MiR-19a Overexpression in FTC-133 Cell Line Induces a More De-Differentiated and Aggressive Phenotype
Abstract
1. Introduction
2. Results
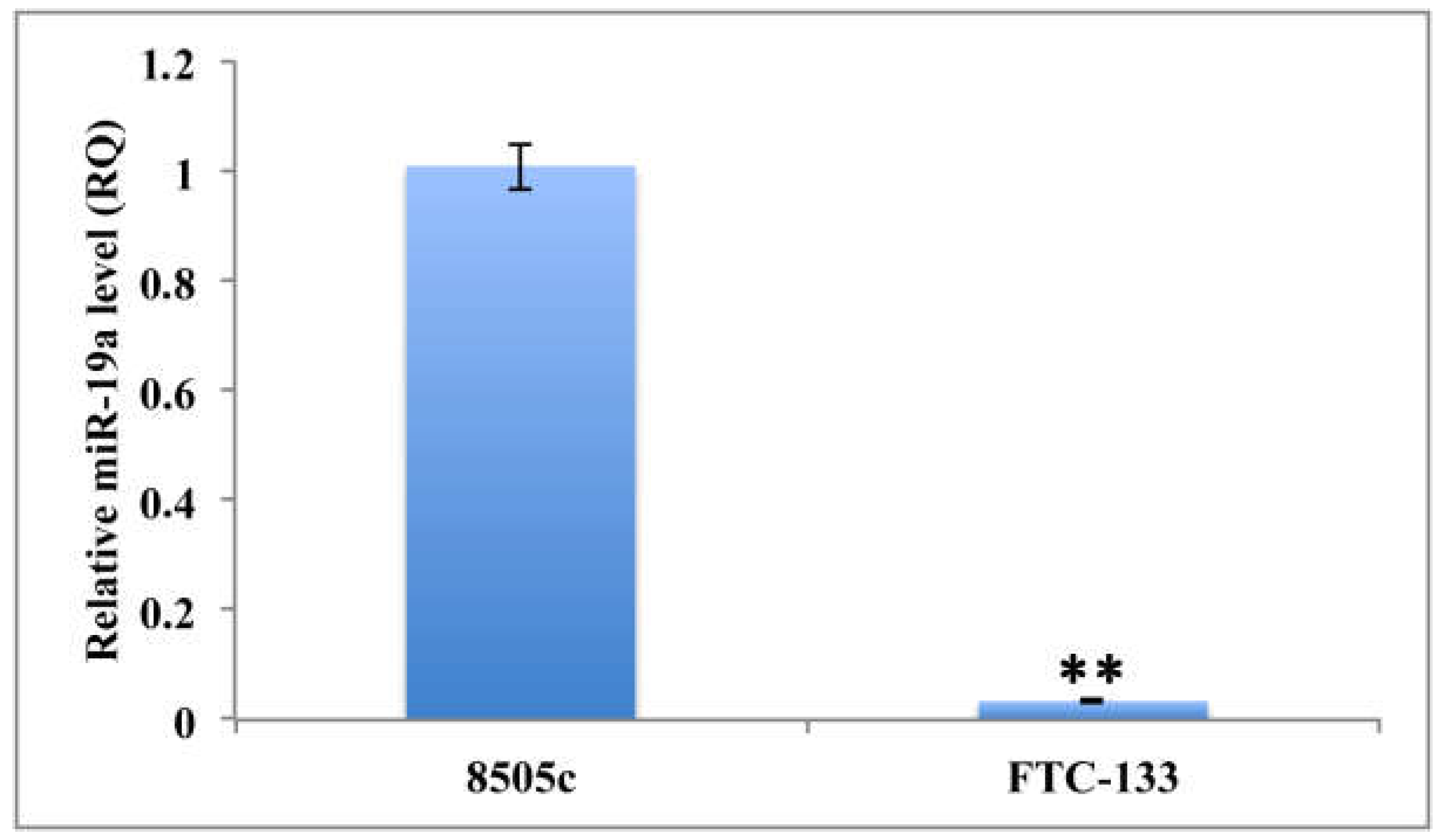
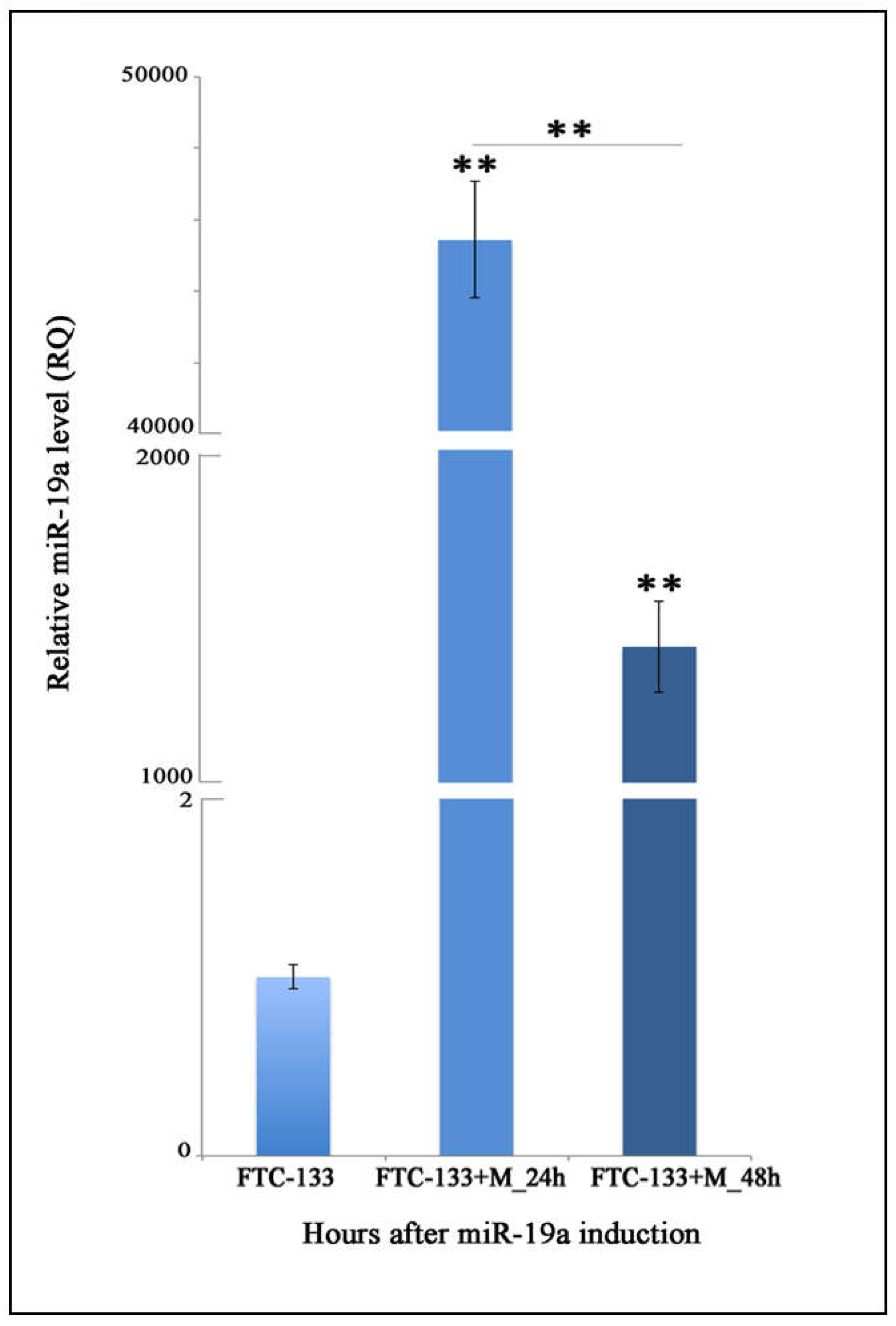
2.1. Quantitation of MiR-19a Level in Basal and Overexpressing Conditions
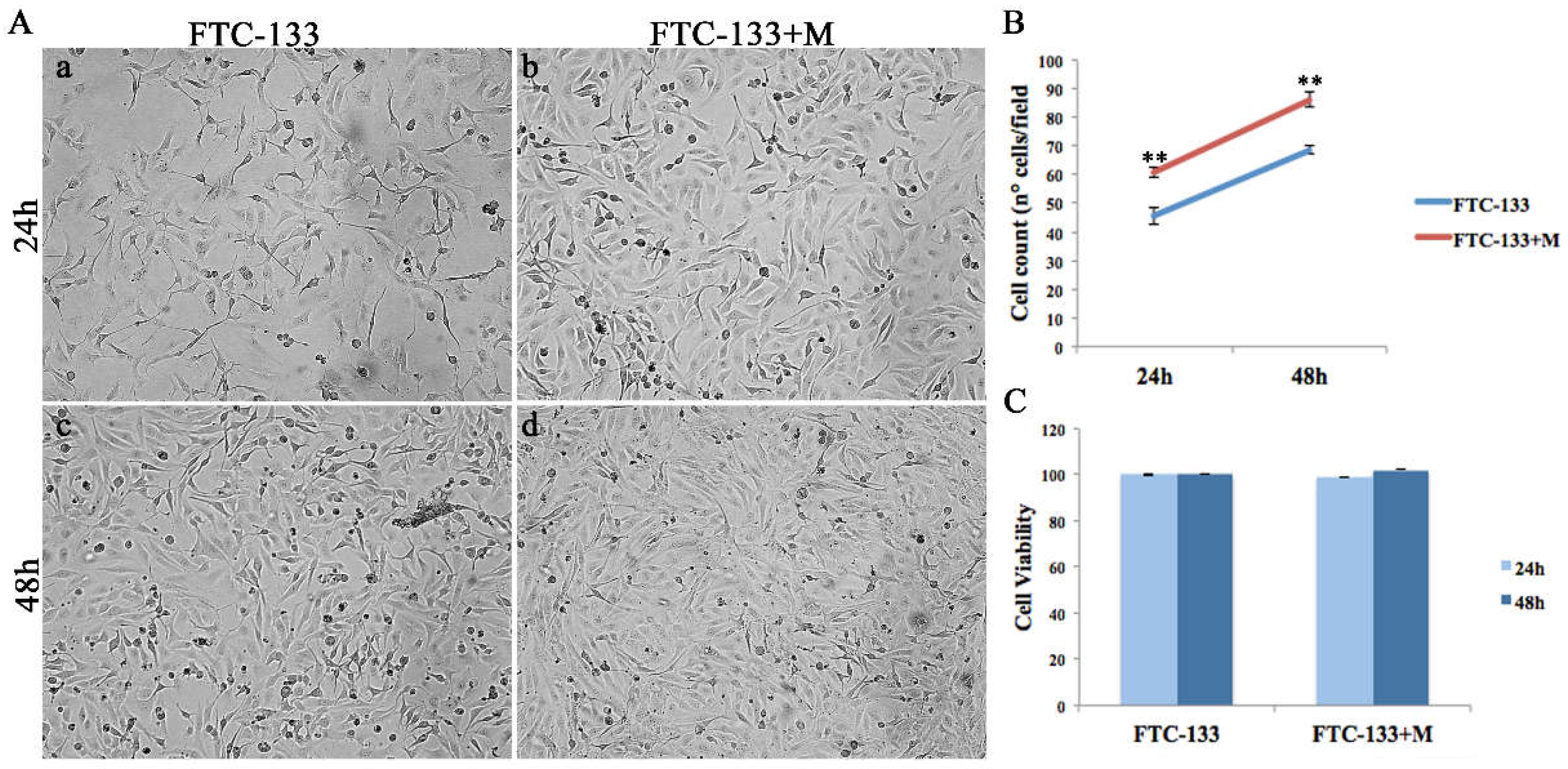
2.2. MiR-19a Overexpression on FTC-133 Induces Phenotypic Changes on Cell Morphology
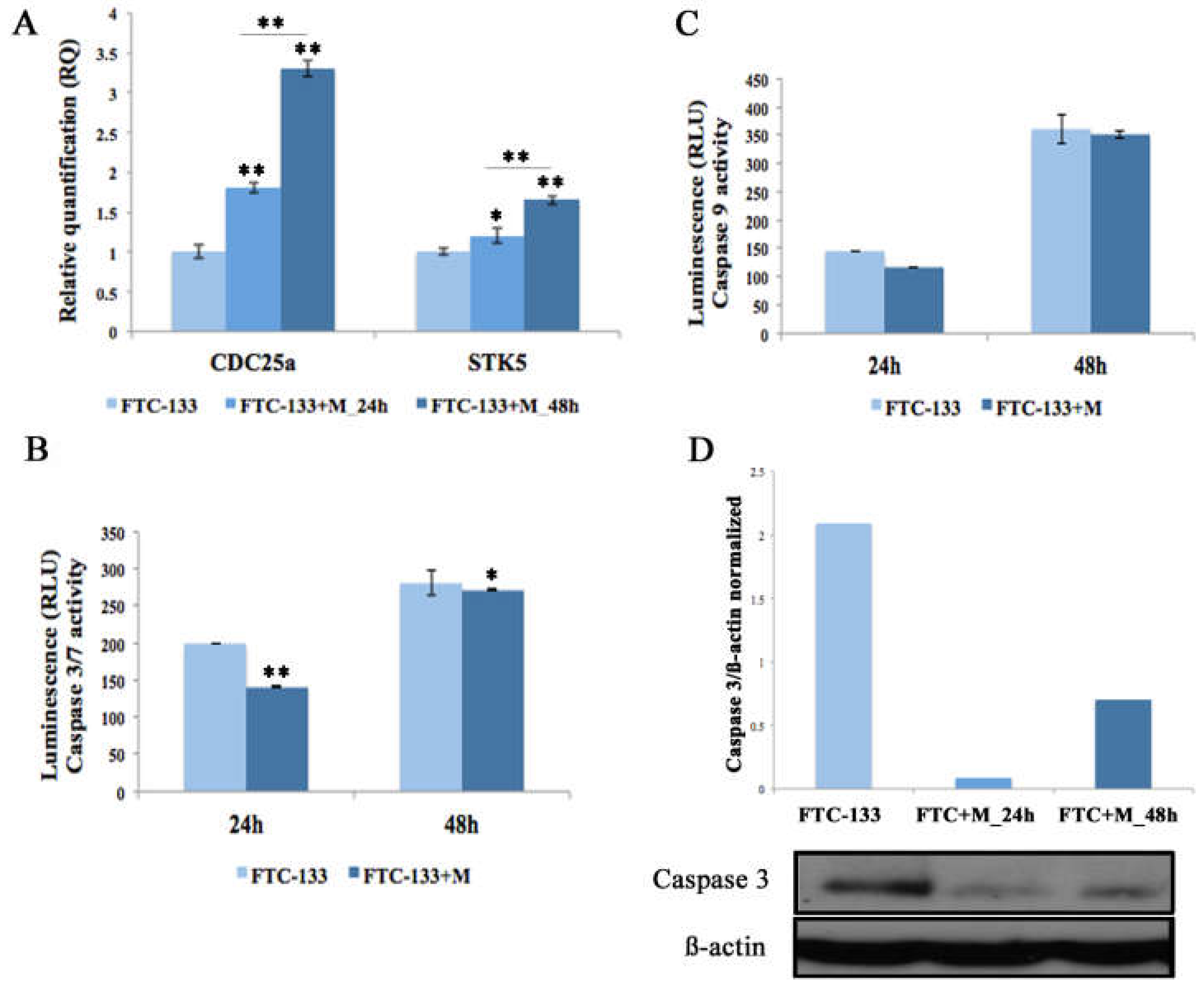
2.3. MiR-19a Overexpression on FTC-133 Promotes Proliferation and Cell Viability and Reduces Apoptosis
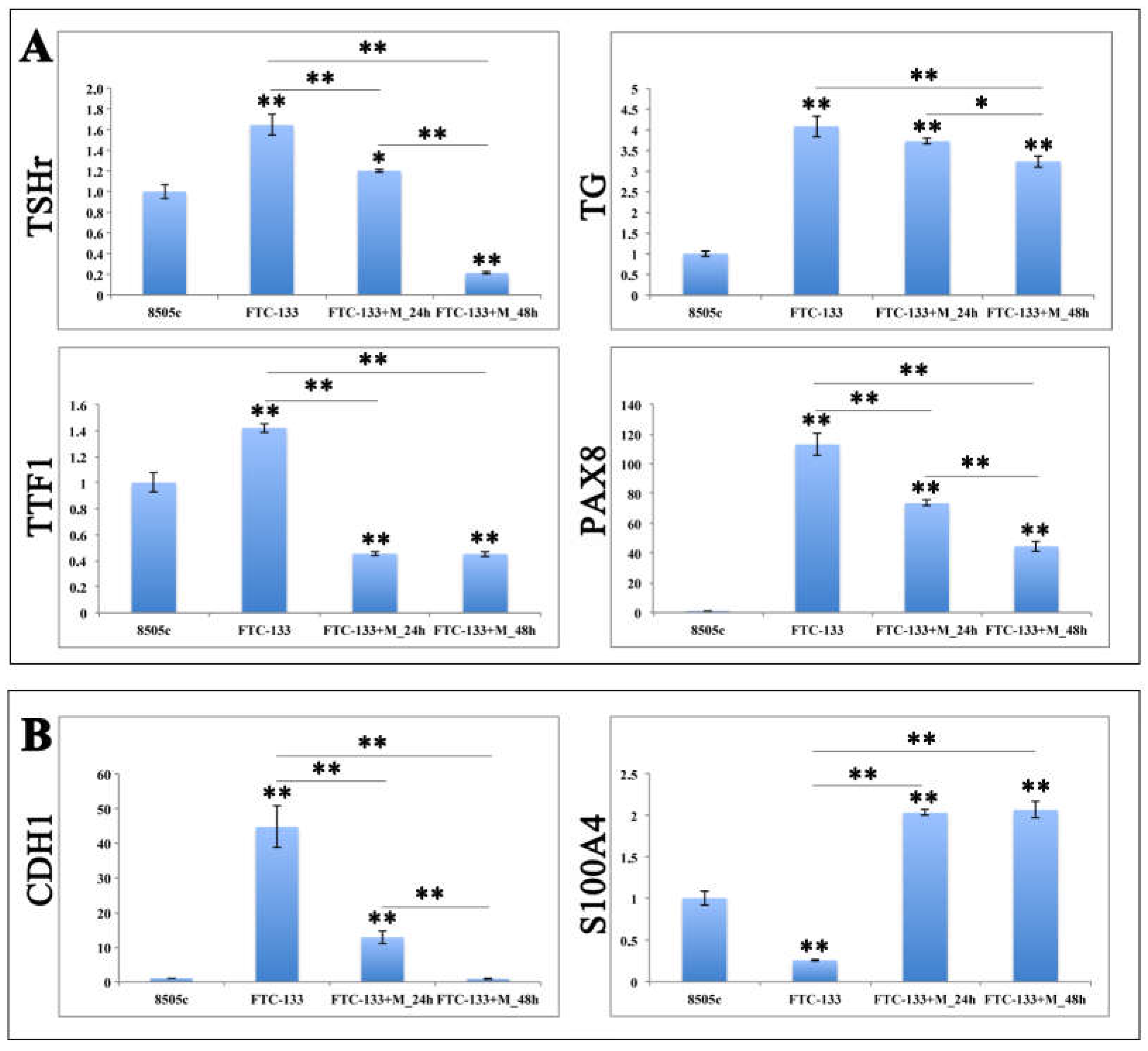
2.4. MiR-19a Overexpression on FTC-133 Alters the Expression of Genes Related to Thyroid Differentiation and Poor Prognosis
3. Discussion
4. Material and Methods
4.1. Cell Culture
4.2. MiRNA Mimic/Inhibitor Transfection on FTC-133 and 8505c Cells
4.3. Cell Proliferation Analysis Through DAPI and Trypan Blue Staining
4.4. Measurement of Cell Viability Through MTT Assay
4.5. Caspase-3/-7 and Caspase-9 Analyses
4.6. qRT-PCR
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kitahara, C.M.; Sosa, J.A. The changing incidence of thyroid cancer. Nat. Rev. Endocrinol. 2016, 12, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Wartofsky, L. Increasing world incidence of thyroid cancer: Increased detection or higher radiation exposure? Hormones (Athens) 2010, 9, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Ezzat, S.; Asa, S.L. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat. Rev. Cancer 2006, 6, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Sherman, S.I.; Tuttle, R.M. American Thyroid Association Guidelines Taskforce. Thyroid 2006, 16, 109–142. [Google Scholar] [CrossRef] [PubMed]
- De Lellis, R.A.; Lloyd, R.V.; Heitz, P.U.; Eng, C. World Health Organization International Classification of Tumors. In Pathology and Genetics of Tumors of Endocrine Organs, 1st ed.; IARC Press: Lyon, France, 2004. [Google Scholar]
- Schneider, D.F.; Chen, H.; Sippel, R.S. Impact of lymph node ratio on survival in papillary thyroid cancer. Ann. Surg. Oncol. 2013, 20, 1906–1911. [Google Scholar] [CrossRef]
- Gillanders, S.L.; O’Neill, J.P. Prognostic markers in well differentiated papillary and follicular thyroid cancer (WDTC). Eur. J. Surg. Oncol. 2018, 44, 286–296. [Google Scholar] [CrossRef]
- Khetrapal, S.; Rana, S.; Jetley, S.; Jairajpuri, Z. Poorly differentiated carcinoma of thyroid: Case report of an uncommon entity. J. Cancer Res. Ther. 2018, 14, 1142–1144. [Google Scholar]
- Parenti, R.; Salvatorelli, L.; Magro, G. Anaplastic Thyroid Carcinoma: Current Treatments and Potential New Therapeutic Options with Emphasis on TfR1/CD71. Int. J. Endocrinol. 2014, 2014, 685396. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Forte, S.; La Rosa, C.; Pecce, V.; Rosignolo, F.; Memeo, L. The Role of MicroRNAs in Thyroid Carcinomas. Anticancer Res. 2015, 35, 2037–2048. [Google Scholar] [PubMed]
- Li, M.; Li, J.; Ding, X.; He, M.; Cheng, S.Y. microRNA and Cancer. AAPS J. 2010, 12, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.B. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Negrini, M. MicroRNAs and Their Role in Cancer. ELS 2012. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Condorelli, G.L.; Croce, C.M.; Condorelli, G. MicroRNAs as regulators of death receptors signaling. Cell Death Differ. 2010, 17, 200–208. [Google Scholar] [CrossRef]
- Garofalo, M.; Quintavalle, C.; Romano, G.; Croce, C.M.; Condorelli, G. miR221/222 in cancer: Their role in tumor progression and response to therapy. Curr. Mol. Med. 2012, 12, 27–33. [Google Scholar] [CrossRef]
- Croce, C.M.; Calin, G.A. miRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Chu, K.-M. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J. Gastroenterol. 2014, 20, 10432–10439. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Li, C.; Liu, C.; Zhao, S.; Wang, Y.; Fu, Z. miRNAs in papillary thyroid carcinoma and their associations with the clinical characteristics of PTC. Cancer Biomark. 2017, 18, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Panoutsopoulou, K.; Avgeris, M.; Scorilas, A. miRNA and long non-coding RNA: Molecular function and clinical value in breast and ovarian cancers. Expert Rev. Mol. Diagn. 2018. [Google Scholar] [CrossRef] [PubMed]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Xia, J.T.; Chen, L.Z.; Jian, W.H.; Wang, K.B.; Yang, Y.Z.; He, W.L.; He, Y.L.; Chen, D.; Li, W. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-KB signaling. J. Transl. Med. 2014, 5, 33. [Google Scholar] [CrossRef]
- Michael, M.Z.; O’Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar]
- Ciafrè, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, C.; Li, C.; Xue, J.; Zhao, S.; Zhan, P.; Lin, Y.; Zhang, P.; Jiang, A.; Chen, W. Effects of microRNA-221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene 2015, 572, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Luzi, E.; Brandi, M.L. MicroRNA Role in Thyroid Cancer Development. J. Thyroid Res. 2011, 2011, 407123. [Google Scholar] [CrossRef] [PubMed]
- Pallante, P.; Visone, R.; Ferracin, M.; Ferraro, A.; Berlingieri, M.T.; Troncone, G.; Chiappetta, G.; Liu, C.G.; Santoro, M.; Negrini, M.; et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer 2006, 13, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Bonauer, A.; Dimmeler, S. The microRNA-17-92 cluster: Still a miRacle? Cell Cycle 2009, 8, 3866–3873. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, J.; Kang, Y.; He, Y.; Liang, B.; Yang, P.; Yu, Z. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J. Exp. Clin. Cancer Res. 2014, 33, 67. [Google Scholar] [CrossRef] [PubMed]
- Lepore, I.; Dell’Aversana, C.; Pilyugin, M.; Conte, M.; Nebbioso, A.; De Bellis, F.; Tambaro, F.P.; Izzo, T.; Garcia-Manero, G.; Ferrara, F.; et al. HDAC inhibitors repress BARD1 isoform expression in acute myeloid leukemia cells via activation of miR-19a and/or b. PLoS ONE 2013, 8, e83018. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, R.; Yang, F.; Cheng, R.; Chen, X.; Cui, S.; Gu, Y.; Sun, W.; You, C.; Liu, Z.; et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol. Cancer 2017, 16, 53. [Google Scholar] [CrossRef]
- Huang, L.; Wang, X.; Wen, C.; Yang, X.; Song, M.; Chen, J.; Wang, C.; Zhang, B.; Wang, L.; Iwamoto, A.; et al. Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α inducedepithelial-to-mesenchymal transition in colorectal cancer. Sci. Rep. 2015, 5, 13350. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, Z.; An, Y.; Hu, H.; Yin, J.; Zhang, P.; Nie, Y.; Wu, K.; Shi, Y.; Fan, D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014, 5, e1144. [Google Scholar] [CrossRef]
- Chen, L.; Li, C.; Zhang, R.; Gao, X.; Qu, X.; Zhao, M.; Qiao, C.; Xu, J.; Li, J. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011, 309, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Jin, P.; Ding, C.; Liu, W. miR-19a/b modulates lung cancer cells metastasis through suppression of MXD1 expression. Oncol Lett. 2016, 12, 1901–1905. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, S.; Zhu, L.; Zhang, Q.; Ren, Y. MiR-19a negatively regulated the expression of PTEN and promoted the growth of ovarian cancer cells. Gene 2018, 670, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Jia, Z.; Li, B.; Zhang, A.; Wang, G.; Pu, P.; Chen, Z.; Wang, Z.; Yang, W. MiR-19 regulates the proliferation and invasion of glioma by RUNX3 via β-catenin/Tcf-4 signalling. Oncotarget 2017, 8, 110785–110796. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Wang, K.; Zhang, A.; Wang, G.; Kang, C.; Han, L.; Pu, P. miR-19a and miR-19b overexpression in gliomas. Pathol. Oncol. Res. 2013, 19, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Ma, X.; Zhang, Y.; Liu, Y.N.; Chen, X.; Gong, H.; Yao, Y.; Liu, K.; Zhang, X. MicroRNA-19a and microRNA-19b promote the malignancy of clear cell renal cell carcinoma through targeting the tumor suppressor RhoB. PLoS ONE 2018, 13, e0192790. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Y.; Zhang, T.H.; Qu, L.M.; Feng, J.P.; Tian, L.L.; Zhang, B.H.; Li, D.D.; Sun, Y.N.; Liu, M. MiR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int. J. Clin. Exp. Pathol. 2013, 7, 56–63. [Google Scholar]
- Yamamoto, K.; Ito, S.; Hanafusa, H.; Shimizu, K.; Ouchida, M. Uncovering direct targets of miR-19a involved in lung cancer progression. PLoS ONE 2015, 10, e0137887. [Google Scholar] [CrossRef]
- Fu, F.; Wan, X.; Wang, D.; Kong, Z.; Zhang, Y.; Huang, W.; Wang, C.; Wu, H.; Li, Y. MicroRNA-19a acts as a prognostic marker and promotes prostate cancer progression via inhibiting VPS37A expression. Oncotarget 2018, 9, 1931–1943. [Google Scholar] [CrossRef]
- Paiva, M.M.; Kimura, T.E.; Coltri, P.P. miR18a and miR19a Recruit Specific Proteins for Splicing in Thyroid Cancer Cells. Cancer Genom. Proteom. 2017, 14, 373–382. [Google Scholar]
- Sadow, P.M.; Faquin, W.C. Poorly differentiated thyroid carcinoma: An incubating entity. Front. Endocrinol. 2012, 3, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Ranganath, R.; Shah, M.A.; Shah, A.R. Anaplastic thyroid cancer. Curr. Opin. Endocrinol. Diabetes Obes. 2015, 22, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, E.; Romei, C.; Biagini, A.; Sabini, E.; Agate, L.; Mazzeo, S.; Materazzi, G.; Sellari-Franceschini, S.; Ribechini, A.; Torregrossa, L.; et al. Anaplastic thyroid carcinoma: From clinicopathology to genetics and advanced therapies. Nat. Rev. Endocrinol. 2017, 13, 644–660. [Google Scholar] [CrossRef] [PubMed]
- Ricarte-Filho, J.C.; Ryder, M.; Chitale, D.A.; Rivera, M.; Heguy, A.; Ladanyi, M.; Janakiraman, M.; Solit, D.; Knauf, J.A.; Tuttle, M.R.; et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009, 69, 4885–4893. [Google Scholar] [CrossRef]
- Smallridge, R.C.; Marlow, L.A.; Copland, J.A. Anaplastic thyroid cancer: Molecular pathogenesis and emerging therapies. Endocr. Relat. Cancer 2009, 16, 17–44. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rostan, G.; Tallini, G.; Herrero, A.; D’Aquila, T.G.; Carcangiu, M.L.; Rimm, D.L. Frequent mutation and nuclear localization of β-catenin in anaplastic thyroid carcinoma. Cancer Res. 1999, 59, 1811–1815. [Google Scholar] [PubMed]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef]
- Fuziwara, C.S.; Kimura, E.T. MicroRNA Deregulation in Anaplastic Thyroid Cancer Biology. Int. J. Endocrinol. 2014, 2014, 743450. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Kimura, E.T.; Gandhi, M.; Biddinger, P.W.; Knauf, J.A.; Basolo, F.; Zhu, Z.; Giannini, R.; Salvatore, G.; Fusco, A.; et al. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J. Clin. Endocrinol. Metab. 2003, 88, 5399–5404. [Google Scholar] [CrossRef]
- Fuziwara, C.S.; Kimura, E. High Iodine Blocks a Notch/miR-19 Loop Activated by the BRAFV600E Oncoprotein and Restores the Response to TGFβ in Thyroid Follicular Cells. Thyroid 2014, 24, 453–462. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, A.; Hao, Y.; Wang, G.; Jia, Z. The emerging role of miR-19 in glioma. J. Cell Mol. Med. 2018, 22, 4611–4616. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.Y.; Kong, J.R.; Zhao, C.S.; Xiao, Y.C.; Peng, T.; Liu, Y.; Wang, W.N. Molecular characterization and function of a PTEN gene from Litopenaeus vannamei after Vibrio alginolyticus challenge. Dev. Comp. Immunol. 2016, 59, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Forte, S.; Pagliuca, A.; Maniscalchi, E.T.; Gulino, R.; Calabrese, G.; Ricci-Vitiani, L.; Pallini, R.; Signore, M.; Parenti, R.; De Maria, R.; et al. Gene Expression Analysis of PTEN Positive Glioblastoma Stem Cells Identifies DUB3 and Wee1 Modulation in a Cell Differentiation Model. PLoS ONE 2013, 8, e81432. [Google Scholar] [CrossRef] [PubMed]
- Opitz, R.; Maquet, E.; Zoenen, M.; Dadhich, R.; Costagliola, S. TSH Receptor Function Is Required for Normal Thyroid Differentiation in Zebrafish. Mol. Endocrinol. 2011, 25, 1579–1599. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Latif, R.; Davies, T.F. Thyroid Follicle Formation and Thyroglobulin expression in multipotent endodermal stem cells. Thyroid 2013, 23, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, D.; Di Loreto, C.; Beltrami, C.A.; Belfiore, A.; Di Lauro, R.; Damante, G. Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Res. 1994, 54, 4744–4749. [Google Scholar] [PubMed]
- Damante, G.; Tell, G.; Di Lauro, R. A unique combination of transcription factors controls differentiation of thyroid cells. Prog. Nucleic Acid Res. Mol. Biol. 2001, 66, 307–356. [Google Scholar]
- Martinez Barbera, J.P.; Clements, M.; Thomas, P.; Rodriguez, T.; Meloy, D.; Kioussis, D.; Beddington, R.S. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development 2000, 127, 2433–2445. [Google Scholar]
- Damante, G.; Di Lauro, R. Thyroid-specific gene expression. Biochim. Biophys. Acta 1994, 1218, 255–266. [Google Scholar] [CrossRef]
- Pasca di Magliano, M.; Di Lauro, R.; Zannini, M. Pax8 has a key role in thyroid cell differentiation. Proc. Natl. Acad. Sci. USA 2000, 97, 13144–13149. [Google Scholar] [CrossRef]
- Ros, P.; Rossi, D.L.; Acebrón, A.; Santisteban, P. Thyroid-specific gene expression in the multi-step process of thyroid carcinogenesis. Biochimie 1999, 81, 389–396. [Google Scholar] [CrossRef]
- van Staveren, W.C.G.; Solís, D.W.; Delys, L.; Duprez, L.; Andry, G.; Franc, B.; Thomas, G.; Libert, F.; Dumont, J.E.; Detours, V.; et al. Human Thyroid Tumor Cell Lines Derived from Different Tumor Types Present a Common Dedifferentiated Phenotype. Cancer Res. 2007, 67, 8113–8120. [Google Scholar] [CrossRef] [PubMed]
- Fei, F.; Qu, J.; Zhang, M.; Li, Y.; Zhang, S. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget 2017, 8, 73219–73239. [Google Scholar] [CrossRef] [PubMed]
- Dahlmann, M.; Kobelt, D.; Walther, W.; Mudduluru, G.; Stein, U. S100A4 in Cancer Metastasis: Wnt Signaling-Driven Interventions for Metastasis Restriction. Cancers (Basel) 2016, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Lee, H.J.; Kim, S.H.; Kim, H.K.; Mok, Y.J.; Park, Y.T.; Choi, J.S.; Moon, H.Y. Expression of protein S100A4 is a predictor of recurrence in colorectal cancer. World J. Gastroenterol. 2010, 16, 3897–3904. [Google Scholar] [CrossRef] [PubMed]
- Yonemura, Y.; Endou, Y.; Kimura, K.; Fushida, S.; Bandou, E.; Taniguchi, E.; Kinoshita, K.; Ninomiya, I.; Sugiyama, K.; Heizmann, C.W.; et al. Inverse Expression of S100A4 and E-Cadherin Is Associated with Metastatic Potential in Gastric Cancer. Clin. Cancer. Res. 2000, 6, 4234–4242. [Google Scholar]
- Montero-Conde, C.; Martín-Campos, J.M.; Lerma, E.; Gimenez, G.; Martínez-Guitarte, J.L.; Combalía, N.; Montaner, D.; Matías-Guiu, X.; Dopazo, J.; de Leiva, A.; et al. Molecular profiling related to poor prognosis in thyroid carcinoma. Combining gene expression data and biological information. Oncogene 2008, 27, 1554–1561. [Google Scholar] [CrossRef]
- Chan, A.O.O. E-cadherin in gastric cancer. World J. Gastroenterol. 2006, 12, 199–203. [Google Scholar] [CrossRef]
- Takeichi, M. Cadherins in cancer: Implications for invasion and metastasis. Curr. Opin. Cell Biol. 1993, 5, 806–811. [Google Scholar] [CrossRef]
- Vicari, L.; La Rosa, C.; Forte, S.; Calabrese, G.; Colarossi, C.; Aiello, E.; Salluzzo, S.; Memeo, L. Differential expression of two activating transcription factor 5 isoforms in papillary thyroid carcinoma. OncoTargets Therapy 2016, 9, 6225–6231. [Google Scholar] [CrossRef]
- Yang, J.; Gao, T.; Tang, J.; Cai, H.; Lin, L.; Fu, S. Loss of microRNA-132 predicts poor prognosis in patients with primary osteosarcoma. Mol. Cell Biochem. 2013, 381, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Caifu, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calabrese, G.; Dolcimascolo, A.; Torrisi, F.; Zappalà, A.; Gulino, R.; Parenti, R. MiR-19a Overexpression in FTC-133 Cell Line Induces a More De-Differentiated and Aggressive Phenotype. Int. J. Mol. Sci. 2018, 19, 3944. https://doi.org/10.3390/ijms19123944
Calabrese G, Dolcimascolo A, Torrisi F, Zappalà A, Gulino R, Parenti R. MiR-19a Overexpression in FTC-133 Cell Line Induces a More De-Differentiated and Aggressive Phenotype. International Journal of Molecular Sciences. 2018; 19(12):3944. https://doi.org/10.3390/ijms19123944
Chicago/Turabian StyleCalabrese, Giovanna, Anna Dolcimascolo, Filippo Torrisi, Agata Zappalà, Rosario Gulino, and Rosalba Parenti. 2018. "MiR-19a Overexpression in FTC-133 Cell Line Induces a More De-Differentiated and Aggressive Phenotype" International Journal of Molecular Sciences 19, no. 12: 3944. https://doi.org/10.3390/ijms19123944
APA StyleCalabrese, G., Dolcimascolo, A., Torrisi, F., Zappalà, A., Gulino, R., & Parenti, R. (2018). MiR-19a Overexpression in FTC-133 Cell Line Induces a More De-Differentiated and Aggressive Phenotype. International Journal of Molecular Sciences, 19(12), 3944. https://doi.org/10.3390/ijms19123944






