Ginsenoside Rg1 Prevents Doxorubicin-Induced Cardiotoxicity through the Inhibition of Autophagy and Endoplasmic Reticulum Stress in Mice
Abstract
1. Introduction
2. Results
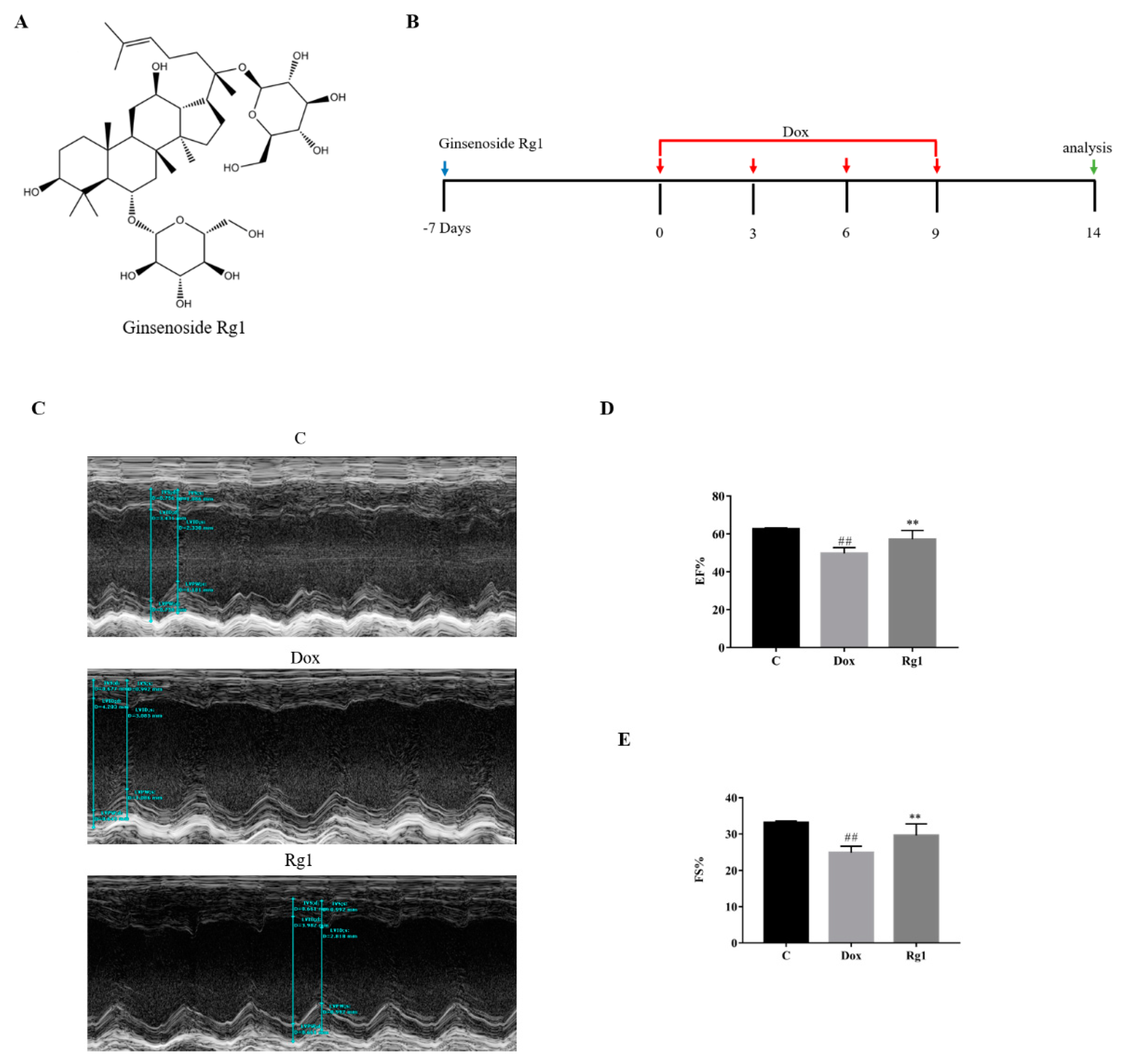
2.1. Ginsenoside Rg1 Improved Cardiac Function in Mice Treated with Doxorubicin
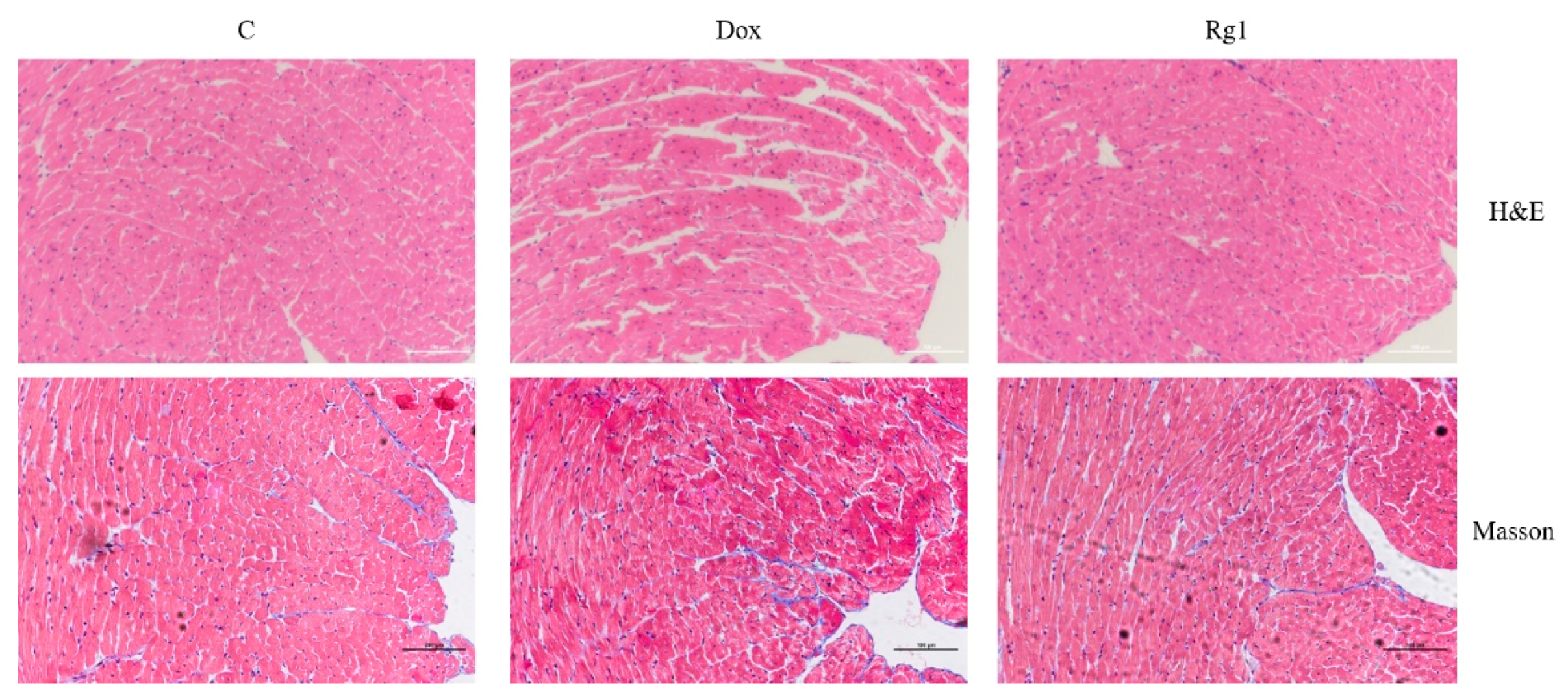
2.2. Ginsenoside Rg1 Alleviated Myocardial Pathological Changes and Cardiac Fibrosis in Mice Treated with Doxorubicin
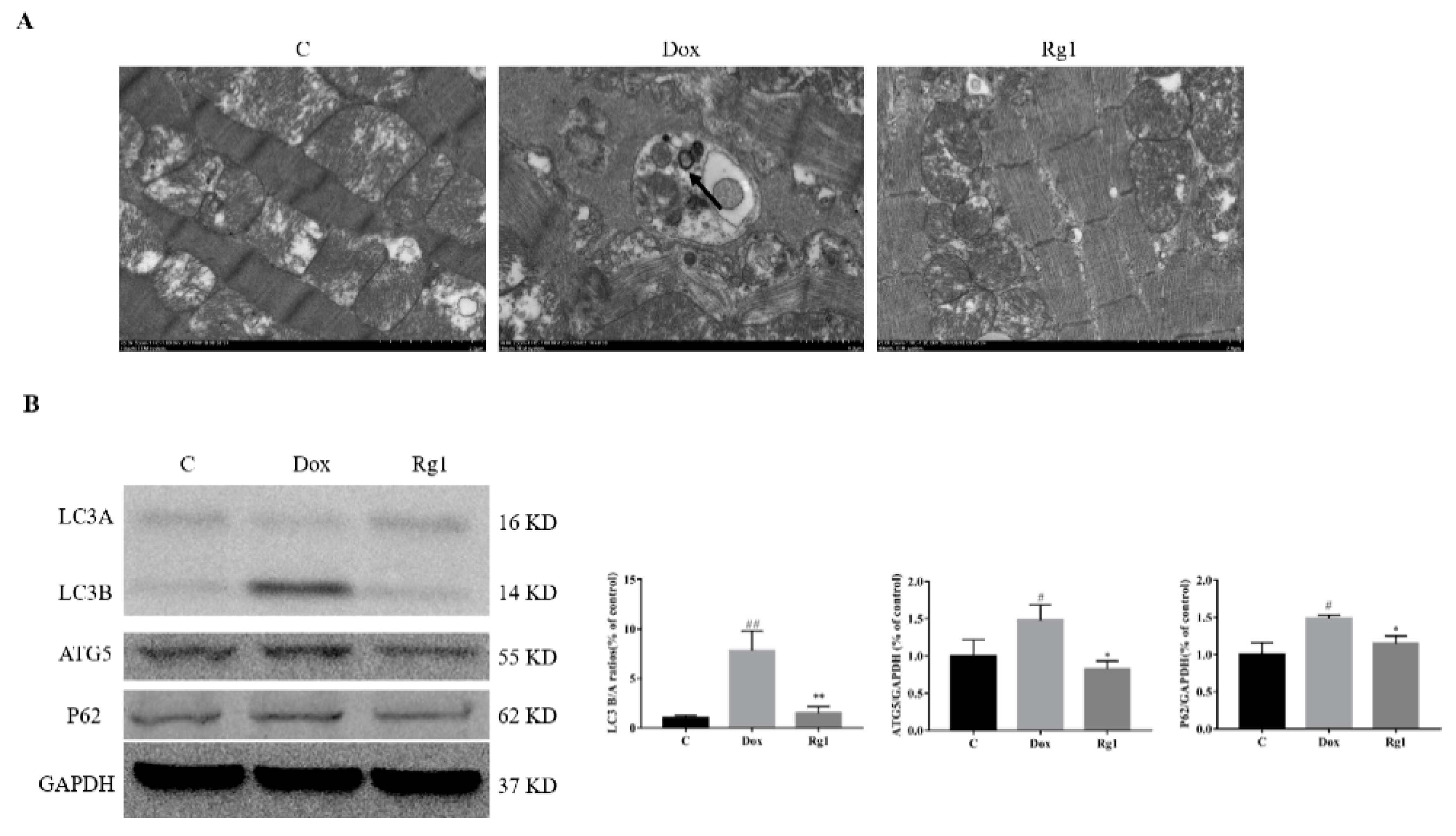
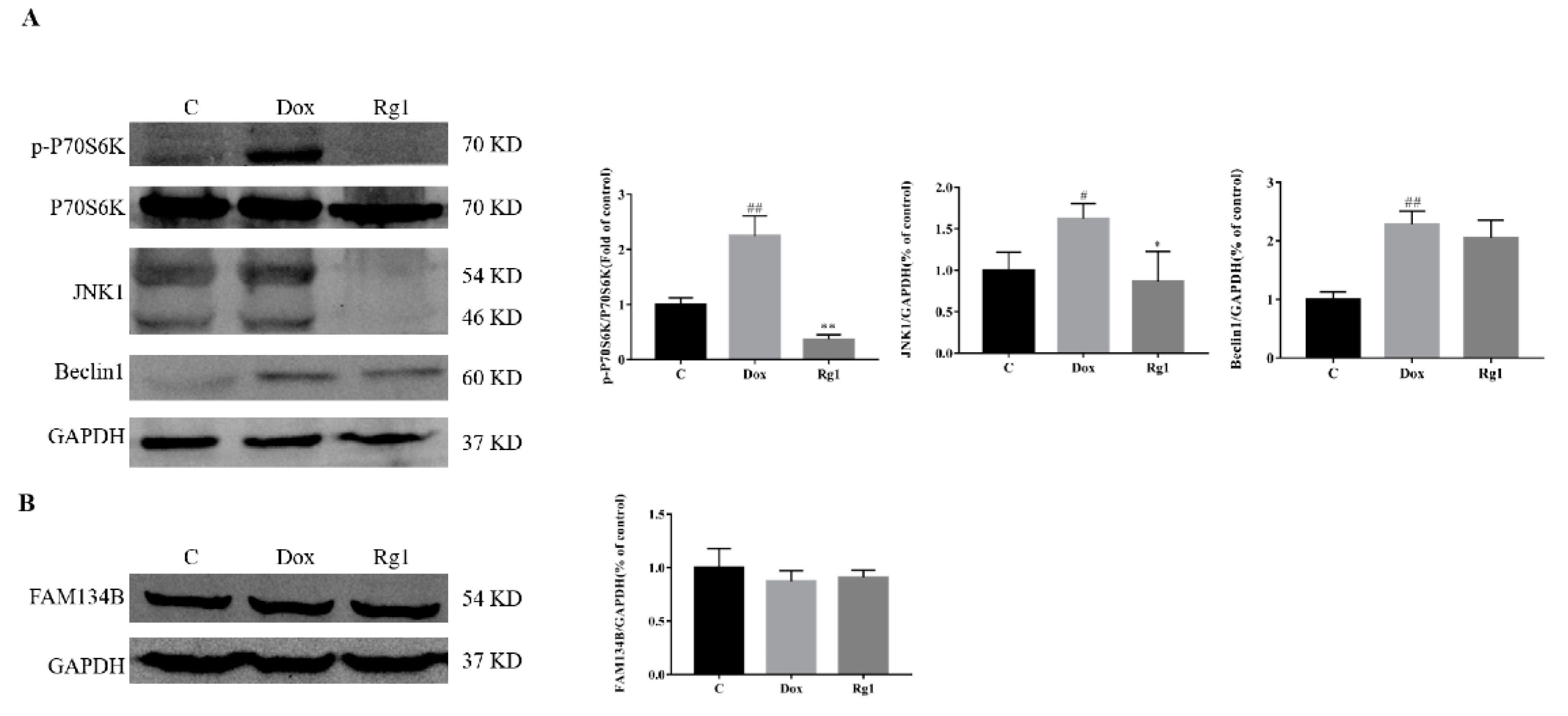
2.3. Ginsenoside Rg1 Inhibits Cardiac Autophagy in Mice Treated with Doxorubicin
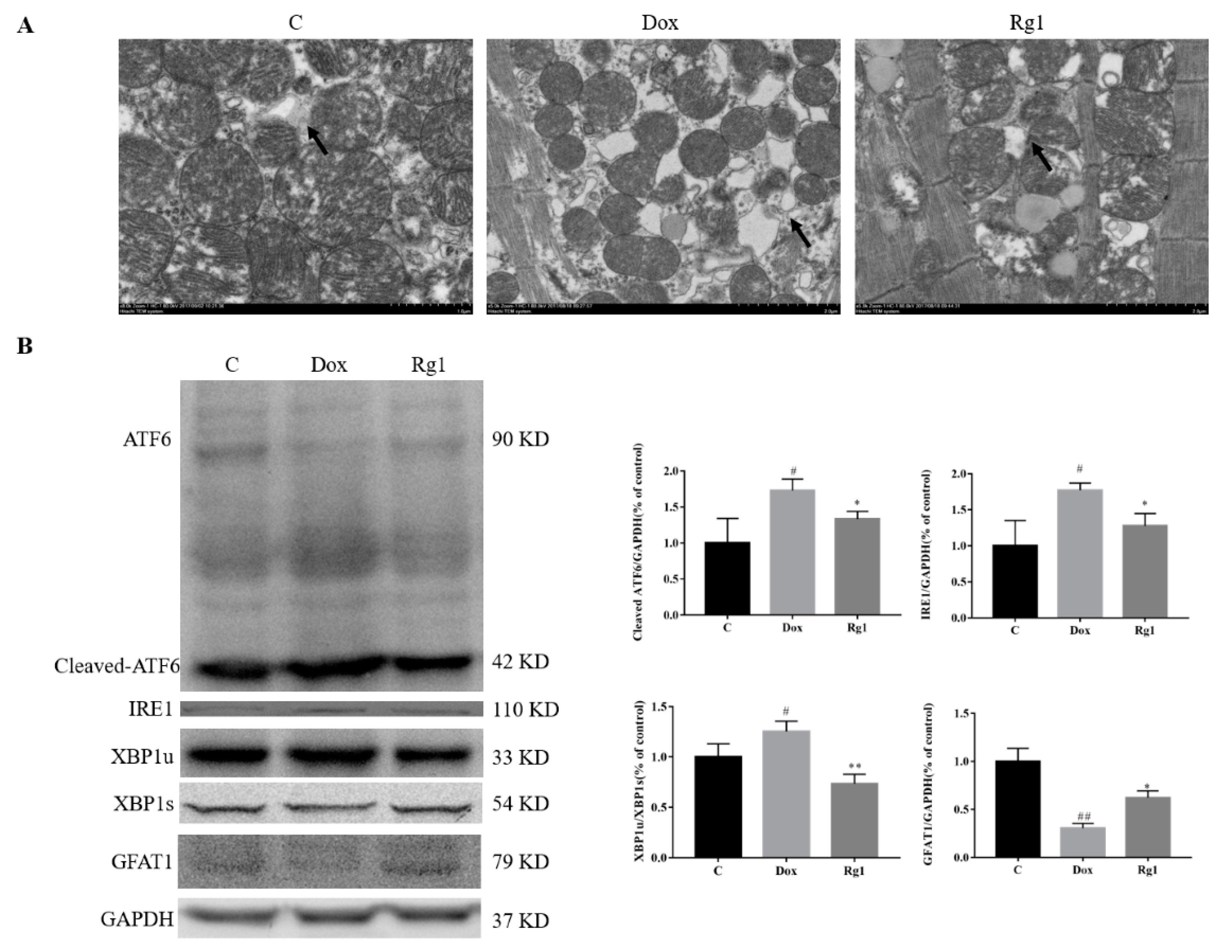
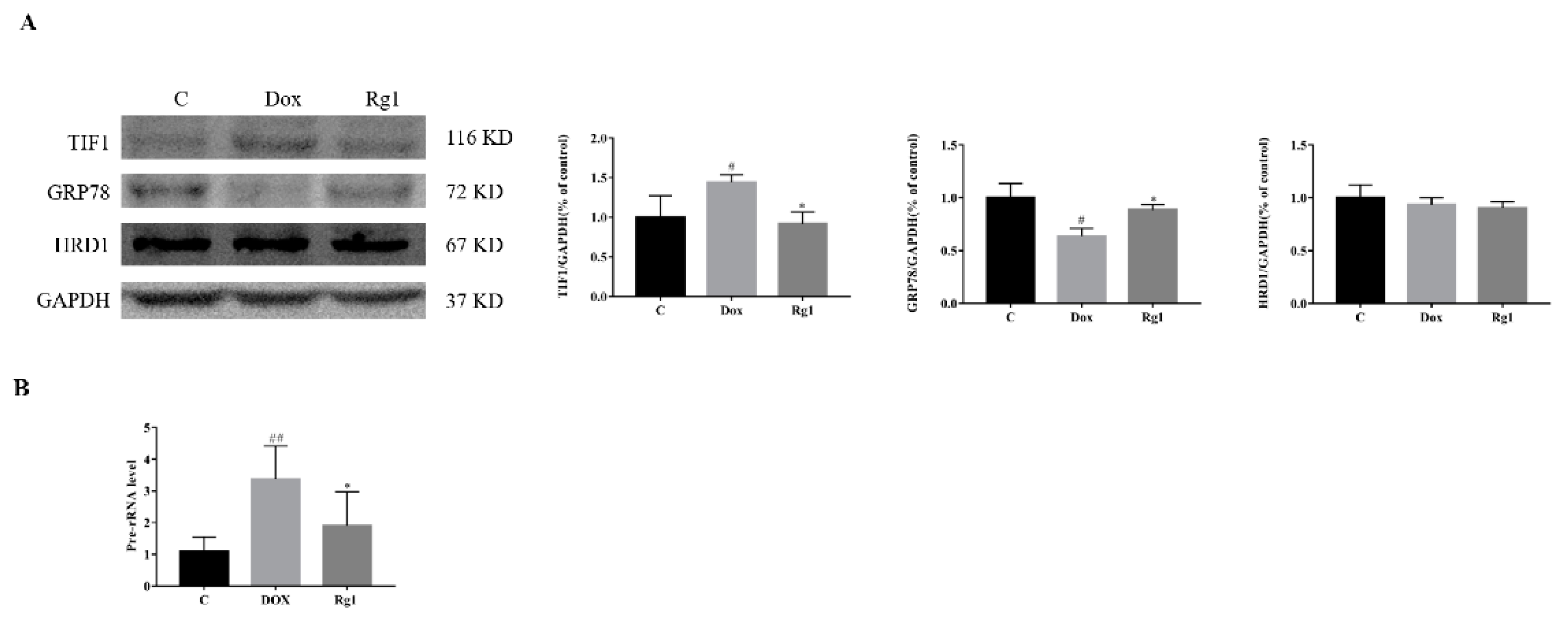
2.4. Ginsenoside Rg1 Improved DOX-Induced Endoplasmic Reticulum Stress in Mice Heart
2.5. The Mechanisms of Rg1 to Improve Endoplasmic Reticulum Stress
2.6. Effects of Rg1 on the Autophagic Pathway Activated by Endoplasmic Reticulum Stress in Mice Hearts
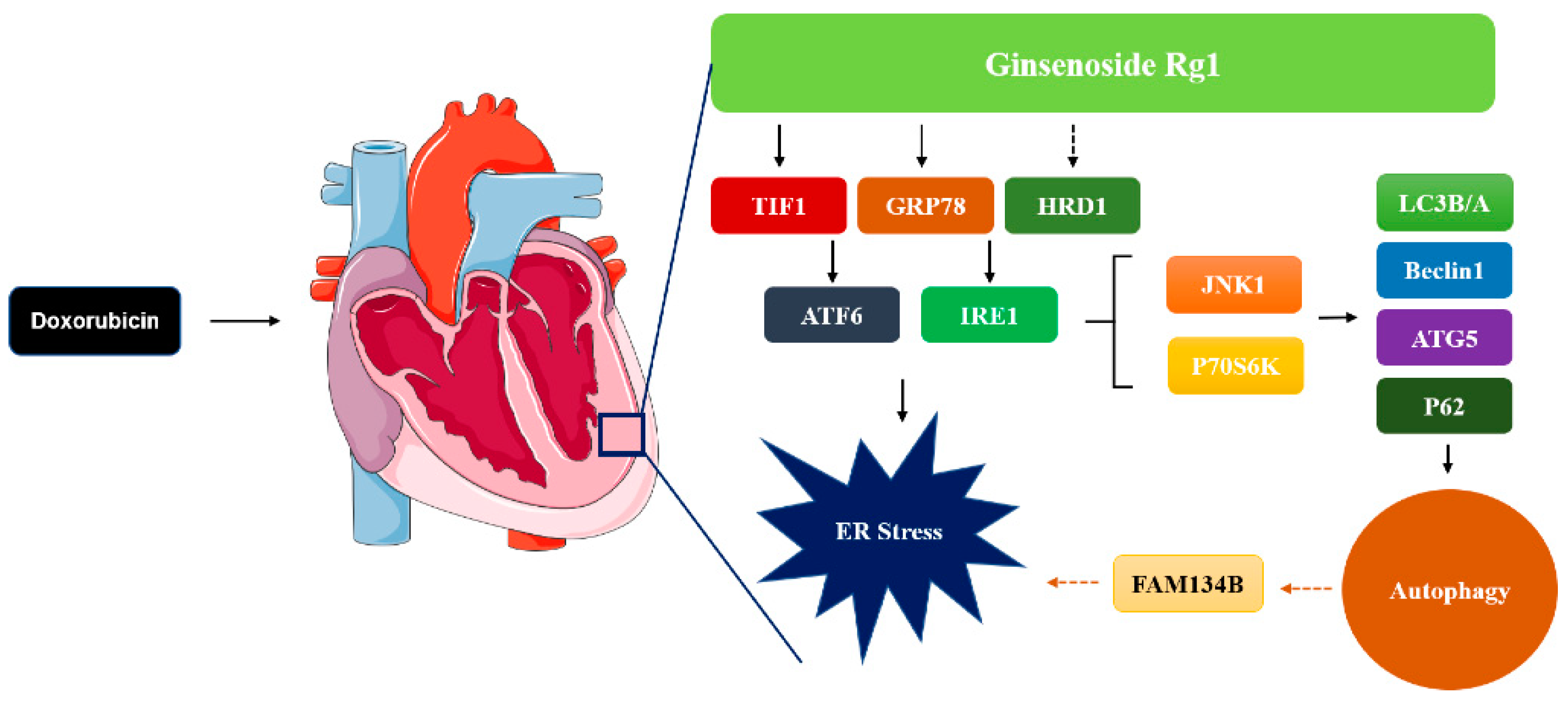
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Animals and Experimental Protocols
4.3. Echocardiography
4.4. Histopathology
4.5. Electron Microscopy
4.6. RNA Extraction and qPCR
4.7. Western Blot
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Carvalho, F.S.; Burgeiro, A.; Garcia, R.; Moreno, A.J.; Carvalho, R.A.; Oliveira, P.J. Doxorubicin-induced cardiotoxicity: From bioenergetic failure and cell death to cardiomyopathy. Med. Res. Rev. 2014, 34, 106–135. [Google Scholar] [CrossRef] [PubMed]
- Octavia, Y.; Tocchetti, C.G.; Gabrielson, K.L.; Janssens, S.; Crijns, H.J.; Moens, A.L. Doxorubicin-induced cardiomyopathy: From molecular mechanisms to therapeutic strategies. J. Mol. Cell. Cardiol. 2012, 52, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Thompson, M.D.; Cohen, R.A.; Tong, X. Autophagy and oxidative stress in cardiovascular diseases. Biochim. Biophys. Acta 2015, 1852, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Koleini, N.; Kardami, E. Autophagy and mitophagy in the context of doxorubicin-induced cardiotoxicity. Oncotarget 2017, 8, 46663–46680. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, L.; Ma, J.; Lu, L.; Wang, X.; Ren, J.; Yang, J. Rutin attenuates doxorubicin-induced cardiotoxicity via regulating autophagy and apoptosis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1904–1911. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Cheng, Y.; Zhang, Y.; Zhang, Q.; Wang, S.; Tian, S.; Zou, Y.; Hu, K.; Ren, J.; Ge, J. Aldehyde dehydrogenase 2 ameliorates doxorubicin-induced myocardial dysfunction through detoxification of 4-HNE and suppression of autophagy. J. Mol. Cell. Cardiol. 2014, 71, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Volden, P.; Timm, D.; Mao, K.; Xu, X.; Liang, Q. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J. Biol. Chem. 2010, 285, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Chevet, E.; Harding, H.P. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 2013, 12, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.Y.; Sanada, S.; Matsuzaki, T.; Liao, Y.; Okuda, K.; Yamato, M.; Tsuchida, S.; Araki, R.; Asano, Y.; Asanuma, H.; et al. Chemical Endoplasmic Reticulum Chaperone Alleviates Doxorubicin-Induced Cardiac Dysfunction. Circ. Res. 2016, 118, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.C.; Sun, G.B.; Ye, J.X.; Wang, J.; Zhang, M.D.; Sun, X.B. Salvianolic acid B attenuates doxorubicin-induced ER stress by inhibiting TRPC3 and TRPC6 mediated Ca2+ overload in rat cardiomyocytes. Toxicol. Lett. 2017, 276, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Lu, M.; Yu, L.; Wang, Q.; Mei, M.; Xu, C.; Han, R.; Hu, J.; Wang, H.; Zhang, Y. Inhibition of TNF-α-mediated NF-κB Activation by Ginsenoside Rg1 Contributes the Attenuation of Cardiac Hypertrophy Induced by Abdominal Aorta Coarctation. J. Cardiovasc. Pharmacol. 2016, 68, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Chen, T.; Ren, X.; Zhang, Z.; Huang, W.; Liu, L.; Luo, P.; Zhou, H. Rg1 prevents myocardial hypoxia/reoxygenation injury by regulating mitochondrial dynamics imbalance via modulation of glutamate dehydrogenase and mitofusin 2. Mitochondrion 2016, 26, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Zhang, X.L.; Li, M.H.; Iqbal, J.; Bourantas, C.V.; Li, J.J.; Su, X.Y.; Muramatsu, T.; Tian, N.L.; Chen, S.L. The ginsenoside Rg1 prevents transverse aortic constriction-induced left ventricular hypertrophy and cardiac dysfunction by inhibiting fibrosis and enhancing angiogenesis. J. Cardiovasc. Pharmacol. 2013, 62, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Fan, Y.; Liu, M.L. Ginsenoside Rg1 inhibits autophagy in H9c2 cardiomyocytes exposed to hypoxia/reoxygenation. Mol. Cell. Biochem. 2012, 365, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.S.; Lin, H.; Ye, J.X.; Lin, M.; Cui, X.P. Rg1 exhibits neuroprotective effects by inhibiting the endoplasmic reticulum stress-mediated c-Jun N-terminal protein kinase apoptotic pathway in a rat model of Alzheimer′s disease. Mol. Med. Rep. 2015, 12, 3862–3868. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhen, J.; Yang, Y.; Gu, J.; Wu, S.; Liu, Q. Ginsenoside Rg1 ameliorates diabetic cardiomyopathy by inhibiting endoplasmic reticulum stress-induced apoptosis in a streptozotocin-induced diabetes rat model. J. Cell. Mol. Med. 2016, 20, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wang, Y.; Liu, H.; Mu, H.; Lu, Y.; Zhang, J.; Huang, J. Oral administration of Ginsenoside Rg1 prevents cardiac toxicity induced by doxorubicin in mice through anti-apoptosis. Oncotarget 2017, 31, 83792–83801. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Chen, A.W.; Varner, J.D. A review of the mammalian unfolded protein response. Biotechnol. Bioeng. 2011, 108, 2777–2793. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.O.; Yadav, R.K.; Kim, H.R.; Chae, H.J. ER stress: Autophagy induction, inhibition and selection. Autophagy 2015, 11, 1956–1977. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Pietrocola, F.; Levine, B.; Kroemer, G. Metabolic control of autophagy. Cell 2014, 159, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Santulli, G. Cardioprotective effects of autophagy: Eat your heart out, heart failure! Sci. Transl. Med. 2018, 10, eaau0462. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zhou, Z.; Liang, B.; Huang, Y.; Yang, Z.; Chen, Y.; Zhang, L.; Yan, C.; Wang, J.; Lu, L.; et al. Inhibiting Receptor of Advanced Glycation End Products Attenuates Pressure Overload-Induced Cardiac Dysfunction by Preventing Excessive Autophagy. Front Physiol. 2018, 9, 1333. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, R.; Li, M.; Yu, Y.; Liang, Y.; Han, F.; Qin, S.; Chen, X.; Su, Y.; Ge, J. Mitochondrial calcium uniporter inhibition provides cardioprotection in pressure overload-induced heart failure through autophagy enhancement. Int. J. Cardiol. 2018, 271, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Brandizzi, F. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013, 23, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, A.; Tomer, Y. Interferon alpha impairs insulin production in human beta cells via endoplasmic reticulum stress. J. Autoimmun. 2017, 80, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.V.; Deng, Y.; Gao, N.; Pedrozo, Z.; Li, D.L.; Morales, C.R.; Criollo, A.; Luo, X.; Tan, W.; Jiang, N.; et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell 2014, 156, 1179–1192. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Bojjireddy, N.; Kim, M.; Li, T.; Zhai, P.; Nagarajan, N.; Sadoshima, J.; Palmiter, R.D.; Tian, R. Activation of γ2-AMPK Suppresses Ribosome Biogenesis and Protects Against Myocardial Ischemia/Reperfusion Injury. Circ. Res. 2017, 121, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kaufman, R.J. The impact of the unfolded protein response on human disease. J. Cell Biol. 2012, 197, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.H.; Ploegh, H.L.; Weissman, J.S. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science 2011, 335, 1086–1090. [Google Scholar] [CrossRef] [PubMed]
- Bordallo, J.; Plemper, R.K.; Finger, A.; Wolf, D.H. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell 1998, 9, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Belmont, P.J.; Tadimalla, A.; Chen, W.J.; Martindale, J.J.; Thuerauf, D.J.; Marcinko, M.; Gude, N.; Sussman, M.A.; Glembotski, C.C. Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (RCAN1), a novel ATF6-inducible gene. J. Biol. Chem. 2008, 283, 14012–14021. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Tan, J.; Miao, Y.; Zhang, Q. Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery. J. Cell Physiol. 2018, 233, 3867–3874. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Wilkinson, S. ER homeostasis and autophagy. Essays Biochem. 2017, 61, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cui, Y.; Parashar, S.; Novick, P.J.; Ferro-Novick, S. ER-phagy requires Lnp1, a protein that stabilizes rearrangements of the ER network. Proc. Natl. Acad. Sci. USA 2018, 115, E6237–E6244. [Google Scholar] [CrossRef] [PubMed]
- Loi, M.; Fregno, I.; Guerra, C.; Molinari, M. Eat it right: ER-phagy and recovER-phagy. Biochem. Soc. Trans. 2018, 46, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Hino, S.; Saito, A.; Morikawa, K.; Kondo, S.; Kanemoto, S.; Murakami, T.; Taniguchi, M.; Tanii, I.; Yoshinaga, K.; et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006, 26, 9220–9231. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Liu, H.; Jiang, C.C.; Fang, L.; Chen, C.; Zhang, X.D.; Jiang, Z.W. Connecting endoplasmic reticulum stress to autophagy through IRE1/JNK/beclin-1 in breast cancer cells. Int. J. Mol. Med. 2014, 34, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Yung, H.W.; Charnock-Jones, D.S.; Burton, G.J. Regulation of AKT phosphorylation at Ser473 and Thr308 by endoplasmic reticulum stress modulates substrate specificity in a severity dependent manner. PLoS ONE 2011, 6, e17894. [Google Scholar] [CrossRef] [PubMed]
- Khaminets, A.; Heinrich, T.; Mari, M.; Grumati, P.; Huebner, A.K.; Akutsu, M.; Liebmann, L.; Stolz, A.; Nietzsche, S.; Koch, N.; et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015, 522, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Grumati, P.; Dikic, I.; Stolz, A. ER-phagy at a glance. J. Cell. Sci. 2018, 131, jcs217364. [Google Scholar] [CrossRef] [PubMed]
- Bernales, S.; Schuck, S.; Walter, P. ER-phagy: Selective autophagy of the endoplasmic reticulum. Autophagy 2007, 3, 285–287. [Google Scholar]
- Santulli, G.; Xie, W.; Reiken, S.R.; Marks, A.R. Mitochondrial calcium overload is a key determinant in heart failure. Proc. Natl. Acad. Sci. USA 2015, 112, 11389–11394. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, M.; Ciccarelli, M.; Fiordelisi, A.; Iaccarino, G.; Pinto, A.; Popolo, A. Diazoxide Improves Mitochondrial Connexin 43 Expression in a Mouse Model of Doxorubicin-Induced Cardiotoxicity. Int. J. Mol. Sci. 2018, 19, 757. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chen, Z.; Chen, A.; Fu, M.; Dong, Z.; Hu, K.; Yang, X.; Zou, Y.; Sun, A.; Qian, J.; et al. LCZ696 improves cardiac function via alleviating Drp1-mediated mitochondrial dysfunction in mice with doxorubicin-induced dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2017, 108, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Putti, R.; Della Rossa, A. PeptideYY and insulin coexist in beta-granules in B cells of the Madagascan lizard, Zonosaurus laticaudatus. Gen. Comp. Endocrinol. 1996, 103, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Y.; Liu, H.; Li, P.; Zhang, H.; Cheng, G. Fortunellin protects against high fructose-induced diabetic heart injury in mice by suppressing inflammation and oxidative stress via AMPK/Nrf-2 pathway regulation. Biochem. Biophys. Res. Commun. 2017, 490, 552–559. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.-M.; Li, C.-B.; Liu, Q.-L.; Li, P.; Yang, H. Ginsenoside Rg1 Prevents Doxorubicin-Induced Cardiotoxicity through the Inhibition of Autophagy and Endoplasmic Reticulum Stress in Mice. Int. J. Mol. Sci. 2018, 19, 3658. https://doi.org/10.3390/ijms19113658
Xu Z-M, Li C-B, Liu Q-L, Li P, Yang H. Ginsenoside Rg1 Prevents Doxorubicin-Induced Cardiotoxicity through the Inhibition of Autophagy and Endoplasmic Reticulum Stress in Mice. International Journal of Molecular Sciences. 2018; 19(11):3658. https://doi.org/10.3390/ijms19113658
Chicago/Turabian StyleXu, Zhi-Meng, Cheng-Bin Li, Qing-Ling Liu, Ping Li, and Hua Yang. 2018. "Ginsenoside Rg1 Prevents Doxorubicin-Induced Cardiotoxicity through the Inhibition of Autophagy and Endoplasmic Reticulum Stress in Mice" International Journal of Molecular Sciences 19, no. 11: 3658. https://doi.org/10.3390/ijms19113658
APA StyleXu, Z.-M., Li, C.-B., Liu, Q.-L., Li, P., & Yang, H. (2018). Ginsenoside Rg1 Prevents Doxorubicin-Induced Cardiotoxicity through the Inhibition of Autophagy and Endoplasmic Reticulum Stress in Mice. International Journal of Molecular Sciences, 19(11), 3658. https://doi.org/10.3390/ijms19113658





