Recruitment of 53BP1 Proteins for DNA Repair and Persistence of Repair Clusters Differ for Cell Types as Detected by Single Molecule Localization Microscopy
Abstract
1. Introduction
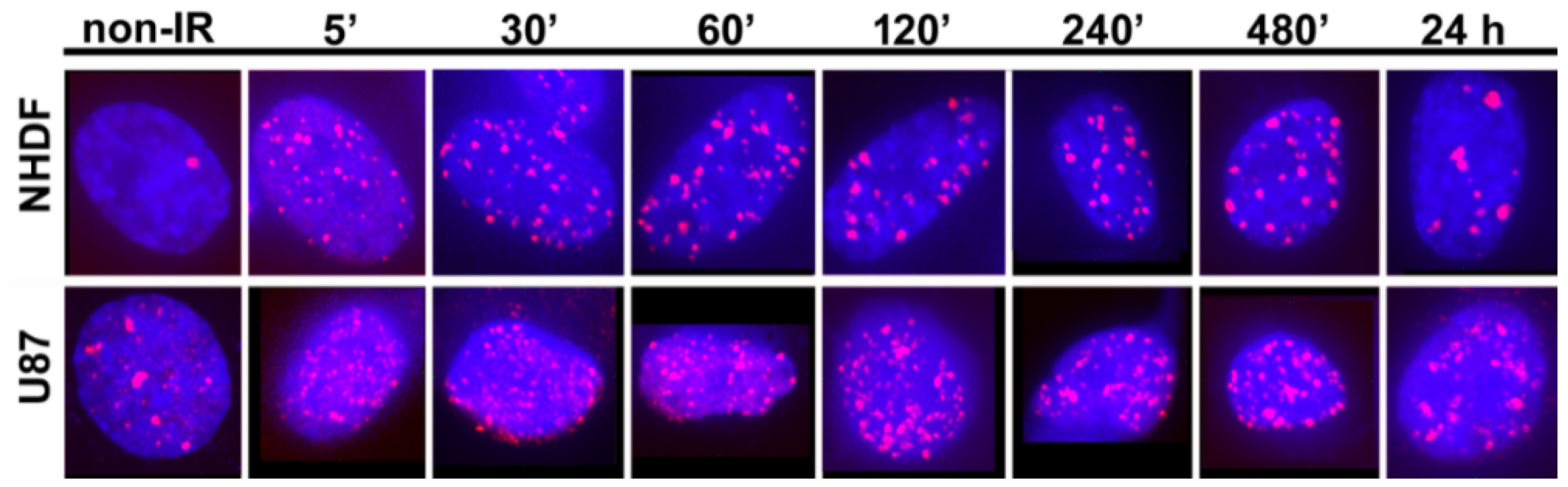
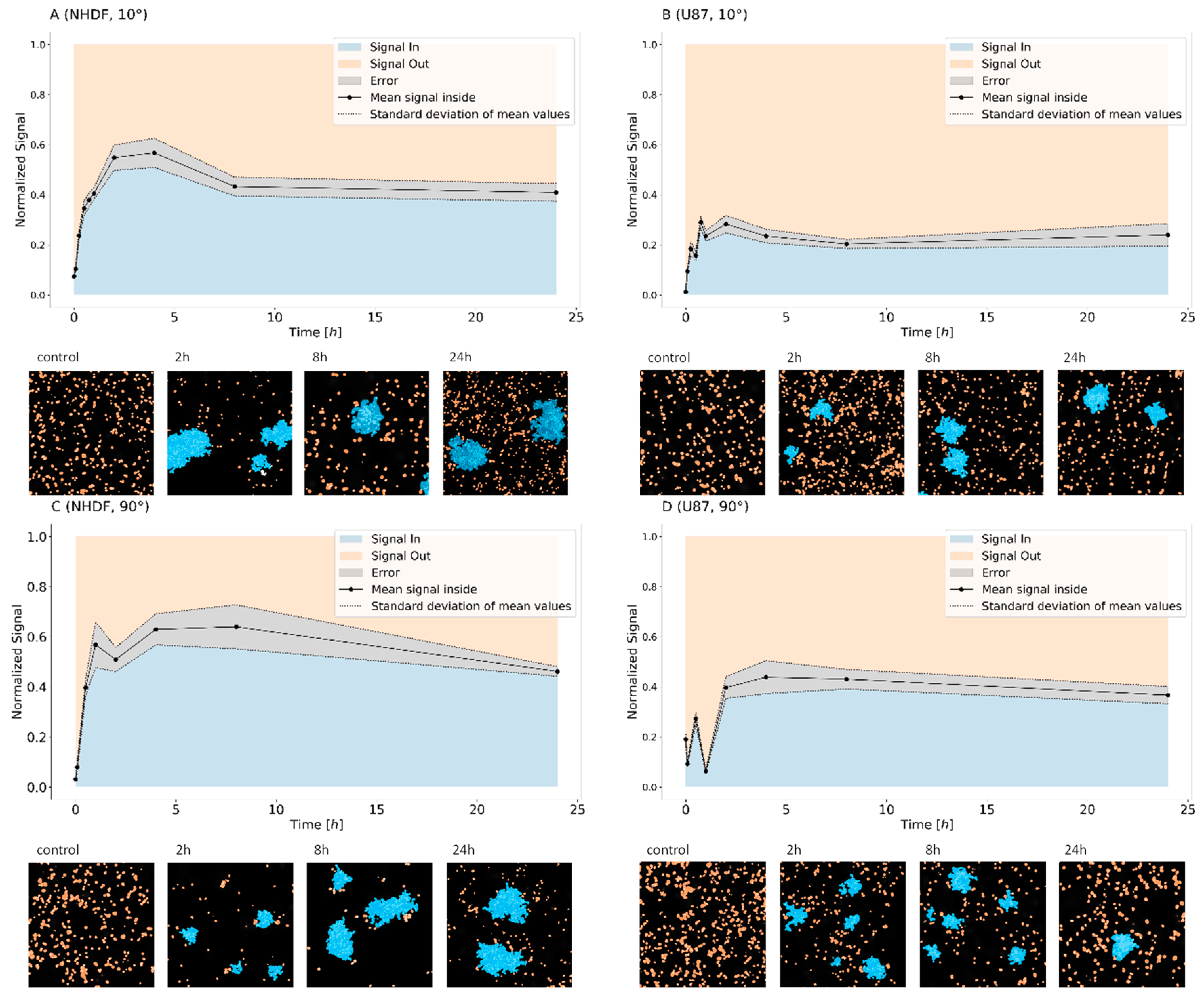
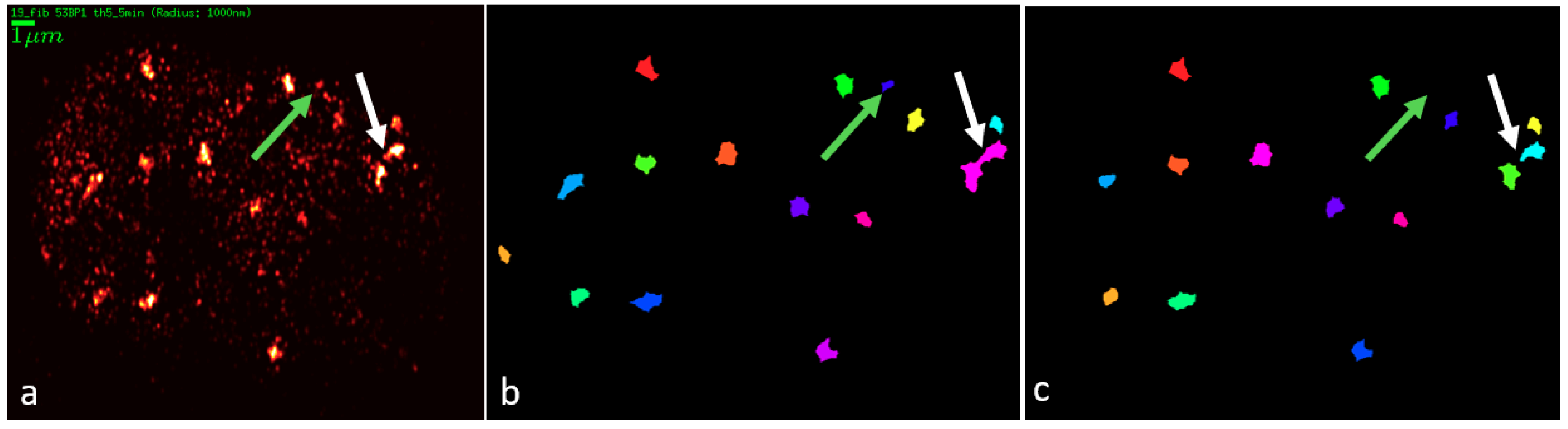
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Ion Irradiation
4.3. Immunofluorescence Staining
4.4. Confocal Microscopy
4.5. Single Molecule Localization Microscopy (SMLM)
4.6. Data Analysis for Single Molecule Localization Microscopy
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jezkova, L.; Zadneprianetc, M.; Kulikova, E.; Smirnova, E.; Bulanova, T.; Depes, D.; Falkova, I.; Boreyko, A.; Krasavin, E.; Davidkova, M.; et al. Particles with similar LET values generate DNA breaks of different complexity and reparability: A high-resolution microscopy analysis of γH2AX/53BP1 foci. Nanoscale 2018, 10, 1162–1179. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.; Hausmann, M.; Lukášová, E.; Biswas, A.; Hildenbrand, G.; Davídková, M.; Krasavin, E.; Kleibl, Z.; Falková, I.; Ježková, L.; et al. Determining OMICS spatiotemporal dimensions using exciting new nanoscopy techniques to asses complex cell responses to DNA damage—PART A (Radiomics). Crit. Rev. Eukaryot. Gene Exp. 2014, 24, 205–223. [Google Scholar] [CrossRef]
- Falk, M.; Hausmann, M.; Lukášová, E.; Biswas, A.; Hildenbrand, G.; Davídková, M.; Krasavin, E.; Kleibl, Z.; Falková, I.; Ježková, L.; et al. Determining OMICS spatiotemporal dimensions using exciting new nanoscopy techniques to asses complex cell responses to DNA damage—PART B (Structuromics). Crit. Rev. Eukaryot. Gene Exp. 2014, 24, 225–247. [Google Scholar] [CrossRef]
- Nikitaki, Z.; Nikolov, V.; Mavragani, I.V.; Mladenov, E.; Mangelis, A.; Laskaratou, D.A.; Fragkoulis, G.I.; Hellweg, C.E.; Martin, O.A.; Emfietzoglou, D.; et al. Measurement of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET). Free Radic. Res. 2016, 50, S64–S78. [Google Scholar] [CrossRef] [PubMed]
- Schipler, A.; Iliakis, G. DNA double-strand–break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Magin, S.; Soni, A.; Iliakis, G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Semin. Cancer Biol. 2016, 37–38, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; Andrea, A.D.D. Repair pathway choices and consequences at the double-strand break. Trends Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Iliakis, G. Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer. Transl. Cancer Res. 2013, 2, 163–177. [Google Scholar]
- Mladenov, E.; Magin, S.; Soni, A.; Iliakis, G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front. Oncol. 2013, 3, 113. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Nandi, S. Synthetic lethality in DNA repair network: A novel avenue in targeted cancer therapy and combination therapeutics. IUBMB Life 2017, 69, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Jeggo, P.A.; Loebrich, M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem. J. 2015, 471, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.; Orecchia, R.; Loeffler, J.S. Charged-particle therapy in cancer: Clinical uses and future perspectives. Nat. Rev. Clin. Oncol. 2017, 14, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.R. A Review: Biological effects of space radiation and developments of effective countermeasures. Life Sci. Space Res. 2014, 1, 10–43. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-starnd breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Iliakis, G.; Murmann, T.; Soni, A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 793, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013, 5, a012740. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.; Lukasova, E.; Stefancikova, L.; Baranova, E.; Falkova, I.; Jezkova, L.; Davidkova, M.; Bacikova, A.; Vachelova, J.; Michaelidesova, A.; et al. Heterochromatinization associated with cell differentiation as a model to study DNA double strand break induction and repair in the context of higher-order chromatin structure. Appl. Radiat. Isotopes 2014, 83, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Yousefi, B. DNA repair and damage pathways in breast cancer development and therapy. DNA Repair 2017, 54, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Wei, F.; Wu, Y.; He, Y.; Shi, L.; Xiong, F.; Gong, Z.; Guo, C.; Li, X.; Deng, H.; et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J. Exp. Clin. Cancer Res. 2018, 37, 87. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, F.; Campa, A.; Esposito, G.; Giardullo, P.; Belli, M.; Dini, V.; Meschini, S.; Simone, G.; Sorrentino, E.; Gerardi, S.; et al. Induction and repair of DNA DSB as revealed by H2AX phosphorylation foci in human fibroblasts exposed to low- and high-LET radiation: Relationship with early and delayed reproductive cell death. Radiat. Res. 2015, 183, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Hildenbrand, G.; Metzler, P.; Pilarczyk, G.; Bobu, V.; Kriz, W.; Hosser, H.; Fleckenstein, J.; Krufczik, M.; Bestvater, F.; Wenz, F.; et al. Dose enhancement effects of gold nanoparticles specifically targeting RNA in breast cancer cells. PLoS ONE 2018, 13, e0190183. [Google Scholar] [CrossRef] [PubMed]
- Rothkamm, K.; Barnard, S.; Moquet, J.; Ellender, M.; Rana, Z.; Burdak-Rothkamm, S. DNA damage foci: Meaning and significance. Environ. Mol. Mutagen. 2015, 56, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Van Veelen, L.R.; Cervelli, T.; van de Rakt, M.W.; Theil, A.F.; Essers, J.; Kanaar, R. Analysis of ionizing radiation-induced foci of DNA damage repair proteins. Mutat. Res. 2005, 574, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Eberlein, U.; Peper, M.; Fernandez, M.; Lassmann, M.; Scherthan, H. Calibration of the γ-H2AX DNA double strand break focus assay for internal radiation exposure of blood lymphocytes. PLoS ONE 2015, 10, e0123174. [Google Scholar] [CrossRef] [PubMed]
- Hauptner, A.; Friedland, W.; Dietzel, S.; Drexler, G.A.; Greubel, C.; Hable, V.; Strickfaden, H.; Cremer, T.; Friedl, A.A.; Krücken, R.; et al. Spatial distribution of DNA double-strand breaks from ion tracks. In Ion Beam Science: Solved and Unsolved Problems; Royal Danish Academy of Sciences and Letters: Copenhagen, Denmark, 2006; pp. 59–85. [Google Scholar]
- Costes, S.V.; Boissière, A.; Ravani, S.; Romano, R.; Parvin, B.; Barcellos-Hoff, M.H. Imaging features that discriminate between foci induced by high- and low-LET radiation in human fibroblasts. Radiat. Res. 2006, 165, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Natale, F.; Rapp, A.; Yu, W.; Maiser, A.; Harz, H.; Schall, A.; Grulich, S.; Anton, T.; Hörl, D.; Chen, W.; et al. Identification of the elementary structural units of the DNA damage response. Nat. Commun. 2017, 8, 15760. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Wagner, E.; Lee, J.-H.; Schrock, G.; Schaufler, W.; Krufczik, M.; Papenfuß, F.; Port, M.; Bestvater, F.; Scherthan, H. Super-resolution microscopy of radiation-induced histone H2AX phosphorylation in relation to H3K9-trimethylation in HeLa cells. Nanoscale 2018, 10, 4320–4331. [Google Scholar] [CrossRef] [PubMed]
- Lorat, Y.; Bunner, C.U.; Schanz, S.; Jacob, B.; Taucher-Scholz, G.; Rübe, C.E. Nanoscale analysis of clustered DNA damage after high-LET irradiation by quantitative electron microscopy—The heavy burden to repair. DNA Repair 2015, 28, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Lorat, Y.; Timm, S.; Jacob, B.; Taucher-Scholz, G.; Rübe, C.E. Clustered double-strand breaks in heterochromatin perturb DNA repair after high linear energy transfer irradiation. Radiother. Oncol. 2016, 121, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Lopez Perez, R.; Best, G.; Nicolay, N.H.; Greubel, C.; Rossberger, S.; Reindl, J.; Dollinger, G.; Weber, K.-J.; Cremer, C.; Huber, P.E. Superresolution light microscopy shows nanostructure of carbon ion radiation-induced DNA double-strand break repair foci. FASEB J. 2016, 30, 2767–2776. [Google Scholar] [CrossRef] [PubMed]
- Sisario, D.; Memmel, S.; Doose, S.; Neubauer, J.; Zimmermann, H.; Flentje, M.; Djuzenova, C.S.; Sauer, M.; Sukhorukov, V.L. Nanostructure of DNA repair foci revealed by superresolution microscopy. FASEB J. 2018, 12, fj201701435. [Google Scholar] [CrossRef] [PubMed]
- Eryilmaz, M.; Schmitt, E.; Krufczik, M.; Theda, F.; Lee, J.-H.; Cremer, C.; Bestvater, F.; Schaufler, W.; Hausmann, M.; Hildenbrand, G. Localization microscopy analyses of MRE11 clusters in 3D-conserved cell nuclei of different cell lines. Cancers 2018, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Reindl, J.S.; Girst, S.; Walsh, D.W.M.; Geubel, C.; Schwarz, B.; Siebenwirth, C.; Drexler, G.A.; Friedl, A.A.; Dollinger, G. Chromatin organization revealed by nanostructure of irradiation induced γH2AX, 53BP1 and Rad51 foci. Sci. Rep. 2017, 7, 40616. [Google Scholar] [CrossRef] [PubMed]
- Hable, V.; Drexler, G.A.; Brüning, T.; Burgdorf, C.; Greubel, C.; Derer, A.; Seel, J.; Strickfaden, H.; Cremer, T.; Friedl, A.A.; et al. Recruitment kinetics of DNA repair proteins Mdc1 and Rad52 but not 53BP1 depend on damage complexity. PLoS ONE 2012, 7, e41943. [Google Scholar] [CrossRef] [PubMed]
- Iliakis, G. The Biological Foundations of Risks from Ionizing Radiation Exposures: How an Understanding of Associated Effects Will Help Their Quantification and Mitigation. In Sustainable Risk Management; Springer: Berlin, Germany, 2018; pp. 149–158. [Google Scholar]
- Hofmann, A.; Krufczik, M.; Heermann, D.W.; Hausmann, M. Using persistent homology as a new approach for super-resolution localization microscopy data analysis and classification of γH2AX foci/clusters. Int. J. Mol. Sci. 2018, 19, 2263. [Google Scholar] [CrossRef] [PubMed]
- Kakarougkas, A.; Ismail, A.; Klement, K.; Goodarzi, A.A.; Conrad, S.; Freire, R.; Shibata, A.; Löbrich, M.; Jeggo, P.A. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013, 41, 9719–9731. [Google Scholar] [CrossRef] [PubMed]
- Depes, D.; Lee, J.-H.; Bobkova, E.; Jezkova, L.; Falkova, I.; Bestvater, F.; Pagacova, E.; Kopecna, O.; Zadneprianetc, M.; Bacikova, A.; et al. Single molecule localization microscopy as a promising tool for γH2AX/53BP1 foci exploration. Eur. Phys. J. D 2018, 72, 158. [Google Scholar] [CrossRef]
- Ochs, F.; Somyajit, K.; Altmeyer, M.; Rask, M.B.; Lukas, J.; Lukas, C. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 2016, 23, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Sossick, A.J.; Boulton, S.J.; Jackson, S.P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 2012, 125, 3529–3534. [Google Scholar] [CrossRef] [PubMed]
- Reindl, J.; Drexler, G.A.; Girst, S.; Greubel, C.; Siebenwirth, C.; Drexler, S.E.; Dollinger, G.; Friedl, A.A. Nanoscopic exclusion between Rad51 and 53BP1 after ion irradiation in human HeLa cells. Phys. Biol. 2015, 12, 066005. [Google Scholar] [CrossRef] [PubMed]
- Pilarczyk, G.; Nesnidal, I.; Gunkel, M.; Bach, M.; Bestvater, F.; Hausmann, M. Localisation microscopy of breast epithelial ErbB-2 receptors and gap junctions: Trafficking after gamma-irradiation, Neuregulin-1b and Herceptin application. Int. J. Mol. Sci. 2017, 18, 362. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Ilić, N.; Pilarczyk, G.; Lee, J.-H.; Logeswaran, A.; Borroni, A.P.; Krufczik, M.; Theda, F.; Waltrich, N.; Bestvater, F.; et al. Challenges for super-resolution localization microscopy and biomolecular fluorescent nano-probing in cancer research. Int. J. Mol. Sci. 2017, 18, 2066. [Google Scholar] [CrossRef] [PubMed]
- Lemmer, P.; Gunkel, M.; Baddeley, D.; Kaufmann, R.; Urich, A.; Weiland, Y.; Reymann, J.; Müller, P.; Hausmann, M.; Cremer, C. SPDM—Light microscopy with single molecule resolution at the nanoscale. Appl. Phys. B 2018, 93, 1–12. [Google Scholar] [CrossRef]
- Lemmer, P.; Gunkel, M.; Weiland, Y.; Müller, P.; Baddeley, D.; Kaufmann, R.; Urich, A.; Eipel, H.; Amberger, R.; Hausmann, M.; et al. Using conventional fluorescent markers for far-field fluorescence localization nanoscopy allows resolution in the 10 nm range. J. Microsc. 2009, 235, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Bach, M.; Savini, C.; Krufczik, M.; Cremer, C.; Rösl, F.; Hausmann, M. Super-resolution localization microscopy of γ-H2AX and heterochromatin after folate deficiency. Int. J. Mol. Sci. 2017, 18, 1726. [Google Scholar] [CrossRef] [PubMed]
- Cremer, C.; Kaufmann, R.; Gunkel, M.; Pres, S.; Weiland, Y.; Müller, P.; Ruckelshausen, T.; Lemmer, P.; Geiger, F.; Degenhard, M.; et al. Superresolution imaging of biological nanostructures by Spectral Precision Distance Microscopy (SPDM). Biotechnol. J. 2011, 6, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.R.; Gridley, D.S.; Slater, J.M. Advances in the Biology, Imaging and Therapies for Glioblastoma. In Radiobiology of Radioresistant Glioblastoma; Cheng, C., Ed.; IntechOPEN: London, UK, 2011; pp. 3–22. [Google Scholar]
- Krufczik, M.; Sievers, A.; Hausmann, A.; Lee, J.-H.; Hildenbrand, G.; Schaufler, W.; Hausmann, M. Combining low temperature fluorescence DNA-hybridization, immunostaining, and super-resolution localization microscopy for nano-structure analysis of ALU elements and their influence on chromatin structure. Int. J. Mol. Sci. 2017, 18, 1005. [Google Scholar] [CrossRef] [PubMed]
- Deckbar, D.; Jeggo, P.A.; Löbrich, M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Scholz, M.; Kraft-Weyrather, W.; Ritter, S.; Kraft, G. Cell cycle delays induced by heavy ion irradiation of synchronous mammalian cells. Int. J. Radiat. Biol. 1994, 66, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Sora, S.; Hamada, N.; Hara, T.; Funayama, T.; Sakashita, T.; Yokota, Y.; Nakano, T.; Kobayashi, Y. Exposure of normal human fibroblasts to heavy-ion radiation promotes their morphological differentiation. Biol. Sci. Space 2008, 22, 54–58. [Google Scholar] [CrossRef]
- Tsuboi, K.; Moritake, T.; Tsuchida, Y.; Tokuuye, K.; Matsumura, A.; Ando, K. Cell cycle checkpoint and apoptosis induction in glioblastoma cells and fibroblasts irradiated with carbon beam. J. Radiat. Res. 2007, 48, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, N.I.; Brunton, H.; Watanabe, R.; Shrikhande, A.; Hirayama, R.; Matsufuji, N.; Fujimori, A.; Murakami, T.; Okayasu, R.; Jeggo, P.; et al. Visualisation of γH2AX foci caused by heavy ion particle traversal; distinction between core track versus non-track damage. PLoS ONE 2013, 8, e70107. [Google Scholar] [CrossRef] [PubMed]
- Lobachevsky, P.; Leong, T.; Daly, P.; Smith, J.; Best, N.; Tomaszewski, J.; Thompson, E.R.; Li, N.; Campbell, I.G.; Martin, R.F.; et al. Compromized DNA repair as a basis for identification of cancer radiotherapy patients with extreme radiosensitivity. Cancer Lett. 2016, 383, 212–219. [Google Scholar] [CrossRef] [PubMed]
- DiBiase, S.J.; Zeng, Z.C.; Chen, R.; Hyslop, T.; Curran, W.J., Jr.; Iliakis, G. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000, 60, 1245–1253. [Google Scholar] [PubMed]
- Schwarz-Finsterle, J.; Scherthan, H.; Huna, A.; González, P.; Müller, P.; Schmitt, E.; Erenpreisa, J.; Hausmann, M. Volume increase and spatial shifts of chromosome territories in nuclei of radiation-induced polyploidizing tumour cells. Mutat. Res. 2013, 756, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Tsao, D.; Kalogerinis, P.; Tabrizi, I.; Dingfelder, M.; Stewart, R.D.; Georgakilas, A.G. Induction and processing of oxidative clustered DNA lesions in 56Fe-ion-irradiated human monocytes. Radiat. Res. 2007, 168, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Short, S.C.; Martindale, C.; Bourne, S.; Brand, G.; Woodcock, M.; Johnston, P. DNA repair after irradiation in glioma cells and normal human astrocytes. Neuro Oncol. 2007, 9, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Bezbakh, A.A.; Zager, V.B.; Kaminski, G.; Krylov, A.I.; Krylov, V.A.; Teterev, Y.G.; Timoshenko, G.N. Upgrading the genome facility for radiobiological experiments with heavy-ion beams. Phys. Part. Nuclei Lett. 2013, 10, 175–178. [Google Scholar] [CrossRef]
- Tarasov, O.B.; Bazin, D. LISE++: Radioactive beam production with in-flight separators. Nucl. Instrum. Meth. B 2008, 266, 4657–4666. [Google Scholar] [CrossRef]
- Stuhlmüller, M.; Schwarz-Finsterle, J.; Fey, E.; Lux, J.; Bach, M.; Cremer, C.; Hinderhofer, K.; Hausmann, M.; Hildenbrand, G. In situ optical sequencing and nano-structure analysis of a trinucleotide expansion region by localization microscopy after specific COMBO-FISH labelling. Nanoscale 2015, 7, 17938–17946. [Google Scholar] [CrossRef] [PubMed]



| Irradiation Angle | Energy (MeV/n) | LET (keV/µm) | Fluence (106/cm) Per 1 Gy | Mean Number of Particles/Nucleus |
|---|---|---|---|---|
| 10° | 13.1 | 181.4 | 3.40 | 2.1 |
| 90° | 13.0 | 182.9 | 3.41 | 25.4 |
| Column-Number | Type of Data |
|---|---|
| 1 | Signal-amplitude in photoelectrons |
| 2 | Lateral y-coordinates in nm |
| 3 | Lateral x-coordinates in nm |
| 4 | Lateral y-coordinates error in nm |
| 5 | Lateral x-coordinates error in nm |
| 6 | Standard-deviation σy in nm |
| 7 | Standard-deviation σx in nm |
| 8 | Number of photoelectrons in the signal |
| 9 | Number of the picture where the signal was detected |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobkova, E.; Depes, D.; Lee, J.-H.; Jezkova, L.; Falkova, I.; Pagacova, E.; Kopecna, O.; Zadneprianetc, M.; Bacikova, A.; Kulikova, E.; et al. Recruitment of 53BP1 Proteins for DNA Repair and Persistence of Repair Clusters Differ for Cell Types as Detected by Single Molecule Localization Microscopy. Int. J. Mol. Sci. 2018, 19, 3713. https://doi.org/10.3390/ijms19123713
Bobkova E, Depes D, Lee J-H, Jezkova L, Falkova I, Pagacova E, Kopecna O, Zadneprianetc M, Bacikova A, Kulikova E, et al. Recruitment of 53BP1 Proteins for DNA Repair and Persistence of Repair Clusters Differ for Cell Types as Detected by Single Molecule Localization Microscopy. International Journal of Molecular Sciences. 2018; 19(12):3713. https://doi.org/10.3390/ijms19123713
Chicago/Turabian StyleBobkova, Elizaveta, Daniel Depes, Jin-Ho Lee, Lucie Jezkova, Iva Falkova, Eva Pagacova, Olga Kopecna, Mariia Zadneprianetc, Alena Bacikova, Elena Kulikova, and et al. 2018. "Recruitment of 53BP1 Proteins for DNA Repair and Persistence of Repair Clusters Differ for Cell Types as Detected by Single Molecule Localization Microscopy" International Journal of Molecular Sciences 19, no. 12: 3713. https://doi.org/10.3390/ijms19123713
APA StyleBobkova, E., Depes, D., Lee, J.-H., Jezkova, L., Falkova, I., Pagacova, E., Kopecna, O., Zadneprianetc, M., Bacikova, A., Kulikova, E., Smirnova, E., Bulanova, T., Boreyko, A., Krasavin, E., Wenz, F., Bestvater, F., Hildenbrand, G., Hausmann, M., & Falk, M. (2018). Recruitment of 53BP1 Proteins for DNA Repair and Persistence of Repair Clusters Differ for Cell Types as Detected by Single Molecule Localization Microscopy. International Journal of Molecular Sciences, 19(12), 3713. https://doi.org/10.3390/ijms19123713







