Comparative Transcriptome Analysis between a Resistant and a Susceptible Wild Tomato Accession in Response to Phytophthora parasitica
Abstract
1. Introduction
2. Results
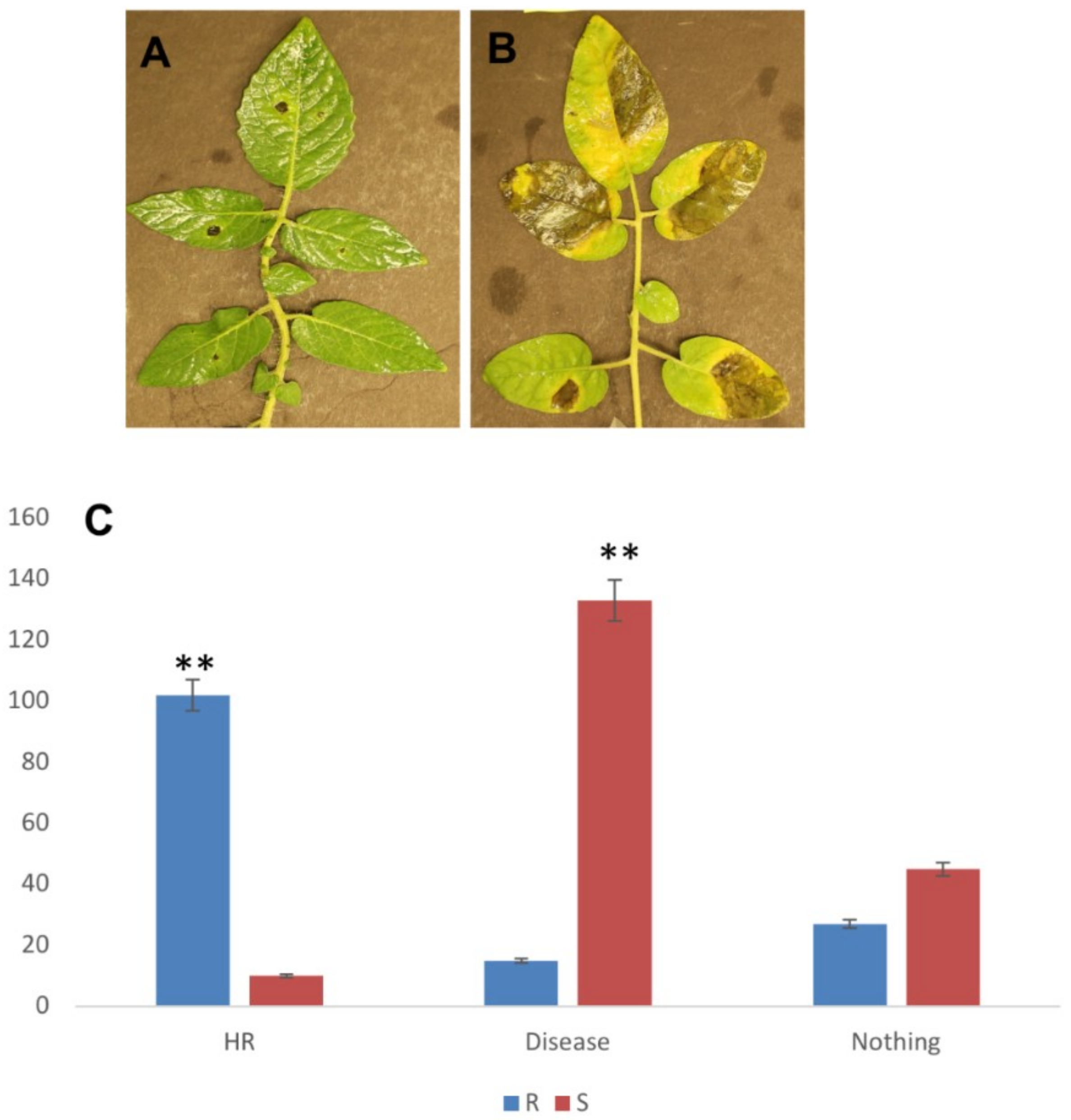
2.1. P. parasitica Infection on S. Pimpinellifolium Accessions
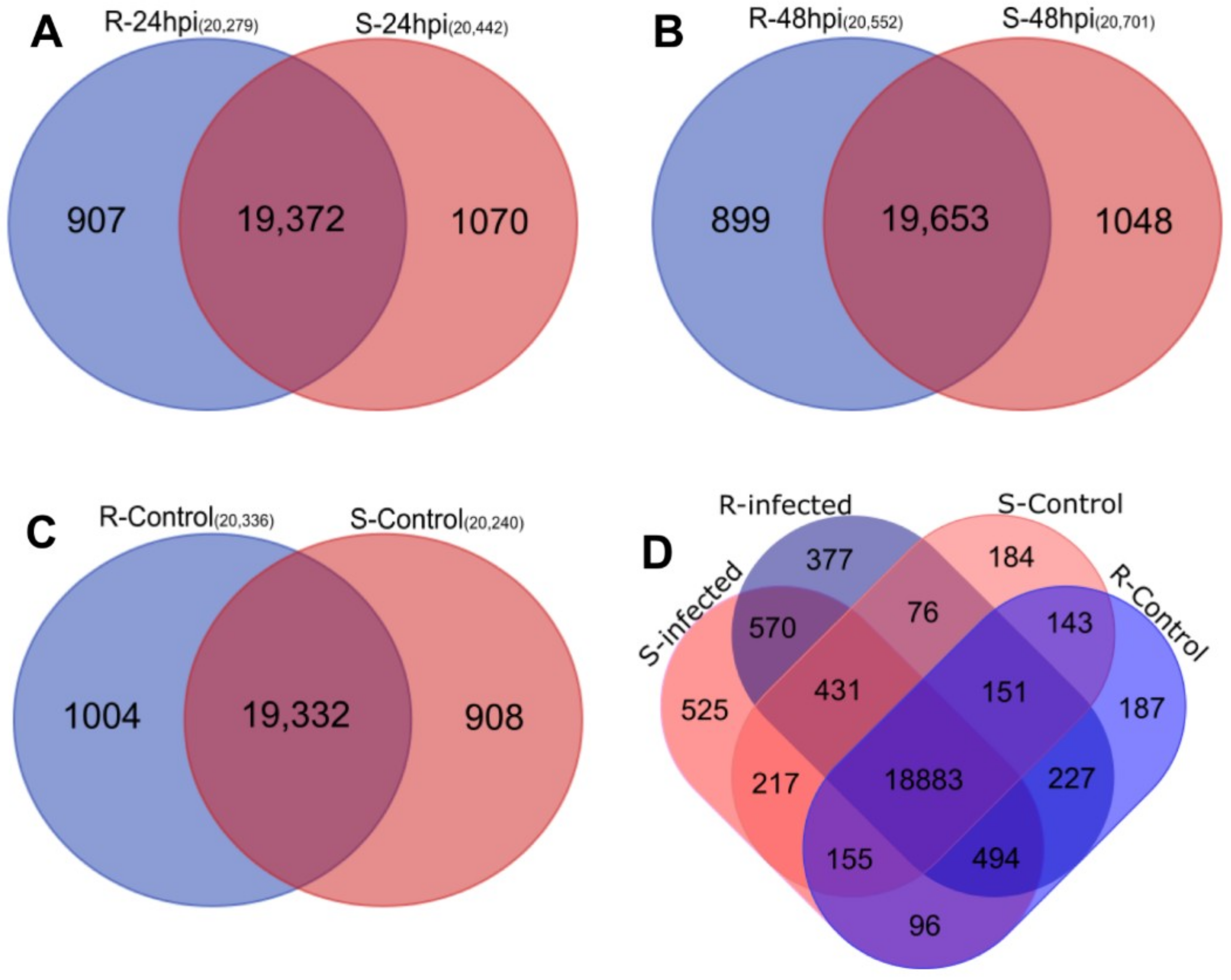
2.2. Analysis of RNA-Seq Data
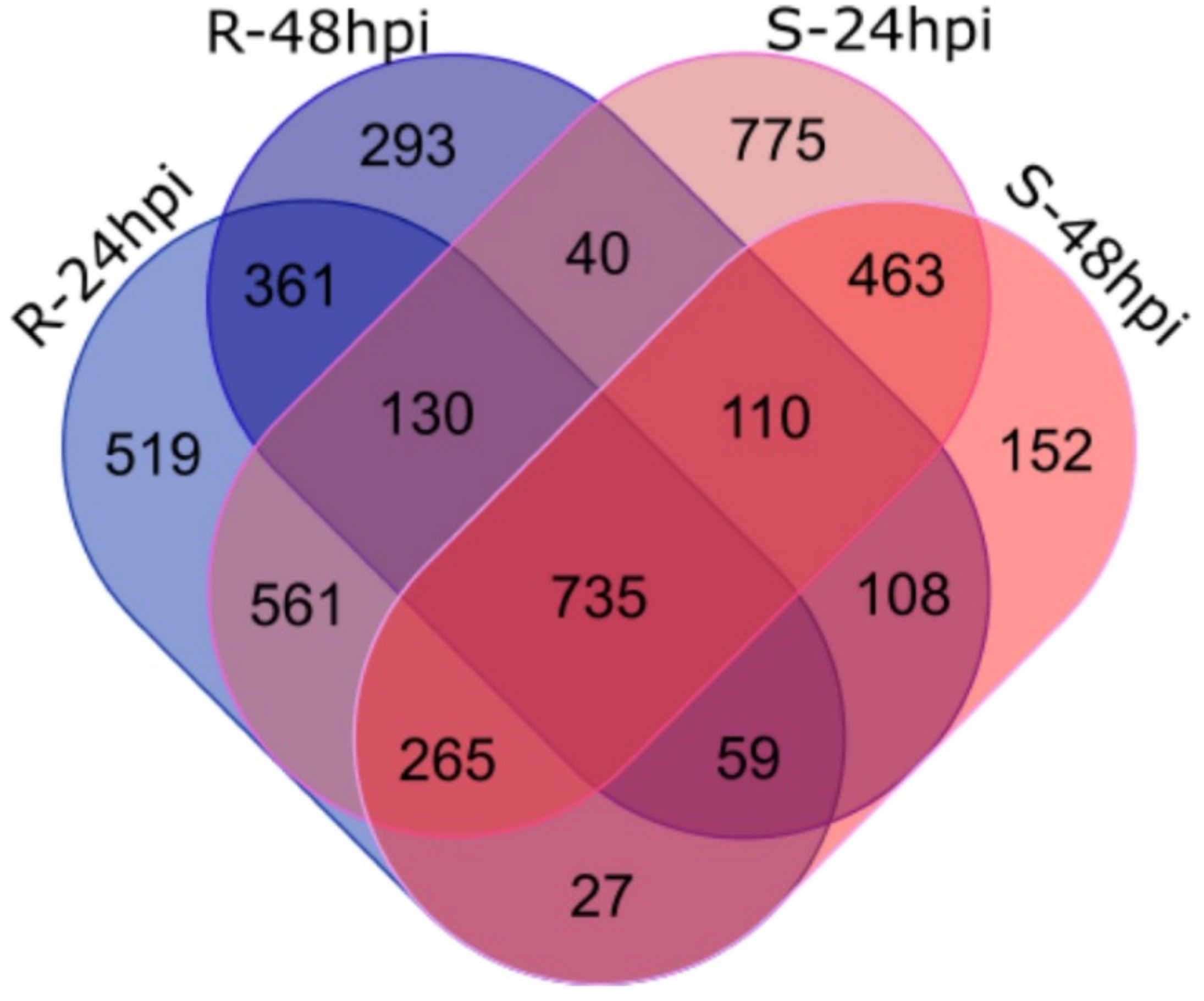
2.3. Differential Expression Analysis
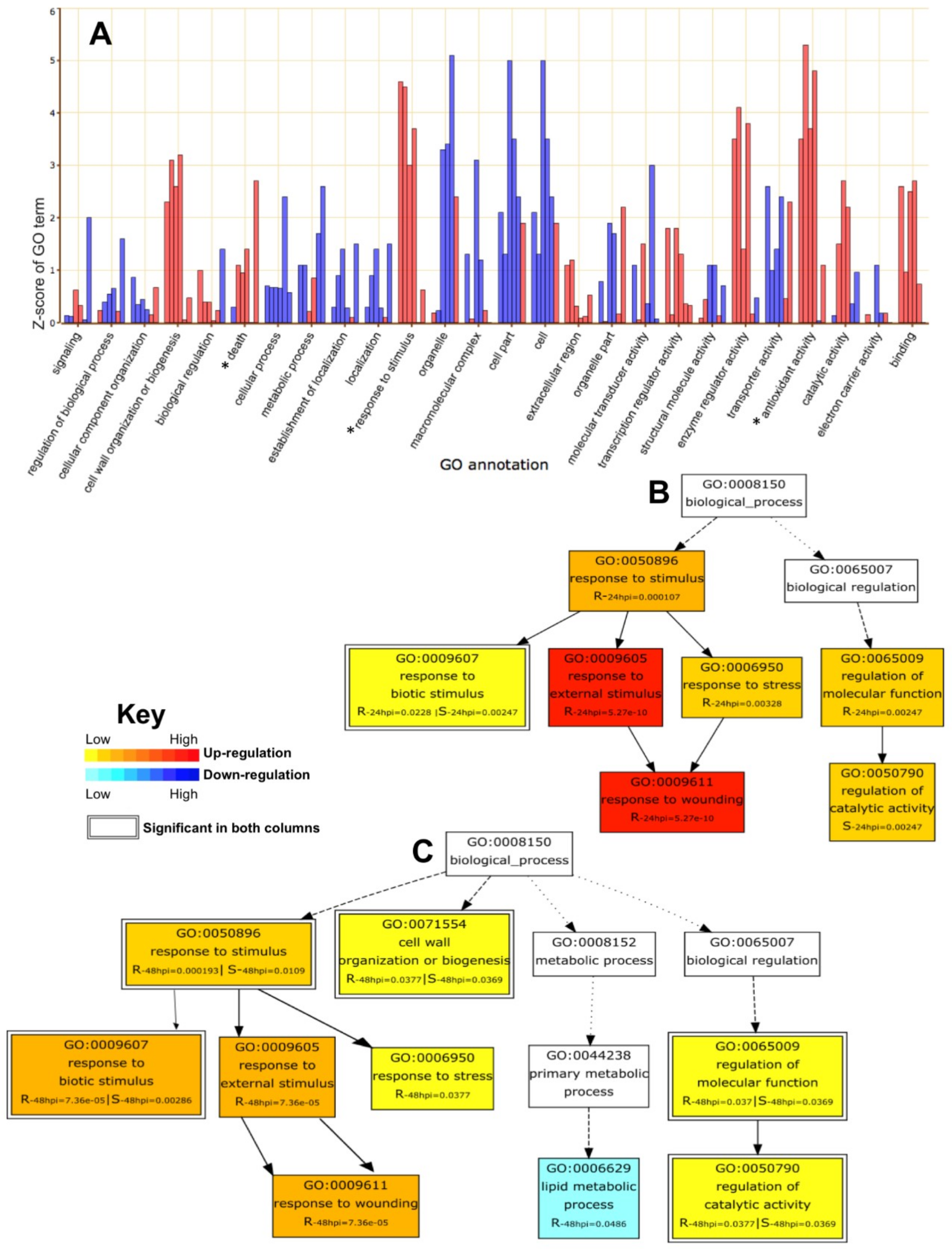
2.4. GO Enrichment Analysis of DEGs
2.5. Parametric Gene Set Enrichment Analysis Based on GO
2.6. KEGG Annotations
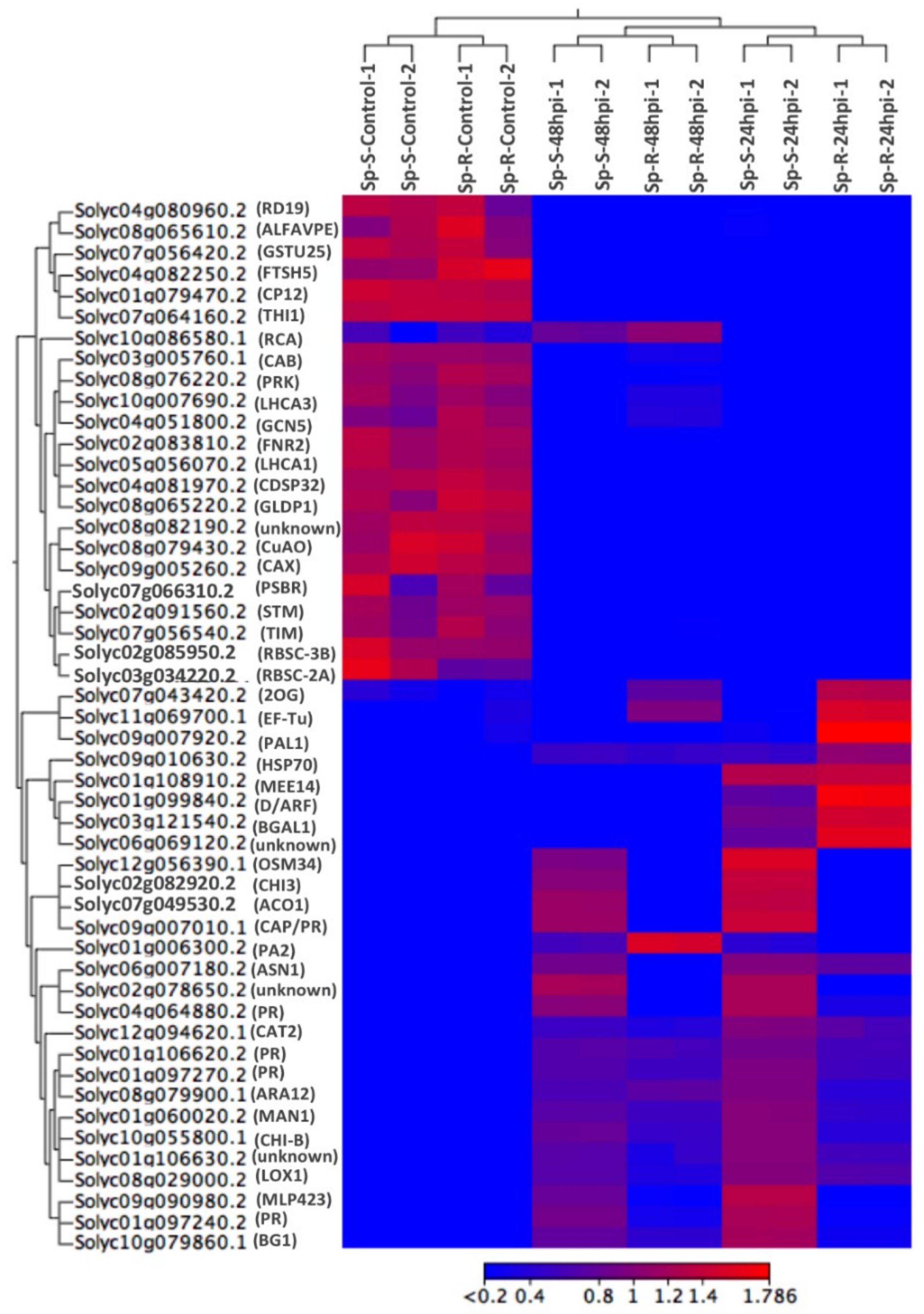
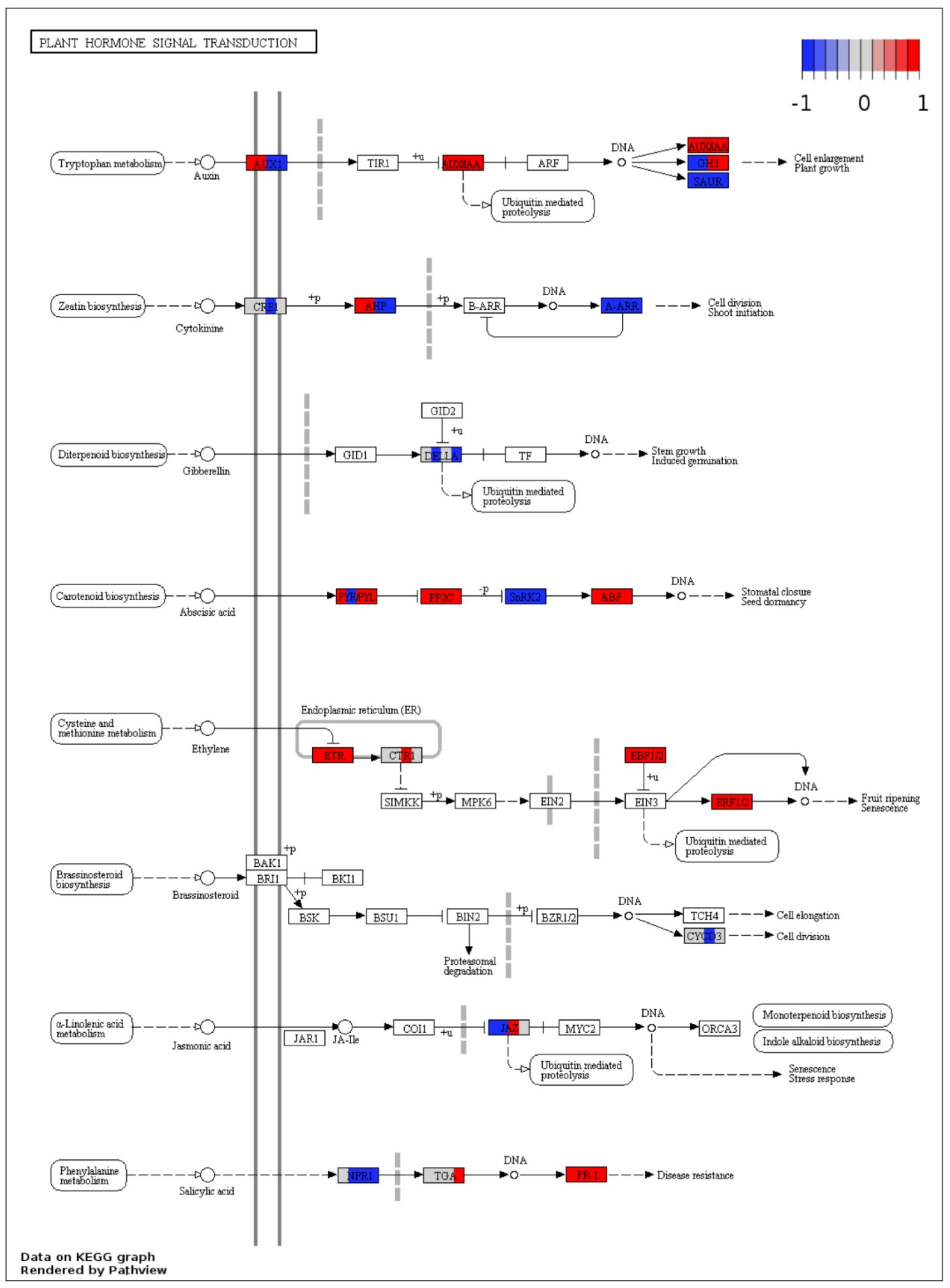
2.7. Visualization of DEGs in Plant Pathways
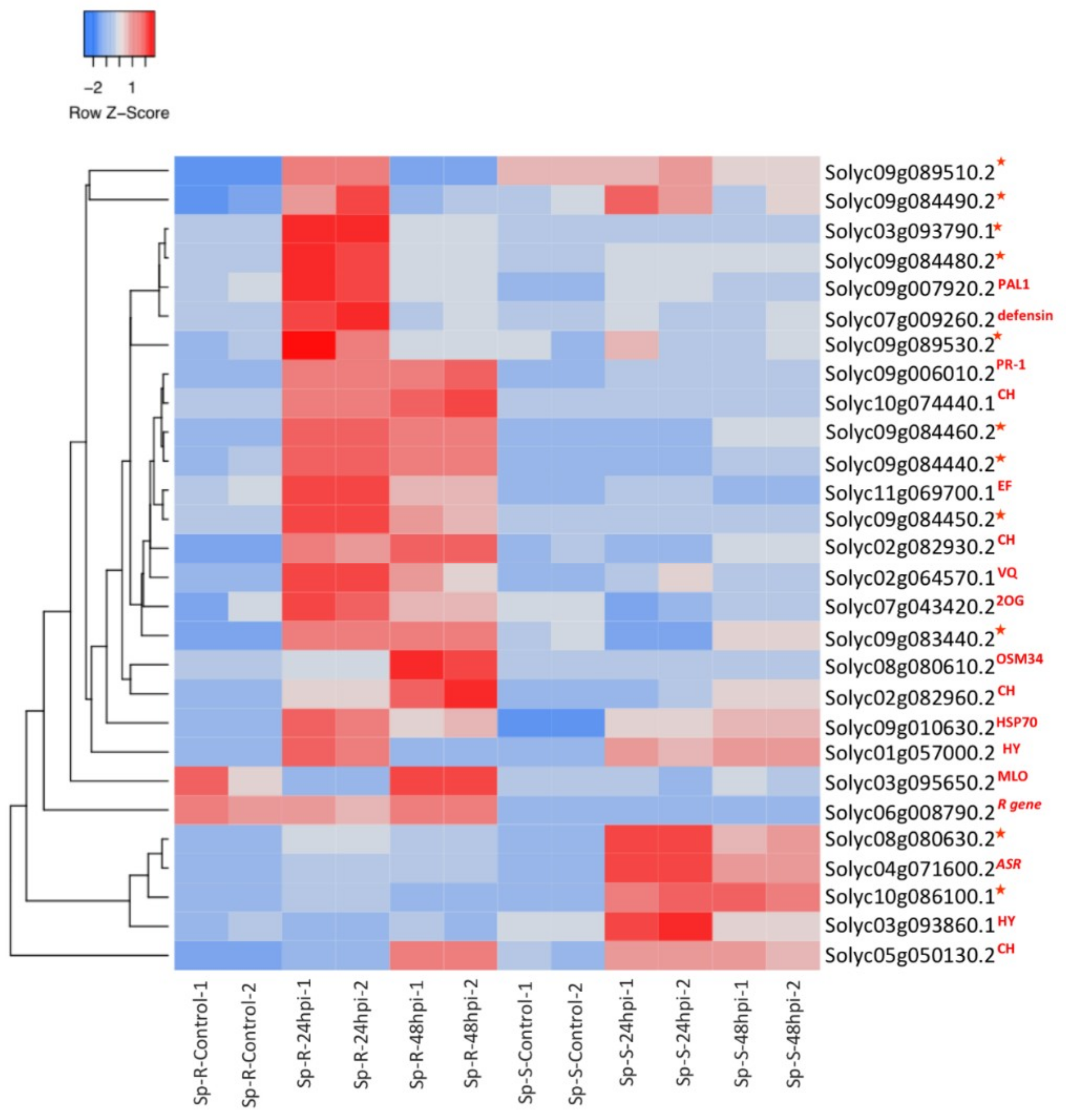
2.8. Validation of RNA-Seq Results with QRTPCR
3. Discussion
3.1. Antimicrobial Genes in Relevance to Resistance Response against P. parasitica
3.2. Contrasting Expression of Downy Mildew Susceptibility Factor in Response to P. parasitica
3.3. Induction of Plant Heat Stress Tolerance Related Genes
3.4. R Genes
4. Materials and Methods
4.1. P. parasitica Inoculation, RNA Extraction and Sequencing
4.2. RNA-Seq Analysis
4.3. Validation of RNA-Seq Results
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| cwt | Hundredweight |
| RPKM | Reads per kilobase million |
| hpi | Hours post inoculation |
| FDR | False discovery rate |
| GTP | Guanosine triphosphate |
| PHE | Phenylalanine |
| NADP | Nicotinamide adenine dinucleotide phosphate |
| KEGG | Kyoto encyclopedia of genes and genomes |
| KO | KEGG ontology |
| PR | Pathogenesis related |
| PR1 | Pathogenesis related protein 1 |
| ERF1 | Eukaryotic release factor 1 |
| EBF1/2 | Early B cell factor ½ |
| PP2C | Protein phosphatase 2C |
| ETR/ERS | Ethylene receptors |
| FLS2 | Flagellin sensing 2 |
| CAT1 | Cationic amino acid transporter 1 |
| PYR/PYL | ABA receptors |
| ChiB | Chitinase B |
| RTE1 | Reversion to ethylene sensitivity |
| ACS6 | 1-aminocyclopropane-1-carboxylate synthase 6 |
| OX1 | Oxidase 1 |
| JAZ | Jasmonate zim domain protein |
| NB- | Nucleotide binding- |
| LRRs | Leucine rich repeats |
| ITAG | International tomato annotation group |
References
- Anderson, J.P.; Gleason, C.A.; Foley, R.C.; Thrall, P.H.; Burdon, J.B.; Singh, K.B. Plants versus pathogens: An evolutionary arms race. Funct. Plant Biol. 2010, 37, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Doughari, J. An Overview of Plant Immunity. J. Plant Pathol. Microbiol. 2015, 6, 10–4172. [Google Scholar]
- USDA. Vegetables Annual Summary 2017. National Agricultural Statistics Service 2018. Available online: http://usda.mannlib.cornell.edu/usda/current/VegeSumm/VegeSumm-02-13-2018.pdf (accessed on 26 June 2018).
- Rodewald, J.; Trognitz, B. Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol. Plant Pathol. 2013, 14, 740–757. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, L.; Wang, X.; Vossen, J.; Li, G.; Li, T.; Zheng, Z.; Gao, J.; Guo, Y.; Visser, R.G.; et al. The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor. Appl. Genet. 2014, 127, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Xu, P.; Meng, J.; Li, J.; Jiang, N.; Luan, Y. Transcriptome signatures of tomato leaf induced by Phytophthora infestans and functional identification of transcription factor SpWRKY3. Theor. Appl. Genet. 2018, 131, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Luan, Y.; Jiang, N.; Bao, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lnc RNA 16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-B.; Luan, Y.-S.; Liu, Z. Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 2015, 155, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Le Berre, J.Y.; Engler, G.; Panabieres, F. Exploration of the late stages of the tomato Phytophthora parasitica interactions through histological analysis and generation of expressed sequence tags. New Phytol. 2008, 177, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Attard, A.; Gourgues, M.; Callemeyn-Torre, N.; Keller, H. The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol. 2010, 187, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Park, S.-C.; Hwang, I.; Cheong, H.; Nah, J.-W.; Hahm, K.-S.; Park, Y. Protease inhibitors from plants with antimicrobial activity. Int. J. Mol. Sci. 2009, 10, 2860–2872. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Cammue, B.P.; Thevissen, K. Plant defensins. Planta 2002, 216, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Le Berre, J.-Y.; Gourgues, M.; Samans, B.; Keller, H.; Panabieres, F.; Attard, A. Transcriptome dynamic of Arabidopsis roots infected with Phytophthora parasitica identifies VQ29, a gene induced during the penetration and involved in the restriction of infection. PLoS ONE 2017, 12, e0190341. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstdler, A.; Lotti, C.; de Giovanni, C.; Ricciardi, L.; Lindhout, P.; Visser, R.; et al. Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol. Plant-Microbe Interact. 2008, 21, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, C.; Rickauer, M.; Fournier, J.; Pouenat, M.-L.; Esquerre-Tugaye, M.-T. Lipoxygenase gene expression in the tobacco-Phytophthora parasitica nicotianae interaction. Plant Physiol. 1996, 112, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.; Van Strien, E. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- George, K.J.; Babu, O.R.; Kumar, I.V.; Eapen, J.S.; Anandaraj, M. Interplay of genes in plant-pathogen interactions: In planta expression and docking studies of a β 1, 3 glucanase gene from Piper colubrinum and a glucanase inhibitor gene from Phytophthora capsici. Physiol. Mol. Biol. Plants 2016, 22, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Schoina, C.; Bouwmeester, K.; Govers, F. Infection of a tomato cell culture by Phytophthora infestans; a versatile tool to study Phytophthora-host interactions. Plant Methods 2017, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Baebler, Å.P.; Stare, K.; Kova, M.; Blejec, A.; Prezelj, N.; Stare, T.A.; Kogovšek, P.; Pompe-Novak, M.; Rosahl, S.; Ravnikar, M.; et al. Dynamics of responses in compatible potato-Potato virus Y interaction are modulated by salicylic acid. PLoS ONE 2011, 6, e29009. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Saitoh, H.; Ito, A.; Fujisawa, S.; Kamoun, S.; Katou, S.; Yoshioka, H.; Terauchi, R. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 2003, 4, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X.; Zhang, F.; Dong, L.; Wu, J.; Cheng, Q.; Qi, D.; Yan, X.; Jiang, L.; Fan, S.; et al. Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 2017, 7, 7242. [Google Scholar] [PubMed]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.J.; Zhang, K. S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Env. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Hatano, K.; Miyauchi, Y.; Suwa, Y.; Sawano, Y.; Tanokura, M. A Secreted Protein with Plant-Specific Cysteine-Rich Motif Functions as a Mannose-Binding Lectin That Exhibits Antifungal Activity. Plant Physiol. 2014, 166, 766–778. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B.N.; Savory, E.A.; Vaillancourt, B.; Childs, K.L.; Hamilton, J.P.; Day, B.; Robin Buell, C. Expression profiling of Cucumis sativus in response to infection by Pseudoperonospora cubensis. PLoS ONE 2012, 7, e34954. [Google Scholar] [CrossRef] [PubMed]
- Judelson, H.S.; Ah-Fong, A.M.; Aux, G.; Avrova, A.O.; Bruce, C.; Cakir, C.; da Cunha, L.; Grenville-Briggs, L.; Latijnhouwers, M.; Ligterink, W.; et al. Gene expression profiling during asexual development of the late blight pathogen Phytophthora infestans reveals a highly dynamic transcriptome. Mol. Plant-Microbe Interact. 2008, 21, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Kunjeti, S.G.; Evans, T.A.; Marsh, A.G.; Gregory, N.F.; Kunjeti, S.; Meyers, B.C.; Kalavacharla, V.S.; Donofrio, N.M. RNA-Seq reveals infection-related global gene changes in Phytophthora phaseoli, the causal agent of lima bean downy mildew. Mol. Plant Pathol. 2012, 13, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Tu, Z.J.; Millett, B.P.; Bradeen, J.M. Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions. BMC Genom. 2013, 14, 340. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-R.; Zhang, B.-Y.; Xing, Y.-P.; Li, Q.-Y.; Li, Y.-P.; Tong, Y.-H.; Xu, J.Y. Transcriptomic analysis of the phytopathogenic oomycete Phytophthora cactorum provides insights into infection-related effectors. BMC genomics 2014, 15, 980. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.; Goodman, R.M.; Gut-Rella, M.; Glascock, C.; Weymann, K.; Friedrich, L.; Maddox, D.; Ahl-Goy, P.; Luntz, T.; Ward, E. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 1993, 90, 7327–7331. [Google Scholar] [CrossRef] [PubMed]
- Riviere, M.-P.; Marais, A.; Ponchet, M.; Willats, W.; Galiana, E. Silencing of acidic pathogenesis-related PR-1 genes increases extracellular-glucanase activity at the onset of tobacco defence reactions. J. Exp. Bot. 2008, 59, 1225–1239. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; de Wit, P.J. The battle in the apoplast: Further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci. 2015, 6, 584. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shi, L.; Yang, S.; Lin, Y.; Weng, Y.; Li, X.; Hussain, A.; Noman, A.; He, S. Functional and promoter analysis of CHIIV3, a chitinase of pepper plant, in response to Phytophthora capsici infection. Int. J. Mol. Sci. 2017, 18, 1661. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Hernández, E.; Hernández-Morales, J.; Conde-Martínez, V.; Michel-Aceves, A.C.; Lopez-Santillan, J.A.; Torres-Castillo, J.A. In vitro activities of Trichoderma species against Phytophthora parasitica and Fusarium oxysporum. Afr. J. Microbiol. Res. 2016, 10, 521–527. [Google Scholar]
- Zainal, Z.; Marouf, E.; Ismail, I.; Fei, C. Expression of the Capsicuum annum (chili) defensin gene in transgenic tomatoes confers enhanced resistance to fungal pathogens. Am. J. Plant Physiol. 2009, 4, 70–79. [Google Scholar] [CrossRef]
- Portieles, R.; Ayra, C.; Gonzalez, E.; Gallo, A.; Rodriguez, R.; Chacon, O.; López, Y.; Rodriguez, M.; Castillo, J.; Pujol, M.; et al. NmDef02, a novel antimicrobial gene isolated from Nicotiana megalosiphon confers high-level pathogen resistance under greenhouse and field conditions. Plant Biotechnol. J. 2010, 8, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, M.; Huibers, R.P.; Elberse, J.; van den Ackerveken, G. Arabidopsis DMR6 encodes a putative 2OG-Fe (II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008, 54, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cai, C.; Wang, Z.; Fan, B.; Zhu, C.; Chen, Z. Plastid Translation Elongation Factor Tu is Prone to Heat-Induced Aggregation Despite Its Critical Role in Plant Heat Tolerance. Plant Physiol. 2018, 176, 3027–3045. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S.E.; Oh, S.; Seo, E.; Choi, D. HSP70s Enhance a Phytophthora infestans Effector-Induced Cell Death via an MAPK Cascade in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 2018, 31, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Wolters, A.-M.A.; Vossen, J.H.; Rouwet, M.E.; Loonen, A.E.; Jacobsen, E.; Visser, R.G.; Bai, Y. Silencing of six susceptibility genes results in potato late blight resistance. Transgen. Res. 2016, 25, 731–742. [Google Scholar]
- Boevink, P.C.; McLellan, H.; Gilroy, E.M.; Naqvi, S.; He, Q.; Yang, L.; Wang, X.; Turnbull, D.; Armstrong, M.R.; Tian, Z.; et al. Oomycetes seek help from the plant: Phytophthora infestans effectors target host susceptibility factors. Mol. Plant 2016, 9, 636–638. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard Jr, S.G.; Brouwer, C. Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Blasing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Kruer, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.; Yassin, M.A.; Ali, G.S. Transcriptional and proteomic profiling of Aspergillus flavipes in response to sulfur starvation. PLoS ONE 2015, 10, e0144304. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.S.; Brennan, M.S.; Khan, A.; Ali, G.S. Implementation of loop-mediated isothermal amplification methods in lateral flow devices for the detection of Rhizoctonia solani. Can. J. Plant Pathol. 2015, 37, 118–129. [Google Scholar] [CrossRef]




| Group | Sample Name | Raw Reads | Clean Reads | Mapped Reads (%) | Uniquely Mapped (%) | No: of Mapped Genes |
|---|---|---|---|---|---|---|
| 1 | R-mock (Rep 1) | 27,886,208 | 23,047,994 | 97.93 | 81.06 | 21,436 |
| R-mock (Rep 2) | 29,938,504 | 24,872,121 | 97.93 | 77.35 | 21,374 | |
| 2 | R-24 hpi (Rep 1) | 25,731,844 | 21,627,411 | 97.97 | 85.44 | 21,222 |
| R-24 hpi (Rep 2) | 27,210,026 | 22,621,023 | 97.96 | 79.42 | 21,175 | |
| 3 | R-48 hpi (Rep 1) | 27,175,390 | 22,690,068 | 97.67 | 76.36 | 21,356 |
| R-48 hpi (Rep 2) | 29,441,742 | 24,775,792 | 97.82 | 80.97 | 21,533 | |
| 4 | S-mock (Rep 1) | 19,181,092 | 16,133,709 | 97.59 | 76.95 | 20,966 |
| S-mock (Rep 2) | 27,938,220 | 23,227,317 | 97.07 | 84.29 | 21,381 | |
| 5 | S-24 hpi (Rep 1) | 30,978,162 | 26,197,946 | 97.31 | 79.66 | 21,429 |
| S-24 hpi (Rep 2) | 28,387,338 | 23,713,599 | 97.53 | 79.46 | 21,280 | |
| 6 | S-48 hpi (Rep 1) | 28,563,832 | 23,854,528 | 96.27 | 76.36 | 21,631 |
| S-48 hpi (Rep 2) | 26,601,354 | 21,881,482 | 97.36 | 77.45 | 21,565 | |
| Total | 329,033,712 | 274,642,990 |
| GO Term | Description | Color Model | No: of DEGs | ||
|---|---|---|---|---|---|
| R | S | R | S | ||
| GO:0003824 | catalytic activity | 899 | 974 | ||
| GO:0004866 | endopeptidase inhibitor activity | 19 | --- | ||
| GO:0030414 | peptidase inhibitor activity | 20 | --- | ||
| GO:0019825 | oxygen binding | 52 | 57 | ||
| GO:0016740 | transferase activity | 337 | 344 | ||
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | 47 | 53 | ||
| GO:0016491 | oxidoreductase activity | 188 | 214 | ||
| GO:0004364 | glutathione transferase activity | --- | 18 | ||
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | 35 | 42 | ||
| GO:0016758 | transferase activity, transferring hexosyl groups | 75 | 64 | ||
| GO:0046527 | glucosyltransferase activity | 49 | 41 | ||
| GO:0016757 | transferase activity, transferring glycosyl groups | 91 | 82 | ||
| GO:0016168 | chlorophyll binding | 15 | 16 | ||
| GO:0008194 | UDP-glycosyltransferase activity | 45 | 40 | ||
| GO:0046906 | tetrapyrrole binding | 21 | 24 | ||
| GO:0035251 | UDP-glucosyltransferase activity | 34 | 32 | ||
| GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 46 | 45 | ||
| GO:0004091 | carboxylesterase activity | 62 | --- | ||
| GO:0016759 | cellulose synthase activity | 15 | --- | ||
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | 37 | 41 | ||
| GO:0015103 | inorganic anion transmembrane transporter activity | 22 | 25 | ||
| GO:0005372 | water transmembrane transporter activity | 14 | 15 | ||
| GO:0015250 | water channel activity | 14 | 15 | ||
| GO:0022891 | substrate-specific transmembrane transporter activity | --- | 127 | ||
| GO:0022892 | substrate-specific transporter activity | --- | 149 | ||
| GO:0005215 | transporter activity | --- | 178 | ||
| GO:0022857 | transmembrane transporter activity | --- | 147 | ||
| GO:0022804 | active transmembrane transporter activity | --- | 86 | ||
| GO:0015291 | secondary active transmembrane transporter activity | --- | 51 | ||
| GO:0019203 | carbohydrate phosphatase activity | --- | 11 | ||
| GO:0008509 | anion transmembrane transporter activity | --- | 34 | ||
| GO:0015171 | amino acid transmembrane transporter activity | --- | 19 | ||
| GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | --- | 34 | ||
| GO:0005275 | amine transmembrane transporter activity | --- | 20 | ||
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naveed, Z.A.; Ali, G.S. Comparative Transcriptome Analysis between a Resistant and a Susceptible Wild Tomato Accession in Response to Phytophthora parasitica. Int. J. Mol. Sci. 2018, 19, 3735. https://doi.org/10.3390/ijms19123735
Naveed ZA, Ali GS. Comparative Transcriptome Analysis between a Resistant and a Susceptible Wild Tomato Accession in Response to Phytophthora parasitica. International Journal of Molecular Sciences. 2018; 19(12):3735. https://doi.org/10.3390/ijms19123735
Chicago/Turabian StyleNaveed, Zunaira Afzal, and Gul Shad Ali. 2018. "Comparative Transcriptome Analysis between a Resistant and a Susceptible Wild Tomato Accession in Response to Phytophthora parasitica" International Journal of Molecular Sciences 19, no. 12: 3735. https://doi.org/10.3390/ijms19123735
APA StyleNaveed, Z. A., & Ali, G. S. (2018). Comparative Transcriptome Analysis between a Resistant and a Susceptible Wild Tomato Accession in Response to Phytophthora parasitica. International Journal of Molecular Sciences, 19(12), 3735. https://doi.org/10.3390/ijms19123735




