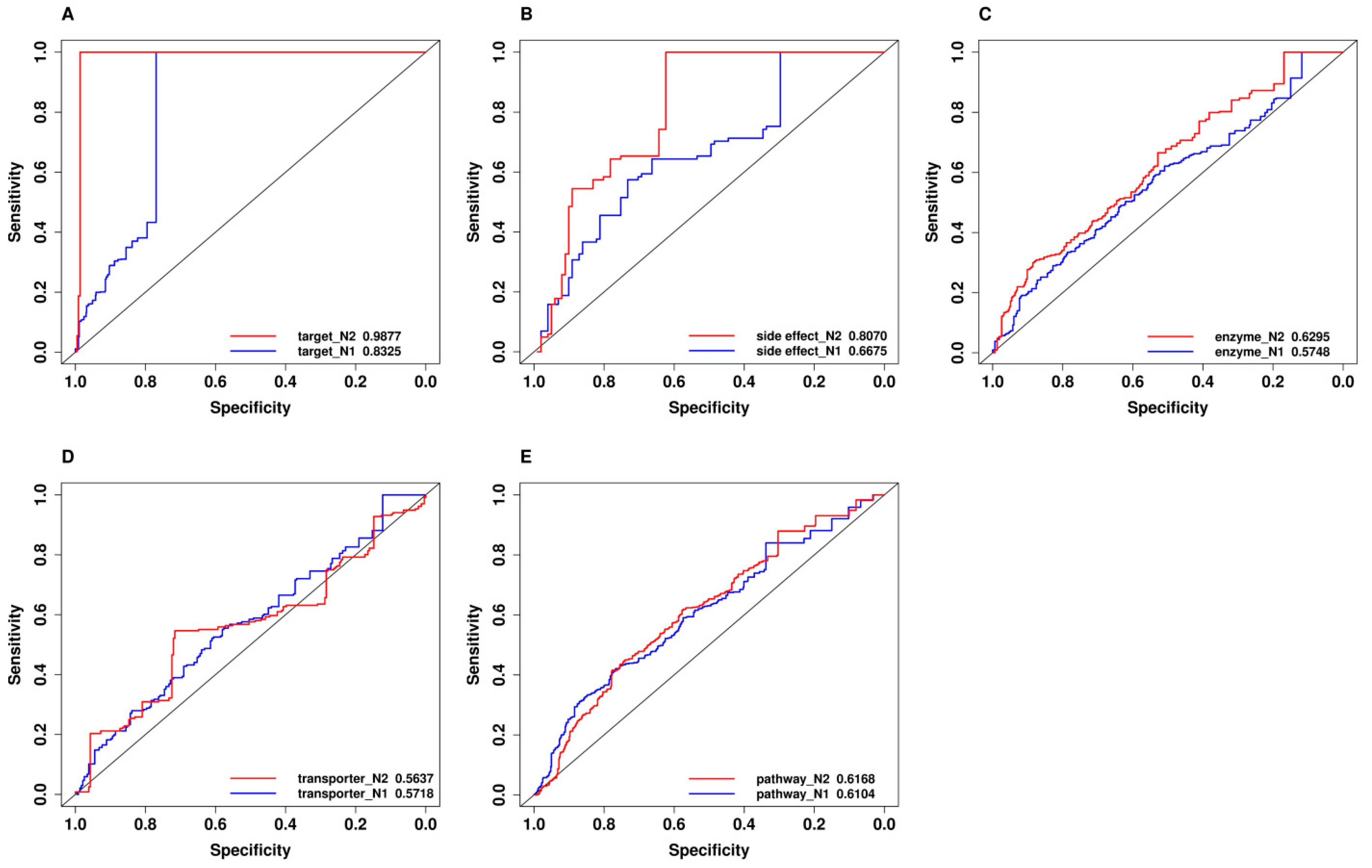
2.7. Comparison of Predictive Power of Individual Features
2.8. Comparison with Previously Published Methods
3. Materials and Methods
3.1. Data Sets
3.1.1. Positive Data Set
3.1.2. Negative Data Sets
3.2. Feature Sets
3.2.1. Feature Representation
3.2.2. Feature Selection
- (1)
- Basic assumptions: In the selection of features, for the convenience of calculation, we set all the features to obey a 0–1 distribution. If a value of the feature element was not 0, no matter how much it was, we took the value as 1.
- (2)
- Basic principles: In the negative samples, we set the frequency for the feature element value to 1; while in the mixed positive and negative samples, we set the frequency for the feature element value to 1. If there was no difference in the positive and negative samples, the frequency and should be approximately equal; while if the difference between and was beyond a certain level of significance, the feature was considered as the important one.
- (3)
- Methods: For all the features, was the abscissa, was the ordinate, and a scatter diagram was drawn.
3.3. Model Construction
3.3.1. Classification Algorithms
3.3.2. Improved Naïve Bayesian Algorithm
- (1)
- We applied normal transformation to the features of all samples, and then obtained a new set of attributes ; all these distributions obey the normal distribution.
- (2)
- According to the new set of attribute values, we calculated the correlation coefficient matrix R and calculated the characteristic values of and corresponding eigenvector of .
- (3)
- We calculated the mean value and the standard deviation of transformed attributes in category and calculated the mean value and the standard deviation of transformed attributes for all samples. Let then, for each sample , calculated as:
- (4)
- If the prior probability is very clear, for each category :is the maximum value, the corresponding category is the predicted category of the sample.
- (5)
- If the prior probability is not clear, then for a certain category , calculated on all samples:
3.4. y-Randomization Test
3.5. Model Evaluation
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jia, J.; Zhu, F.; Ma, X.; Cao, Z.; Li, Y.; Chen, Y.Z. Mechanisms of drug combinations: Interaction and network perspectives. Nat. Rev. Drug Discov. 2009, 8, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Gao, J.; Wu, D.; Zhang, S.; Mamitsuka, H.; Zhu, S. DrugE-Rank: Improving drug-target interaction prediction of new candidate drugs or targets by ensemble learning to rank. Bioinformatics 2016, 32, i18–i27. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Takigawa, I.; Mamitsuka, H.; Zhu, S. Similarity-based machine learning methods for predicting drug-target interactions: A brief review. Brief. Bioinform. 2014, 15, 734–747. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.R.; Lehar, J.; Keith, C.T. Multi-target therapeutics: When the whole is greater than the sum of the parts. Drug Discov. Today 2007, 12, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Xu, K.J.; Song, J.; Zhao, X.M. Exploring drug combinations in genetic interaction network. BMC Bioinform. 2012, 13, S7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.M.; Iskar, M.; Zeller, G.; Kuhn, M.; van Noort, V.; Bork, P. Prediction of drug combinations by integrating molecular and pharmacological data. PLoS Comput. Biol. 2011, 7, e1002323. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, B.Q.; Zheng, M.Y.; Zhang, J.; Feng, K.Y.; Cai, Y.D. Prediction of effective drug combinations by chemical interaction, protein interaction and target enrichment of KEGG pathways. BioMed Res. Int. 2013, 2013, 723780. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xiong, Y.; Xu, Q.; Wei, D. A hadoop-based method to predict potential effective drug combination. BioMed Res. Int. 2014, 2014, 196858. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xiong, Y.; Dai, H.; Kumari, K.M.; Xu, Q.; Ou, H.Y.; Wei, D.Q. PDC-SGB: Prediction of effective drug combinations using a stochastic gradient boosting algorithm. J. Theor. Biol. 2017, 417, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gayvert, K.M.; Aly, O.; Platt, J.; Bosenberg, M.W.; Stern, D.F.; Elemento, O. A Computational Approach for Identifying Synergistic Drug Combinations. PLoS Comput. Biol. 2017, 13, e1005308. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ren, B.; Chen, M.; Wang, Q.; Zhang, L.; Yan, G. NLLSS: Predicting Synergistic Drug Combinations Based on Semi-supervised Learning. PLoS Comput. Biol. 2016, 12, e1004975. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, H.; Lu, P.; Liu, X.; Cao, H. Synergy evaluation by a pathway-pathway interaction network: A new way to predict drug combination. Mol. BioSyst. 2016, 12, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Huang, C.; Fu, Y.; Wang, J.; Wu, Z.; Ru, J.; Zheng, C.; Guo, Z.; Chen, X.; Zhou, W.; et al. Large-scale exploration and analysis of drug combinations. Bioinformatics 2015, 31, 2007–2016. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Sawada, R.; Mizutani, S.; Kotera, M.; Yamanishi, Y. Large-Scale Prediction of Beneficial Drug Combinations Using Drug Efficacy and Target Profiles. J. Chem. Inf. Model. 2015, 55, 2705–2716. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, F.; Sheng, J.; Xia, X.; Ma, J.; Zhan, M.; Wong, S.T. DrugComboRanker: Drug combination discovery based on target network analysis. Bioinformatics 2014, 30, i228–i236. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhang, P.; Qu, X.A.; Sanseau, P.; Yang, L. Systematic prediction of drug combinations based on clinical side-effects. Sci. Rep. 2014, 4, 7160. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, Y.; Cui, H.; Huang, T.; Wang, D.; Lian, B.; Li, W.; Qin, G.; Chen, L.; Xie, L. Prediction of synergistic anti-cancer drug combinations based on drug target network and drug induced gene expression profiles. Artif. Intell. Med. 2017, 83, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Zakharov, A.V.; Varlamova, E.V.; Lagunin, A.A.; Dmitriev, A.V.; Muratov, E.N.; Fourches, D.; Kuz’min, V.E.; Poroikov, V.V.; Tropsha, A.; Nicklaus, M.C. QSAR Modeling and Prediction of Drug-Drug Interactions. Mol. Pharm. 2016, 13, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Mei, H. QSAR modeling and molecular interaction analysis of natural compounds as potent neuraminidase inhibitors. Mol. BioSyst. 2016, 12, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Qiu, K.; Xie, X. 3D QSAR studies, pharmacophore modeling and virtual screening on a series of steroidal aromatase inhibitors. Int. J. Mol. Sci. 2014, 15, 20927–20947. [Google Scholar] [CrossRef] [PubMed]
- Sprous, D.G.; Palmer, R.K.; Swanson, J.T.; Lawless, M. QSAR in the pharmaceutical research setting: QSAR models for broad, large problems. Curr. Top. Med. Chem. 2010, 10, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Nembri, S.; Grisoni, F.; Consonni, V.; Todeschini, R. In Silico Prediction of Cytochrome P450-Drug Interaction: QSARs for CYP3A4 and CYP2C9. Int. J. Mol. Sci. 2016, 17, 914. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, J.; Fan, X.; Sui, X.; Zhang, L.; Wang, Y.; He, Z. Considerations and recent advances in QSAR models for cytochrome P450-mediated drug metabolism prediction. J. Comput. Aided Mol. Des. 2008, 22, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Sahakian, D.C.; de Morais, S.M.; Xu, J.J.; Polzer, R.J.; Winter, S.M. The role of absorption, distribution, metabolism, excretion and toxicity in drug discovery. Curr. Top. Med. Chem. 2003, 3, 1125–1154. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wei, Q.; Yu, G.; Gai, W.; Li, Y.; Chen, X. DCDB 2.0: A major update of the drug combination database. Database (Oxford) 2014, 2014, bau124. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Campillos, M.; Letunic, I.; Jensen, L.J.; Bork, P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010, 6, 343. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wu, Y.; Chen, Y.; Sun, J.; Zhao, Z.; Chen, X.W.; Matheny, M.E.; Xu, H. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J. Am. Med. Inform. Assoc. 2012, 19, e28–e35. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Wan, S.; Ju, Y.; Tang, J.; Zeng, X. Pretata: Predicting TATA binding proteins with novel features and dimensionality reduction strategy. BMC Syst. Biol. 2016, 10, 114. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Xing, P.; Shi, G.; Ji, Z.L.; Zou, Q. Fast prediction of protein methylation sites using a sequence-based feature selection technique. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Rucker, C.; Rucker, G.; Meringer, M. y-Randomization and its variants in QSPR/QSAR. J. Chem. Inf. Model. 2007, 47, 2345–2357. [Google Scholar] [CrossRef] [PubMed]
- Baldi, P.; Brunak, S.; Chauvin, Y.; Andersen, C.A.; Nielsen, H. Assessing the accuracy of prediction algorithms for classification: An overview. Bioinformatics 2000, 16, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qu, Q.L.; Zhang, Y.Q.; Wang, W. The linear neighborhood propagation method for predicting long non-coding RNA—Protein interactions. Neurocomputing 2018, 273, 526–534. [Google Scholar] [CrossRef]
- Li, L.; Xiong, Y.; Zhang, Z.Y.; Guo, Q.; Xu, Q.; Liow, H.H.; Zhang, Y.H.; Wei, D.Q. Improved feature-based prediction of SNPs in human cytochrome P450 enzymes. Interdiscip. Sci. Comput. Life Sci. 2015, 7, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Xiong, Y.; Kihara, D. Large-scale binding ligand prediction by improved patch-based method Patch-Surfer2.0. Bioinformatics 2015, 31, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Zhang, W. Quantitative prediction of drug side effects based on drug-related features. Interdiscip. Sci. Comput. Life Sci. 2017, 9, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Zhang, T.; Xiong, Y.; Li, L.; Huo, J.; Wei, D.Q. Mutation probability of cytochrome P450 based on a genetic algorithm and support vector machine. Biotechnol. J. 2011, 6, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Chen, W.; Lin, H. Identifying Antioxidant Proteins by Using Optimal Dipeptide Compositions. Interdiscip. Sci. Comput. Life Sci. 2016, 8, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, J.; Xiong, Y.; Zhu, L.; Zhou, X. Analysis and classification of DNA-binding sites in single-stranded and double-stranded DNA-binding proteins using protein information. IET Syst. Biol. 2014, 8, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xiong, Y.; Zhao, M.; Zou, H.; Ye, X.; Liu, J. Prediction of conformational B-cell epitopes from 3D structures by random forests with a distance-based feature. BMC Bioinform. 2011, 12, 341. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Xia, J.; Zhang, W.; Liu, J. Exploiting a reduced set of weighted average features to improve prediction of DNA-binding residues from 3D structures. PLoS ONE 2011, 6, e28440. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.Q.; Bi, D.X.; Wei, D.Q.; Ou, H.Y. Prediction of Type II Toxin-Antitoxin Loci in Klebsiella pneumoniae Genome Sequences. Interdiscip. Sci. Comput. Life Sci. 2016, 8, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Liu, J.; Zhang, W.; Zeng, T. Prediction of heme binding residues from protein sequences with integrative sequence profiles. Proteome Sci. 2012, 10, S20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Niu, Y.; Xiong, Y.; Zhao, M.; Yu, R.; Liu, J. Computational prediction of conformational B-cell epitopes from antigen primary structures by ensemble learning. PLoS ONE 2012, 7, e43575. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Liu, J.; Wei, D.Q. An accurate feature-based method for identifying DNA-binding residues on protein surfaces. Proteins 2011, 79, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Zhang, J.; Tang, H.; Chen, W.; Lin, H. Predicting the Organelle Location of Noncoding RNAs Using Pseudo Nucleotide Compositions. Interdiscip. Sci. Comput. Life Sci. 2017, 9, 540–544. [Google Scholar] [CrossRef] [PubMed]
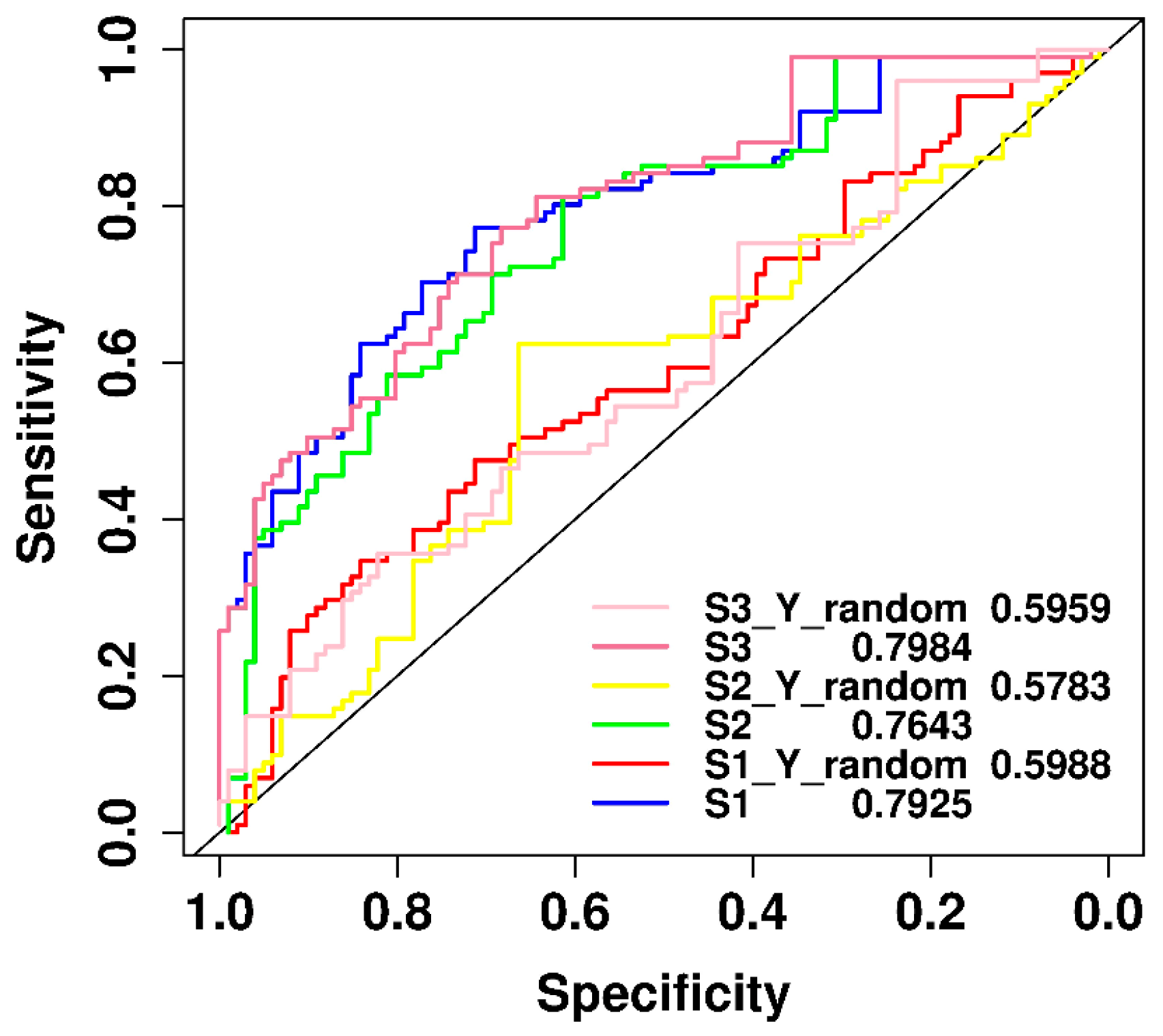
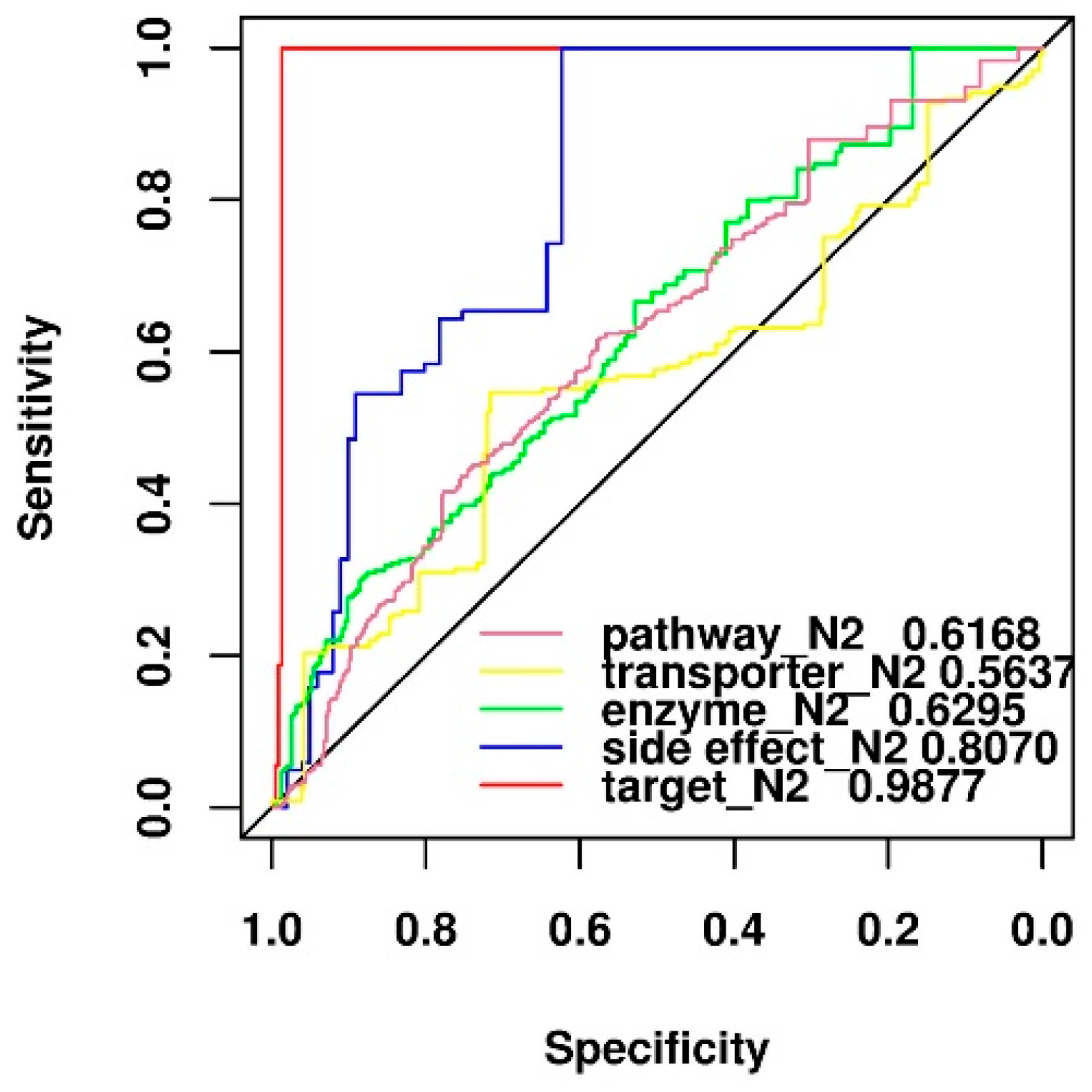
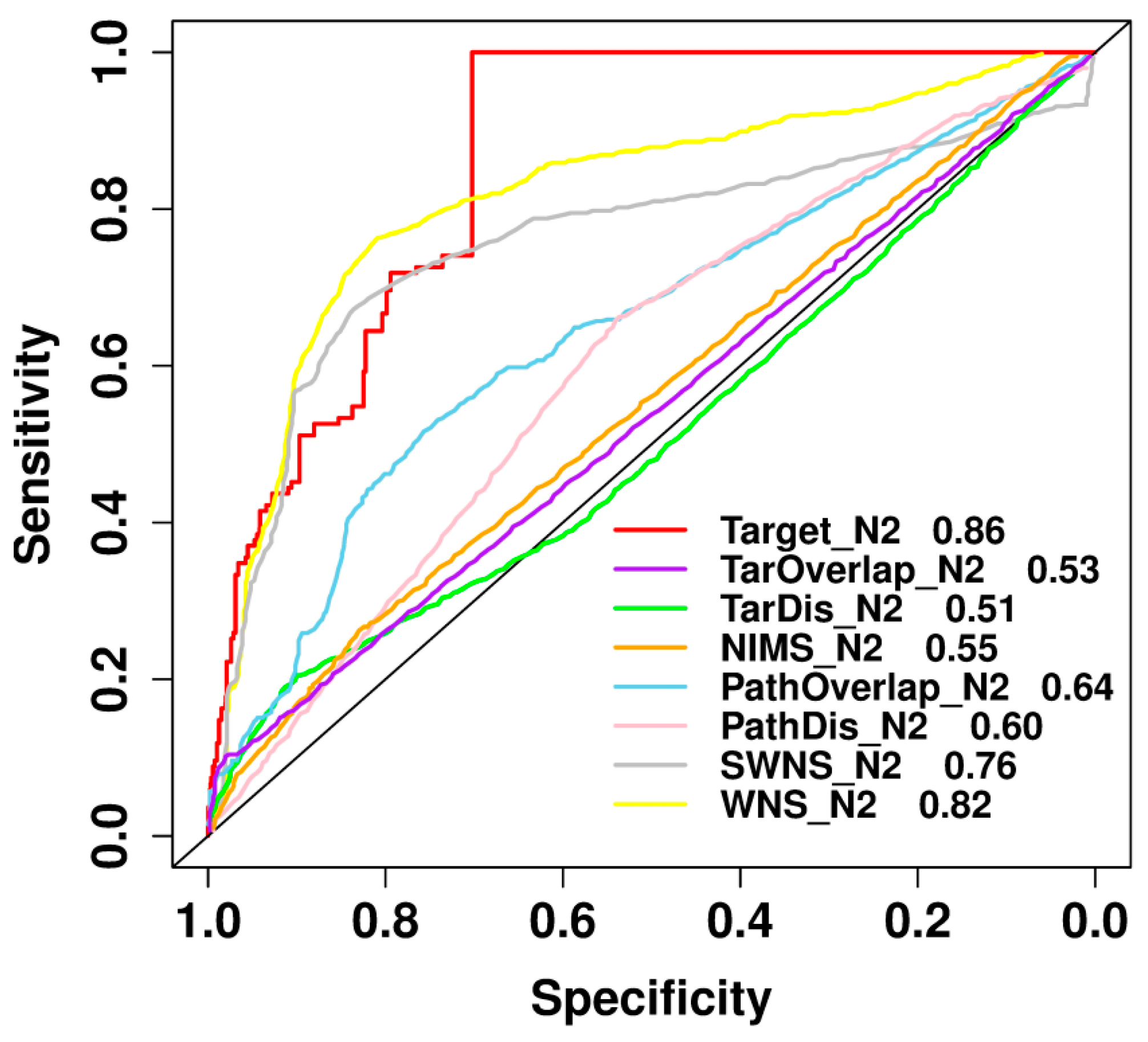
| Positive-to-Negative Ratio | Accuracy | F-Measure | MCC | Recall | Precision |
|---|---|---|---|---|---|
| 1:1 | 0.6800 | 0.6667 | 0.3612 | 0.6400 | 0.6957 |
| 1:2 | 0.6667 | 0.5098 | 0.2638 | 0.5652 | 0.4643 |
| 1:3 | 0.6832 | 0.3043 | 0.0992 | 0.3043 | 0.3043 |
| Feature Type | Accuracy | F-Measure | MCC | Recall | Precision |
|---|---|---|---|---|---|
| Targets | 0.7034 | 0.6431 | 0.4771 | 0.5000 | 0.9008 |
| Side effect | 0.6800 | 0.6667 | 0.3612 | 0.6400 | 0.6957 |
| Pathways | 0.6238 | 0.6174 | 0.2474 | 0.6216 | 0.6133 |
| Enzymes | 0.6115 | 0.6904 | 0.2144 | 0.8095 | 0.6018 |
| Transporters | 0.5339 | 0.5865 | 0.1216 | 0.7500 | 0.4815 |
| Category | Feature | Source | Dimension (N1) | Dimension (N2) |
|---|---|---|---|---|
| Pharmacodynamics | Targets | DrugBank | 681 | 787 |
| Pathways | KEGG | 255 | 263 | |
| Pharmacokinetic | Enzymes | DrugBank | 135 | 146 |
| Transporters | DrugBank | 76 | 86 | |
| Phenotypic | Side effect | SIDER | 3005 | 3889 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, L.-Y.; Dai, H.; Xu, Q.; Junaid, M.; Peng, S.-L.; Zhu, X.; Xiong, Y.; Wei, D.-Q. Prediction of Effective Drug Combinations by an Improved Naïve Bayesian Algorithm. Int. J. Mol. Sci. 2018, 19, 467. https://doi.org/10.3390/ijms19020467
Bai L-Y, Dai H, Xu Q, Junaid M, Peng S-L, Zhu X, Xiong Y, Wei D-Q. Prediction of Effective Drug Combinations by an Improved Naïve Bayesian Algorithm. International Journal of Molecular Sciences. 2018; 19(2):467. https://doi.org/10.3390/ijms19020467
Chicago/Turabian StyleBai, Li-Yue, Hao Dai, Qin Xu, Muhammad Junaid, Shao-Liang Peng, Xiaolei Zhu, Yi Xiong, and Dong-Qing Wei. 2018. "Prediction of Effective Drug Combinations by an Improved Naïve Bayesian Algorithm" International Journal of Molecular Sciences 19, no. 2: 467. https://doi.org/10.3390/ijms19020467
APA StyleBai, L.-Y., Dai, H., Xu, Q., Junaid, M., Peng, S.-L., Zhu, X., Xiong, Y., & Wei, D.-Q. (2018). Prediction of Effective Drug Combinations by an Improved Naïve Bayesian Algorithm. International Journal of Molecular Sciences, 19(2), 467. https://doi.org/10.3390/ijms19020467







