Sensing of Abiotic Stress and Ionic Stress Responses in Plants
Abstract
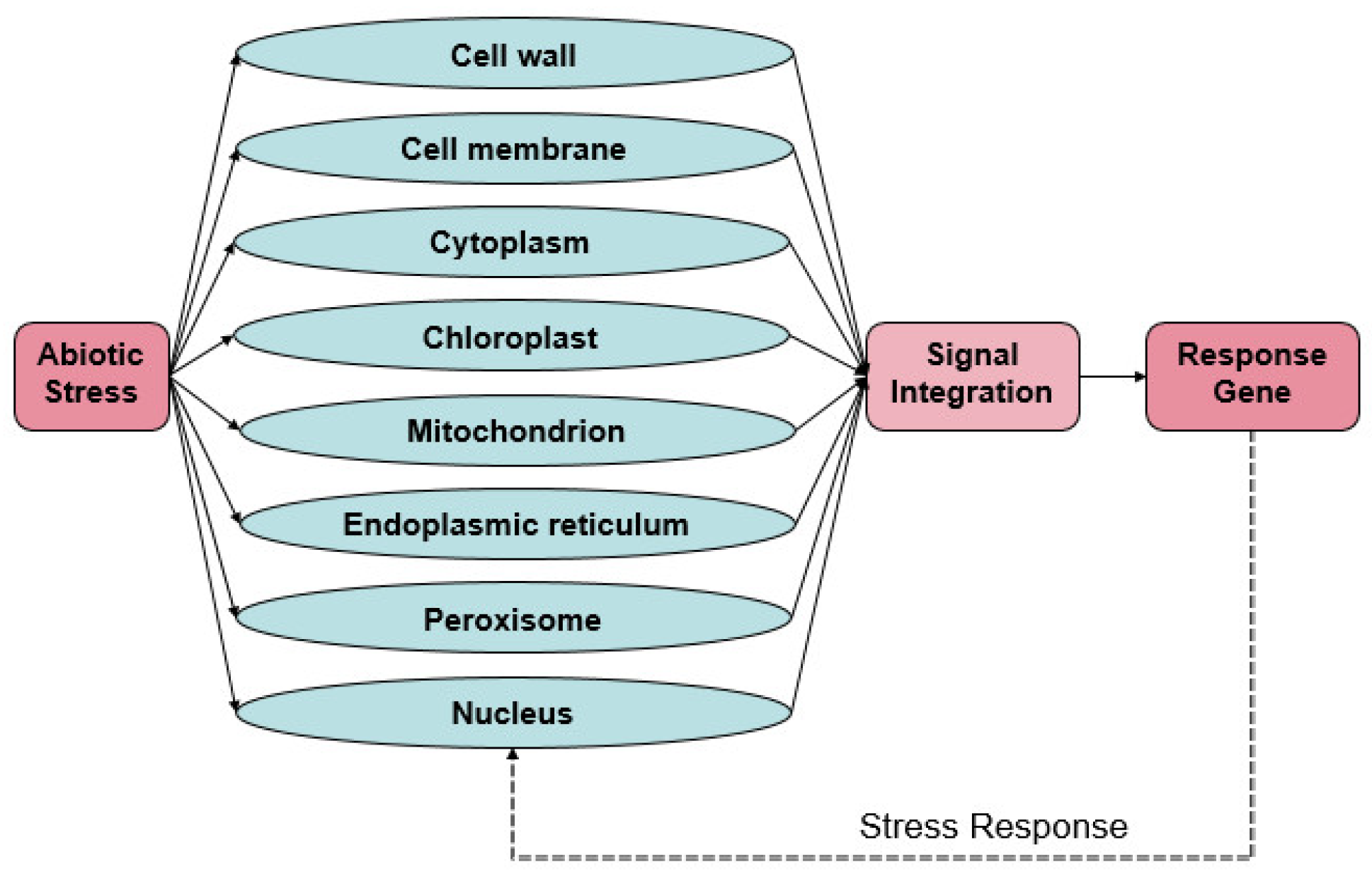
1. Introduction
2. Sensing of Organellar Stress
3. Ionic Stress Signaling
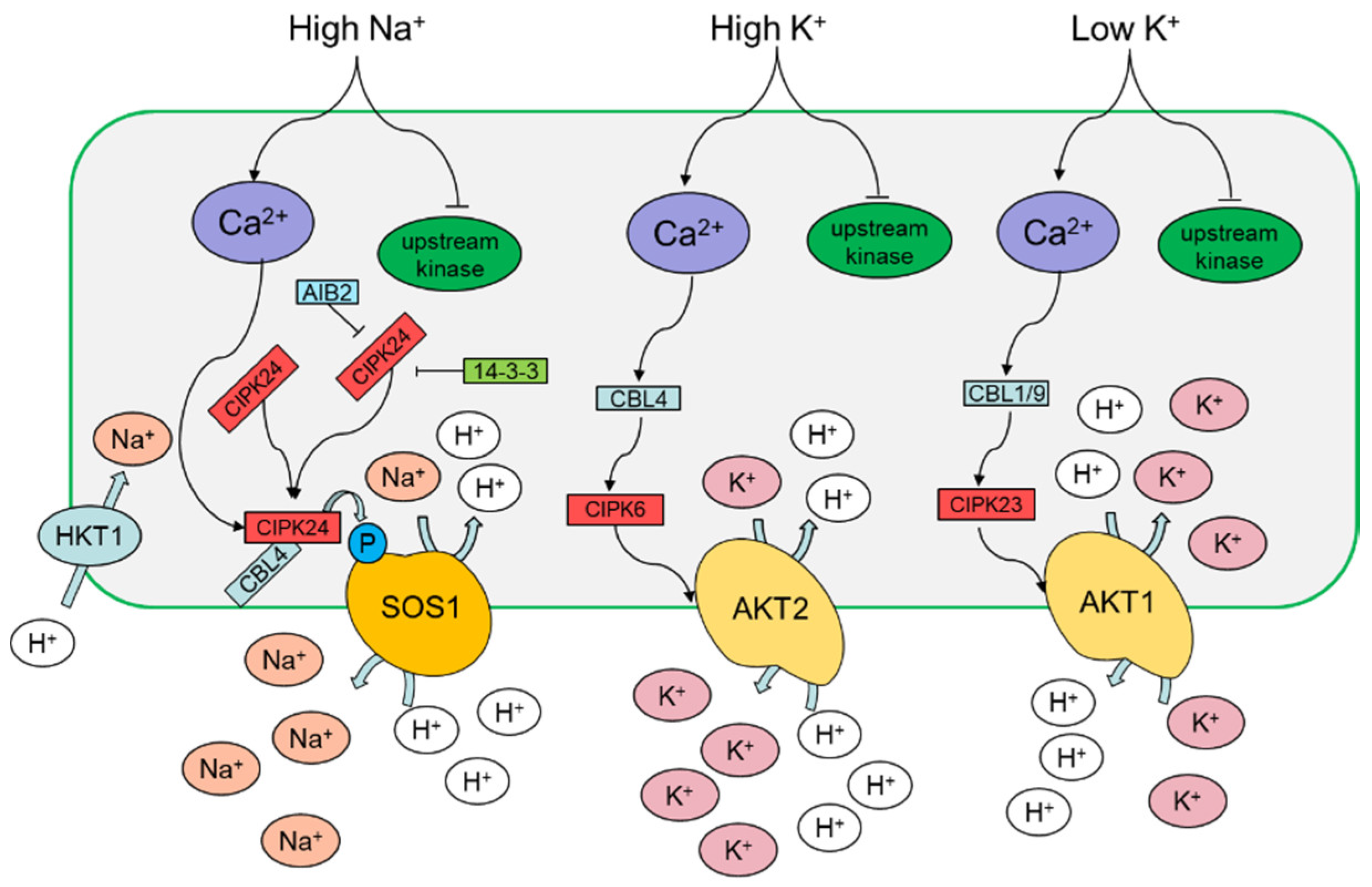
3.1. Sodium
3.2. Potassium
3.3. Nitrate
3.4. Phosphorus
3.5. Magnesium
3.6. Calcium
4. Discussion
Acknowledgments
Conflicts of Interest
References
- Manik, S.M.; Shi, S.; Mao, J.; Dong, L.; Su, Y.; Wang, Q.; Liu, H. The calcium sensor CBL-CIPK is involved in plant’s response to abiotic stresses. Int. J. Genom. 2015, 2015, 493191. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop. J. 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Penna, S.; Nguyen, D.V.; Tran, L.S. Multifaceted roles of aquaporins as molecular conduits in plant responses to abiotic stresses. Crit. Rev. Biotechnol. 2016, 36, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Latif, F.; Ullah, F.; Mehmood, S.; Khattak, A.; Khan, A.U.; Khan, S.; Husain, I. Effects of salicylic acid on growth and accumulation of Phenolics in Zea mays L. under drought stress. Acta Agric. Scand. Sect. B Soil Plant Sci. 2015, 66, 325–332. [Google Scholar]
- Gorji, A.H.; Hajianfar, R.; Rostamforody, B.; Production, P. Effects of cold and other abiotic stress on plants. Int. J. Agron. 2013, 3597–3604. [Google Scholar]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 162, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gao, K.; Ren, H.; Tang, W. Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Berriri, S.; Kumar, S.V. PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Curr. Biol. 2017, 27, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yuan, T.; Tarkowská, D.; Kim, J.; Nam, H.G.; Novák, O.; He, K.; Gou, X.; Li, J. Brassinosteroid biosynthesis is modulated via a transcription factor cascade of COG1, PIF4, and PIF5. Plant Physiol. 2017, 174, 1260–1273. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M. Crop plants and abiotic stresses. J. Biomol. Res. Ther. 2014, 03, e125. [Google Scholar] [CrossRef]
- Kumar, K.; Kumar, M.; Kim, S.R.; Ryu, H.; Cho, Y.G. Insights into genomics of salt stress response in rice. Rice 2013, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Choi, J.; An, G.; Kim, S.R. Ectopic expression of OSSTA2 enhances salt stress tolerance in rice. Front. Plant Sci. 2017, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; et al. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yan, S.; Yang, T.; Zhang, S.; Chen, Y.Q.; Liu, B. Small RNAs in regulating temperature stress response in plants. J. Integr. Plant Biol. 2017, 59, 774–791. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, M.; Zhang, X. The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 2016, 58, 312–327. [Google Scholar] [CrossRef] [PubMed]
- De Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant. Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant. Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (BZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central HUB for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Mignolet-Spruyt, L.; Xu, E.; Idanheimo, N.; Hoeberichts, F.A.; Muhlenbock, P.; Brosche, M.; van Breusegem, F.; Kangasjarvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef] [PubMed]
- Bobik, K.; Burch-Smith, T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015, 6, 781. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Hu, S.; Wu, L.; Ge, C.; Cui, Y.; Chen, P.; Wang, X.; Xu, J.; Ren, D.; Dong, G.; et al. White stripe leaf 12 (WSL12), encoding a nucleoside diphosphate kinase 2 (OsNDPK2), regulates chloroplast development and abiotic stress response in rice (Oryza sativa L.). Mol. Breed. 2016, 36, 57. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; de Clercq, I.; Van Aken, O.; Law, S.R.; Ivanova, A.; Willems, P.; Giraud, E.; van Breusegem, F.; Whelan, J. Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant 2014, 7, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; van Aken, O.; Schwarzlander, M.; Belt, K.; Millar, A.H. The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed]
- Leucci, M.R.; Lenucci, M.S.; Piro, G.; Dalessandro, G. Water stress and cell wall polysaccharides in the apical root zone of wheat cultivars varying in drought tolerance. J. Plant Physiol. 2008, 165, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budinska, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef] [PubMed]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Sensing the environment: Key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 2013, 64, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Mizuno, S.; Tanaka, H.; Maruyama, K.; Osakabe, K.; Todaka, D.; Fujita, Y.; Kobayashi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J. Biol. Chem. 2010, 285, 9190–9201. [Google Scholar] [CrossRef] [PubMed]
- Leister, D.; Wang, L.; Kleine, T. Organellar gene expression and acclimation of plants to environmental stress. Front. Plant Sci. 2017, 8, 387. [Google Scholar] [CrossRef] [PubMed]
- Koster, P.; Wallrad, L.; Edel, K.H.; Faisal, M.; Alatar, A.A.; Kudla, J. The battle of two ions: Ca2+ signalling against Na+ stress. Plant Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Aragon, R.; Rodriguez-Navarro, A. Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J. 2017, 91, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant. Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Wu, H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. N. Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; An, L.; Li, W. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014, 33, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 2000, 97, 3730–3734. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000, 124, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Gilliham, M.; Hrmova, M. Plant high-affinity potassium (HKT) transporters involved in salinity tolerance: Structural insights to probe differences in ion selectivity. Int. J. Mol. Sci. 2013, 14, 7660–7680. [Google Scholar] [CrossRef] [PubMed]
- Rus, A.; Lee, B.H.; Munoz-Mayor, A.; Sharkhuu, A.; Miura, K.; Zhu, J.K.; Bressan, R.A.; Hasegawa, P.M. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 2004, 136, 2500–2511. [Google Scholar] [CrossRef] [PubMed]
- Sunarpi; Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.Y.; Leung, H.Y.; Hattori, K.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 44, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Maser, P.; Eckelman, B.R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimura, M.; Uozumi, N.; Robertson, W. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.T.; Bandillo, N.; Al Shiblawi, F.R.A.; Sharma, S.; Liu, K.; Du, Q.; Schmitz, A.J.; Zhang, C.; Very, A.A.; Lorenz, A.J.; et al. Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLoS Genet. 2017, 13, e1006823. [Google Scholar] [CrossRef] [PubMed]
- Cotsaftis, O.; Plett, D.; Shirley, N.; Tester, M.; Hrmova, M. A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 2012, 7, e39865. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Yamaji, N.; Costa, A.; Okuma, E.; Kobayashi, N.I.; Kashiwagi, T.; Katsuhara, M.; Wang, C.; Tanoi, K.; Murata, Y.; et al. OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 2016, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Wangsawang, T.; Chuamnakthong, S.; Kohnishi, E.; Sripichitt, P.; Sreewongchai, T.; Ueda, A. A salinity-tolerant japonica cultivar has Na+ exclusion mechanism at leaf sheaths through the function of a Na+ transporter OsHKT1;4 under salinity stress. J. Agron. Crop. Sci. 2018, 204, 274–284. [Google Scholar] [CrossRef]
- Oda, Y.; Kobayashi, N.I.; Tanoi, K.; Ma, J.F.; Itou, Y.; Katsuhara, M.; Itou, T.; Horie, T. T-DNA tagging-based gain-of-function of OsHKT1;4 reinforces na exclusion from leaves and stems but triggers Na toxicity in roots of rice under salt stress. Int. J. Mol. Sci. 2018, 19, 235. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Lin, H.; Chen, S.; Wu, Y.; Zhang, J.; Fuglsang, A.T.; Palmgren, M.G.; Wu, W.; Guo, Y. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 2011, 156, 2235–2243. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin, H.; Chen, S.; Becker, K.; Yang, Y.; Zhao, J.; Kudla, J.; Schumaker, K.S.; Guo, Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14–3–3 proteins. Plant Cell 2014, 26, 1166–1182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Wang, J.; Chen, X.; Wang, Z.X.; Wu, J.W. Crystal structure of the kinase and UBA domains of SnRK reveals a distinct UBA binding mode in the AMPK family. Biochem. Biophys. Res. Commun. 2018, 495, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Carraro, D.M.; Lambais, M.R.; Carrer, H. In silico characterization and expression analyses of sugarcane putative sucrose non-fermenting-1 (SNF1) related kinases. Genet. Mol. Biol. 2001, 24, 35–41. [Google Scholar] [CrossRef]
- Dong, X.F.; Cui, N.; Wang, L.; Zhao, X.C.; Qu, B.; Li, T.L.; Zhang, G.L. The SnRK protein kinase family and the function of SnRK1 protein kinase. Int. J. Agric. Biol. 2012, 14, 575–579. [Google Scholar]
- Alderson, A.; Sabelli, P.A.; Dickinson, J.R.; Cole, D.; Richardson, M.; Kreis, M.; Shewry, P.R.; Halford, N.G. Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cdna. Proc. Natl. Acad. Sci. USA 1991, 88, 8602–8605. [Google Scholar] [CrossRef] [PubMed]
- Halford, N.G.; Hardie, D.G. SNF1-related protein kinases: Global regulators of carbon metabolism in plants? Plant Mol. Biol. 1998, 37, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Crozet, P.; Margalha, L.; Confraria, A.; Rodrigues, A.; Martinho, C.; Adamo, M.; Elias, C.A.; Baena-Gonzalez, E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant Sci. 2014, 5, 190. [Google Scholar] [CrossRef] [PubMed]
- Muranaka, T.; Banno, H.; Machida, T. Characterization of tobacco protein kinase NPK5, a homolog of saccharomyces cerevisiae SNF1 that constitutively activates expression of the glucose-repressible SUC2 gene for a secreted invertase of S. cerevisiae. Mol. Cell Biol. 1994, 14, 2958–2965. [Google Scholar] [CrossRef] [PubMed]
- Saha, J.; Chatterjee, C.; Sengupta, A.; Gupta, K.; Gupta, B. Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput. Biol. Chem. 2014, 49, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant. Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Du, W.; Yang, Y.; Schumaker, K.S.; Guo, Y. A calcium-independent activation of the Arabidopsis SOS2-like protein kinase24 by its interacting SOS3-like calcium binding protein1. Plant Physiol. 2014, 164, 2197–2206. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Mian, A.; Maathuis, F.J. Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J. Exp. Bot. 2016, 67, 2689–2698. [Google Scholar] [CrossRef] [PubMed]
- Adams, E.; Shin, R. Transport, signaling, and homeostasis of potassium and sodium in plants. J. Integr. Plant Biol. 2014, 56, 231–249. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, H.D.; Chen, L.Q.; Wang, Y.; Liu, L.L.; He, L.; Wu, W.H. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Cheong, Y.H.; Pandey, G.K.; Grant, J.J.; Batistic, O.; Li, L.; Kim, B.G.; Lee, S.C.; Kudla, J.; Luan, S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007, 52, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Thoday-Kennedy, E.L.; Jacobs, A.K.; Roy, S.J. The role of the CBL–CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 2015, 76, 3–12. [Google Scholar] [CrossRef]
- Pilot, G.; Gaymard, F.; Mouline, K.; Chérel, I.; Sentenac, H. Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 2003, 51, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lv, F.; Han, X.; Xia, X.; Yin, W. The calcium sensor PeCBL1, interacting with PeCIPK24/25 and PeCIPK26, regulates Na+/K+ homeostasis in Populus euphratica. Plant Cell Rep. 2013, 32, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Long, Y.; Qi, G.N.; Li, J.; Xu, Z.J.; Wu, W.H.; Wang, Y. The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 2014, 26, 3387–3402. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.L.; Qi, G.N.; Feng, H.Q.; Zhao, S.; Zhao, S.S.; Wang, Y.; Wu, W.H. Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 2013, 74, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Gajdanowicz, P.; Michard, E.; Sandmann, M.; Rocha, M.; Correa, L.G.; Ramirez-Aguilar, S.J.; Gomez-Porras, J.L.; Gonzalez, W.; Thibaud, J.B.; van Dongen, J.T.; et al. Potassium K+ gradients serve as a mobile energy source in plant vascular tissues. Proc. Natl. Acad. Sci. USA 2011, 108, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, M.; Skłodowski, K.; Gajdanowicz, P.; Michard, E.; Rocha, M.; Gomez-Porras, J.L.; González, W.; Corrêa, L.G.G.; Ramírez-Aguilar, S.J.; Cuin, T.A.; et al. The K+ battery-regulating Arabidopsis K+ channel AKT2 is under the control of multiple post-translational steps. Plant Signal. Behav. 2011, 6, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Deeken, R.; Geiger, D.; Fromm, J.; Koroleva, O.; Ache, P.; Langenfeld-Heyser, R.; Sauer, N.; May, S.T.; Hedrich, R. Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 2002, 216, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Held, K.; Pascaud, F.; Eckert, C.; Gajdanowicz, P.; Hashimoto, K.; Corratge-Faillie, C.; Offenborn, J.N.; Lacombe, B.; Dreyer, I.; Thibaud, J.B.; et al. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011, 21, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Chérel, I.E.M.; Platet, N.; Mouline, K.; Alcon, C.; Sentenac, H.; Thibaud, J.B. Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell Online 2002, 14, 1133–1146. [Google Scholar] [CrossRef]
- Liu, L.L.; Ren, H.M.; Chen, L.Q.; Wang, Y.; Wu, W.H. A protein kinase, CIPK9 interacts with calcium sensor CBL3 and regulates K+ homeostasis under low-K+ stress in Arabidopsis. Plant Physiol. 2013, 161, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.G. Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 817. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, W.H. Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 2013, 64, 451–476. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, Q.; Luo, L.; Yang, T.; Zhang, S.; Hu, Y.; Yu, L.; Xu, G. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 2015, 38, 2747–2765. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, C.; Gao, Z.; Zhang, Y.; Jiang, H.; Zhu, L.; Ren, D.; Yu, L.; Xu, G.; Qian, Q. OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice. Front. Plant Sci. 2017, 8, 1885. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, C.; Gao, Z.; Zhang, Y.; Zhang, A.; Zhu, L.; Hu, J.; Ren, D.; Yu, L.; Xu, G.; et al. Variation in the abundance of OsHAK1 transcript underlies the differential salinity tolerance of an indica and a Japonica rice cultivar. Front. Plant Sci. 2017, 8, 2216. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Sugawara, M.; Okada, T.; Taira, K.; Kaothien-Nakayama, P.; Katsuhara, M.; Shinmyo, A.; Nakayama, H. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 2011, 111, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Taghavi, T.; Folta, K.M. A comparison of wild and cultivated strawberries for nitrogen uptake and reduction. Horticult. Environ. Biotechnol. 2014, 55, 196–206. [Google Scholar] [CrossRef]
- Bouguyon, E.; Gojon, A.; Nacry, P. Nitrate sensing and signaling in plants. Semin. Cell Dev. Biol. 2012, 23, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 2002, 53, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Crawford, N.M.; Coruzzi, G.M.; Tsay, Y.F. Nitrate signaling: Adaptation to fluctuating environments. Curr. Opin. Plant Biol. 2010, 13, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef]
- Guan, P. Dancing with hormones: A current perspective of nitrate signaling and regulation in Arabidopsis. Front. Plant Sci. 2017, 8, 1697. [Google Scholar] [CrossRef] [PubMed]
- De Angeli, A.; Monachello, D.; Ephritikhine, G.; Frachisse, J.M.; Thomine, S.; Gambale, F.; Barbier-Brygoo, H. CLC-mediated anion transport in plant cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 2000, 1465, 219–235. [Google Scholar] [CrossRef]
- Liu, K.; Tsay, Y.; Huang, C. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 1999, 11, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, Y.; Okamoto, M.; Crawford, N.M.; Siddiqi, M.Y.; Glass, A.D. Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 2007, 143, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Araújo, O.J.L.; Pinto, M.S.; Sperandio, M.V.L.; Santos, L.A.; Stark, E.M.L.M.; Fernandes, M.S.; Santos, A.M.D.; Souza, S.R.D. Expression of the genes OsNRT1.1, OsNRT2.1, OsNRT2.2, and kinetics of nitrate uptake in genetically contrasting rice varieties. Am. J. Plant Sci. 2015, 06, 306–314. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.C.; Wang, Y.Y.; Tsay, Y.F. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009, 57, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Bieleski, R.L. Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 1973, 24, 225–252. [Google Scholar] [CrossRef]
- Chen, L.; Ren, F.; Zhou, L.; Wang, Q.Q.; Zhong, H.; Li, X.B. The Brassica napus Calcineurin B-like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 2012, 63, 6211–6222. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Chen, P.; Zhang, Y.; Wang, Z.; Hu, S.; Jin, G.; Tang, C.; Guo, L. Natural variation among Arabidopsis thaliana accessions in tolerance to high magnesium supply. Sci. Rep. 2018, 8, 13640. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.J.; Zhao, F.G.; Garcia, V.J.; Kleist, T.J.; Yang, L.; Zhang, H.X.; Luan, S. Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3134–3139. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhao, Q.; Jiang, L. Vacuoles protect plants from high magnesium stress. Proc. Natl. Acad. Sci. USA 2015, 112, 2931–2932. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; Waadt, R.; Cheong, Y.H.; Pandey, G.K.; Dominguez-Solis, J.R.; SchãLtke, S.; Lee, S.C.; Kudla, J.; Luan, S. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007, 52, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B–like proteins. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Yin, X.; Wang, Q.; Chen, Q.; Xiang, N.; Yang, Y.; Yang, Y. Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in turnip (Brassica rapa var. rapa). Front. Plant Sci. 2017, 8, 1191. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.L.; Zhang, X.H.; Li, R.J.; Shi, H.T.; Lu, Y.T. A calcium sensor-interacting protein kinase negatively regulates salt stress tolerance in rice (Oryza sativa). Funct. Plant Biol. 2011, 38, 441–450. [Google Scholar]
- Piao, H.; Xuan, Y.H.; Suhyun, P.; Byoungil, J.; Soonju, P.; Sunghan, P.; Chulmin, K.; Jin, H.; Wang, G.K.; Kim, M. OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol. Cells 2010, 30, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kurusu, T.; Hamada, J.; Nokajima, H.; Kitagawa, Y.; Kiyoduka, M.; Takahashi, A.; Hanamata, S.; Ohno, R.; Hayashi, T.; Okada, K.; et al. Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol. 2010, 153, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, U. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, B.; Liu, W.Z.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y.Q. Identification and characterization of CBL and CIPK gene families in canola ( Brassica napus L.). BMC Plant Biol. 2014, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F.; Zhi-Min, G.U.; Feng, L.; Bo-Jun, M.A.; Zhang, H.S. Molecular analysis of rice CIPKs involved in both biotic and abiotic stress responses. Rice Sci. 2011, 18, 1–9. [Google Scholar] [CrossRef]
- Yu, Y.; Xia, X.; Yin, W.; Zhang, H. Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 2007, 52, 101–110. [Google Scholar] [CrossRef]
- Zhang, H.; Yin, W.; Xia, X. Calcineurin B-like family in Populus: Comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul. 2008, 56, 129–140. [Google Scholar] [CrossRef]
- Jing, L.; Jiang, M.M.; Li, R.; Yang, L.; Chen, H.Y. Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genom. 2016, 291, 1769–1781. [Google Scholar]
- Niu, L.; Dong, B.; Song, Z.; Meng, D.; Fu, Y. Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple (Malus domestica). Int. J. Mol. Sci. 2018, 19, 2131. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Schumaker, K.; Zhu, J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 2004, 55, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
| Taxonomic Group | Latin Name | Species | TFs | Loci | Families |
|---|---|---|---|---|---|
| Monocots | Oryza sativa subsp. japonica | Rice | 2408 | 1862 | 56 |
| Oryza sativa subsp. indica | 1891 | 1891 | 56 | ||
| Sorghum bicolor | Sorghum | 2654 | 1859 | 56 | |
| Triticum aestivum | Wheat | 3606 | 3606 | 56 | |
| Zea mays | Maize | 3308 | 2289 | 56 | |
| Eudicots | Arabidopsis thaliana | Arabidopsis | 2296 | 1717 | 58 |
| Nicotiana tabacum | Tobacco | 5176 | 3625 | 57 | |
| Solanum lycopersicum | Tomato | 1845 | 1845 | 58 | |
| Solanum tuberosum | Potato | 2405 | 1736 | 56 |
| Species | CBLs | CIPKs | Reference |
|---|---|---|---|
| Arabidopsis thaliana | 10 | 26 | [120,128,129] |
| O. sativa | 10 | 31 | [128] |
| Triticum aestivum L. | 7 | 20 | [130] |
| Z. mays | 8 | 43 | [131] |
| Populus trichocarpa | 10 | 27 | [132,133] |
| B. napus L. | 7 | 23 | [129] |
| S. melongena L. | 5 | 15 | [134] |
| Malus domestica | - | 34 | [135] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Lv, Y.; Jahan, N.; Chen, G.; Ren, D.; Guo, L. Sensing of Abiotic Stress and Ionic Stress Responses in Plants. Int. J. Mol. Sci. 2018, 19, 3298. https://doi.org/10.3390/ijms19113298
Zhang Y, Lv Y, Jahan N, Chen G, Ren D, Guo L. Sensing of Abiotic Stress and Ionic Stress Responses in Plants. International Journal of Molecular Sciences. 2018; 19(11):3298. https://doi.org/10.3390/ijms19113298
Chicago/Turabian StyleZhang, Yu, Yang Lv, Noushin Jahan, Guang Chen, Deyong Ren, and Longbiao Guo. 2018. "Sensing of Abiotic Stress and Ionic Stress Responses in Plants" International Journal of Molecular Sciences 19, no. 11: 3298. https://doi.org/10.3390/ijms19113298
APA StyleZhang, Y., Lv, Y., Jahan, N., Chen, G., Ren, D., & Guo, L. (2018). Sensing of Abiotic Stress and Ionic Stress Responses in Plants. International Journal of Molecular Sciences, 19(11), 3298. https://doi.org/10.3390/ijms19113298








