miRNA-Mediated Interactions in and between Plants and Insects
Abstract
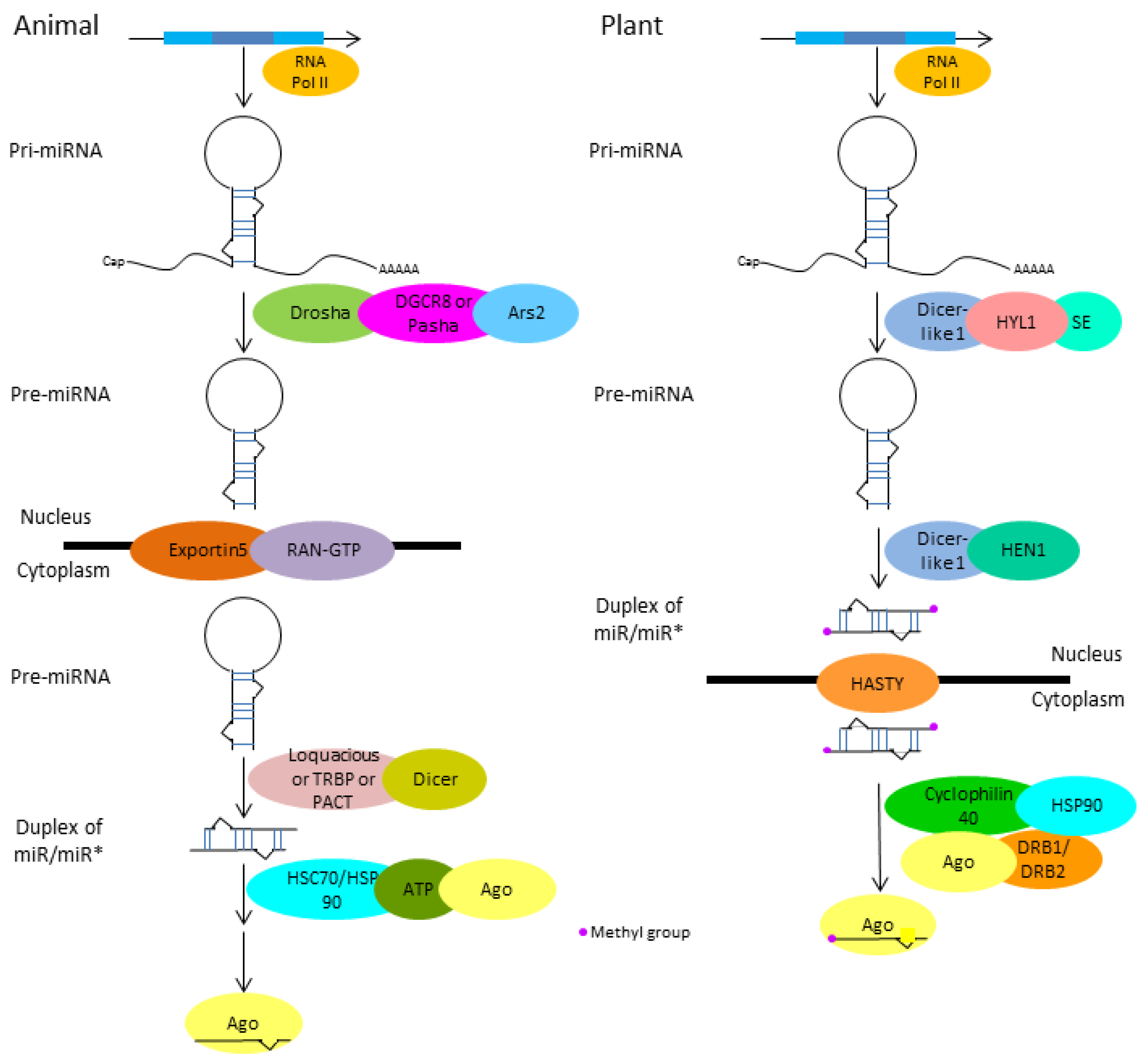
1. MicroRNA (miRNA) Biogenesis and Regulation of Cellular Processes
2. Plant-Insect Interactions
2.1. Plant miRNAs Targeting Plant Defense Responses
2.2. Insect miRNAs Targeting Plant Defense Responses
2.3. Plant miRNA Targeting Herbivorous Insects
2.4. Insect Subversion
2.5. Plant miRNA: Interactions with Beneficial Insects
3. Biotechnological Applications
Funding
Conflicts of Interest
References
- Wang, J.; Meng, X.; Dobrovolskaya, O.B.; Orlov, Y.L.; Chen, M. Non-coding RNAs and Their Roles in Stress Response in Plants. Genom. Proteom. Bioinform. 2017, 15, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, A.; Zielenkiewicz, P. Plant MicroRNAs-Novel Players in Natural Medicine? Int. J. Mol. Sci. 2016, 18, E9. [Google Scholar] [CrossRef] [PubMed]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs as Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Q.; Pan, X. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol. 2007, 210, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Hohn, T. Biogenesis and Biological Activity of Secondary siRNAs in Plants. Scientifica 2013. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.Q.; Tong, W.S.; Yu, H.Y.; Tobe, S.S.; Bendena, W.G.; Hui, J.H.L. The role of microRNAs in Drosophila regulation of insulin-like peptides and ecdysteroid signaling: Where are we now? Adv. Insect Physiol. 2017, 53, 55–85. [Google Scholar] [CrossRef]
- Moran, Y.; Agron, M.; Praher, D.; Technau, U. The evolutionary origin of plant and animal microRNAs. Nat. Ecol. Evol. 2017, 1, 27. [Google Scholar] [CrossRef] [PubMed]
- Guleria, P.; Mahajan, M.; Bhardwaj, J.; Yadav, S.K. Plant Small RNAs: Biogenesis, Mode of Action and Their Roles in Abiotic Stresses. Genom. Proteom. Bioinform. 2011, 9, 183–199. [Google Scholar] [CrossRef]
- Yang, L.; Wu, G.; Poethig, R.S. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Chekulaeva, M.; Mathys, H.; Zipprich, J.T.; Attig, J.; Colic, M.; Parker, R.; Filipowicz, W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011, 18, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Fredman, D.; Praher, D.; Li, X.Z.; Wee, L.M.; Rentzsch, F.; Zamore, P.D.; Technau, U.; Seitz, H. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 2014, 24, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, M.; Miura, S.; Nei, M. Origins and evolution of microRNA genes in plant species. Genome Biol. Evol. 2012, 4, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Valli, A.A.; Santos, B.A.; Hnatova, S.; Bassett, A.R.; Molnar, A.; Chung, B.Y.; Baulcombe, D.C. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 2016, 26, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Barbosa, D. Integrating Studies on Plant–Pollinator and Plant–Herbivore Interactions. Trends Plant Sci. 2016, 21, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Aljbory, Z.; Chen, M.S. Indirect plant defense against insect herbivores: A review. Insect Sci. 2018, 25, 2–23. [Google Scholar] [CrossRef] [PubMed]
- Fürstenberg-Hägg, J.; Zagrobelny, M.; Bak, S. Plant defense against insect herbivores. Int. J. Mol. Sci. 2013, 14, 10242–10297. [Google Scholar] [CrossRef] [PubMed]
- Poelman, E.H. From induced resistance to defence in plant-insect interactions. Entomol. Exp. Appl. 2015, 157, 11–17. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef] [PubMed]
- Zhu-Salzman, K.; Bi, J.; Liu, T. Molecular strategies of plant defense and insect counter-defense. Insect Sci. 2005, 12, 3–15. [Google Scholar] [CrossRef]
- Alba, J.M.; Glas, J.J.; Schimmel, B.C.J.; Kant, M.R. Avoidance and suppression of plant defenses by herbivores and pathogens. J. Plant Interact. 2011, 6, 221–227. [Google Scholar] [CrossRef]
- Bronstein, J.L.; Alarcón, R.; Geber, M. The evolution of plant-insect mutualisms. New Phytol. 2006, 172, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Filella, I.; Primante, C.; Llusià, J.; Martín González, A.M.; Seco, R.; Farré-Armengol, G.; Rodrigo, A.; Bosch, J.; Peñuelas, J. Floral advertisement scent in a changing plant-pollinators market. Sci. Rep. 2013, 3, 3434. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Barbosa, D.; Sun, P.; Hakman, A.; van Beek, T.A.; van Loon, J.J.A.; Dicke, M. Visual and odour cues: Plant responses to pollination and herbivory affect the behaviour of flower visitors. Funct. Ecol. 2016, 30, 431–441. [Google Scholar] [CrossRef]
- Sattar, S.; Thompson, G.A. Small RNA regulators of plant-hemipteran interactions: Micromanagers with versatile roles. Front. Plant Sci. 2016, 7, 1241. [Google Scholar] [CrossRef] [PubMed]
- Bohn, G.W.; Kishaba, A.N.; Toba, H.H. Mechanisms of resistance to melon aphid in a muskmelon line. Hortscience 1972, 7, 281–282. [Google Scholar]
- Sattar, S.; Song, Y.; Anstead, J.A.; Sunkar, R.; Thompson, G.A. Cucumis melo microRNA expression profile during aphid herbivory in a resistant and susceptible interaction. Mol. Plant Microbe Interact. 2012, 25, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraj, A.; Liu, S.; Zhang, X.; Zhang, R.; Shangguan, M.; Wei, C. Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L.). Sci. Rep. 2017, 7, 13634. [Google Scholar] [CrossRef] [PubMed]
- Saedler, R.; Baldwin, I.T. Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuate. J. Exp. Bot. 2004, 55, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Steppuhn, A.; Gase, K.; Krock, B.; Halitschke, R.; Baldwin, I.T. Nicotine’s Defensive Function in Nature. PLoS Boil. 2004, 2, e217. [Google Scholar] [CrossRef] [PubMed]
- Bozorov, T.A.; Pandey, S.P.; Dinh, S.T.; Kim, S.; Heinrich, M.; Gase, K.; Baldwin, I.T. DICER-like Proteins and Their Role in Plant-herbivore Interactions in Nicotiana attenuata. J. Integr. Plant Biol. 2012, 54, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Baldwin, I.T. RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature. Plant J. 2007, 50, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Mithöfer, A.; Boland, W. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Després, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, C.E.; Shan, G.; Ottea, J. Overview of Carboxylesterases and Their Role in the Metabolism of Insecticides. J. Pestic. Sci. 2005, 30, 75–83. [Google Scholar] [CrossRef]
- Liu, N.; Li, M.; Gong, Y.; Liu, F.; Li, T. Cytochrome P450s—Their expression, regulation, and role in insecticide resistance. Pestic. Biochem. Physiol. 2015, 120, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Calla, B.; Noble, K.; Johnson, R.M.; Walden, K.K.O.; Schuler, M.A.; Robertson, H.M.; Berenbaum, M.R. Cytochrome P450 diversification and hostplant utilization patterns in specialist and generalist moths: Birth, death and adaptation. Mol. Ecol. 2017, 26, 6021–6035. [Google Scholar] [CrossRef] [PubMed]
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Pavlidi, N.; Vontas, J.; Van Leeuwen, T. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opin. Insect Sci. 2018, 27, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Li, F.; Liang, P.; Chen, X.; Liu, Y.; Tang, Q.; Gao, X. RNA interference of Dicer-1 and Argonaute-1 increasing the sensitivity of Aphis gossypii Glover (Hemiptera: Aphididae) to plant allelochemical. Pestic. Biochem. Physiol. 2017, 138, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhou, S.; Guan, J. Finding MicroRNA Targets in Plants Current Status and Perspectives. Genom. Proteom. Bioinform. 2012, 10, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.S.; Eamens, A.L.; Waterhouse, P.M. Missing Pieces in the Puzzle of Plant MicroRNAs. Trends Plant Sci. 2015, 20, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Sabin, L.R.; Zhou, R.; Gruber, J.J.; Lukinova, N.; Bambina, S.; Berman, A.; Lau, C.K.; Thompson, C.B.; Cherry, S. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 2009, 138, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.S.; Li, F.; Liu, Y.; Liang, P.Z.; Chen, X.W.; Gao, X.W. Identification of microRNAs and their response to the stress of plant allelochemicals in Aphis gossypii (Hemiptera: Aphididae). BMC Mol. Biol. 2017, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Pan, Y.; Gao, X.; Xi, J.; Zhang, L.; Ma, K.; Wu, Y.; Zhang, J.; Shang, Q. Reduced abundance of the CYP6CY3-targeting let-7 and miR-100 miRNAs accounts for host adaptation of Myzus persicae nicotianae. Insect Biochem. Mol. Biol. 2016, 75, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Heidel-Fischer, H.M.; Vogel, H. Molecular mechanisms of insect adaptation to plant secondary compounds. Curr. Opin. Insect Sci. 2015, 8, 8–14. [Google Scholar] [CrossRef]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 2013, 58, 517–541. [Google Scholar] [CrossRef] [PubMed]
- Etebari, K.; Afrad, M.H.; Tang, B.; Silva, R.; Furlong, M.J.; Asgari, S. Involvement of microRNA miR-2b-3p in regulation of metabolic resistance to insecticides in Plutella xylostella. Insect Mol. Biol. 2018, 27, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Lummen, P.; Khajehali, J.; Luther, K.; Leeuwen, T.V. The cyclic keto-enol insecticide spirotetramat inhibits insect and spider mite acetyl-CoA carboxylases by interfering with the carboxyltransferase partial reaction. Insect Biochem. Mol. Biol. 2014, 55. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zheng, C.; Peng, T.F.; Pan, Y.O.; Xi, J.H.; Chen, X.W.; Zhang, J.H.; Yang, S.; Gao, X.W.; Shang, Q.L. miR-276 and miR-3016-modulated expression of acetyl-CoA carboxylase accounts for spirotetramat resistance in Aphis gossypii Glover. miR-276 and miR-3016-modulated expression of acetyl-CoA carboxylase accounts for spirotetramat resistance in Aphis gossypii Glover. Insect Biochem. Mol. Boil. 2016, 79, 57–65. [Google Scholar] [CrossRef]
- Yu, T.; Li, X.; Coates, B.S.; Zhang, Q.; Siegfried, B.D.; Zhou, X. microRNA profiling between Bacillus thuringiensis Cry1Ab-susceptible and -resistant European corn borer, Ostrinia nubilalis (Hübner). Insect Mol. Biol. 2018, 27, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Carrière, Y.; Crowder, D.W.; Tabashnik, B.E. Evolutionary ecology of insect adaptation to Bt crops. Evol. Appl. 2010, 3, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.L.; Soccol, V.T.; Soccol, C.R. Bacillus thuringiensis: Mechanism of action, resistance, and new applications: A review. Crit. Rev. Biotechnol. 2016, 36, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Belles, X. MicroRNAs and the evolution of insect metamorphosis. Annu. Rev. Entomol. 2017, 62, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Sattar, S.; Addo-Quaye, C.; Song, Y.; Anstead, J.A.; Sunkar, R.; Thompson, G.A. Expression of small RNA in Aphis gossypii and its potential role in the resistance interaction with melon. PLoS ONE 2012, 7, e48579. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, H.; Kosaka, N.; Ochiya, T. Secretory microRNAs as a versatile communication tool. Commun. Integr. Biol. 2010, 3, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Wei, Y.; Wang, D.; Zhang, C.Y.; Zen, K.; Li, L. Argonaute 2 in Cell-Secreted Microvesicles Guides the Function of Secreted miRNAs in Recipient Cells. PLoS ONE 2014, 9, e103599. [Google Scholar] [CrossRef] [PubMed]
- Fabris, L.; Calin, G.A. Circulating free xeno-microRNAs—The new kids on the block. Mol. Oncol. 2016, 10, 503–508. [Google Scholar] [CrossRef] [PubMed]
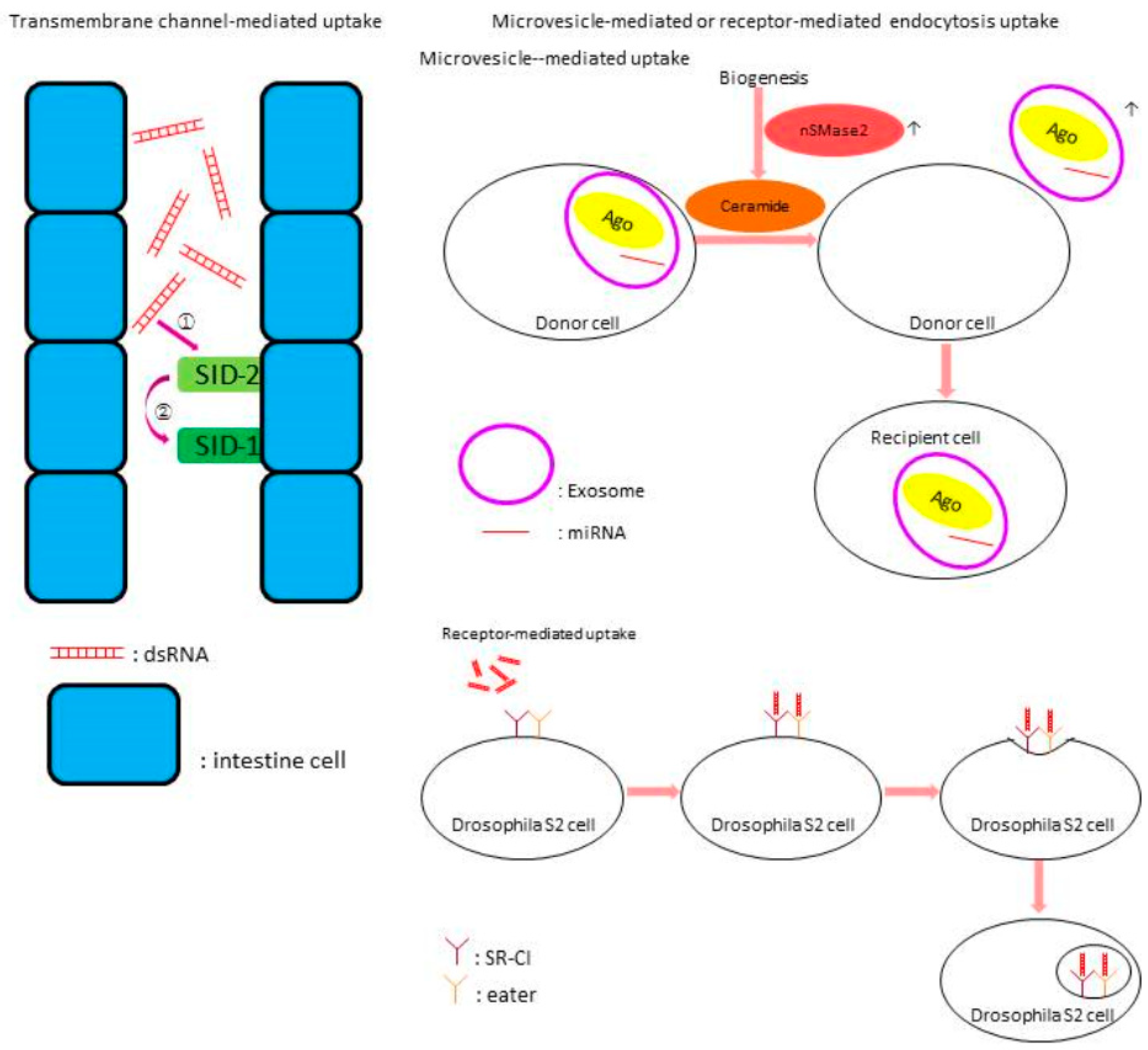
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Sang, X.; Hong, Z. Beyond nutrients: Food-derived microRNAs provide cross-kingdom regulation. Bioessays 2012, 34, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Ulvila, J.; Parikka, M.; Kleino, A.; Sormunen, R.; Ezekowitz, R.A.; Kocks, C.; Rämet, M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem. 2006, 281, 14370–14375. [Google Scholar] [CrossRef] [PubMed]
- Perge, P.; Nagy, Z.; Decmann, Á.; Igaz, I.; Igaz, P. Potential relevance of microRNAs in inter-species epigenetic communication, and implications for disease pathogenesis. RNA Biol. 2017, 14, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Hinas, A.; Wright, A.J.; Hunter, C.P. SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr. Biol. 2012, 22, 1938–1943. [Google Scholar] [CrossRef] [PubMed]
- Jose, A.M.; Smith, J.J.; Hunter, C.P. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc. Natl. Acad. Sci. USA 2009, 106, 2283–2288. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.; Bleackley, M.; Anderson, M.; Mathivanan, S. Extracellular vesicles including exosomes in cross kingdom regulation: A viewpoint from plant-fungal interactions. Front. Plant Sci. 2015, 6, 766. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Huang, L.; Cao, J.; Zen, K.; Chen, X.; Zhang, C.Y. Regulation of mammalian gene expression by exogenous microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, C.; Dou, Y.; Yu, B.; Liu, Y.; Heng-Moss, T.M.; Lu, G.; Wachholtz, M.; Bradshaw, J.D.; Twigg, P.; et al. Insect and plant-derived miRNAs in greenbug (Schizaphis graminum) and yellow sugarcane aphid (Sipha flava) revealed by deep sequencing. Gene 2017, 599, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Völkl, W.; Woodring, J.; Fischer, M.; Lorenz, M.W.; Hoffmann, K.H. Ant-aphid mutualisms: The impact of honeydew production and honeydew sugar composition on ant preferences. Oecologia 1999, 118, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Zhang, D.; Xiang, Z.; He, N. Nonfunctional ingestion of plant miRNAs in silkworm revealed by digital droplet PCR and transcriptome analysis. Sci. Rep. 2015, 5, 12290. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the Mechanism of Action of Double-Stranded RNA Activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef] [PubMed]
- Ivashuta, S.; Zhang, Y.; Wiggins, B.E.; Ramaseshadri, P.; Segers, G.C.; Johnson, S.; Meyer, S.E.; Kerstetter, R.A.; McNulty, B.C.; Bolognesi, R.; et al. Environmental RNAi in herbivorous insects. RNA 2015, 21, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Dow, J.A. pH Gradients in Lepidopteran Midgut. J. Exp. Biol. 1992, 172 Pt 1, 355–375. [Google Scholar]
- Li, Y.; Breaker, R.R. Kinetics of RNA Degradation by Specific Base Catalysis of Transesterification Involving the 2′-Hydroxyl Group. J. Am. Chem. Soc. 1999, 121, 5364–5372. [Google Scholar] [CrossRef]
- Terra, W.R. The origin and functions of the insect peritrophic membrane and peritrophic gel. Arch. Insect Biochem. Physiol. 2001, 47, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Varsani, S.; Louis, J. Altering Plant Defenses: Herbivore-Associated Molecular Patterns and Effector Arsenal of Chewing Herbivores. Mol. Plant Microbe Interact. 2018, 31, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Shahi, P.; Gase, K.; Baldwin, I.T. Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuate. Proc. Natl. Acad. Sci. USA 2008, 105, 4559–4564. [Google Scholar] [CrossRef] [PubMed]
- Diezel, C.; von Dahl, C.C.; Gaquerel, E.; Baldwin, I.T. Different Lepidopteran Elicitors Account for Cross-Talk in Herbivory-Induced Phytohormone Signaling. Plant Physiol. 2009, 150, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Bozorov, T.A.; Baldwin, I.T.; Kim, S.G. Identification and profiling of miRNAs during herbivory reveals jasmonate-dependent and -independent patterns of accumulation in Nicotiana attenuata. BMC Plant Biol. 2012, 12, 209. [Google Scholar] [CrossRef] [PubMed]
- Groen, S.C.; Jiang, S.; Murphy, A.M.; Cunniffe, N.J.; Westwood, J.H.; Davey, M.P.; Bruce, T.J.; Caulfield, J.C.; Furzer, O.J.; Reed, A.; et al. Virus Infection of Plants Alters Pollinator Preference: A Payback for Susceptible Hosts? PLoS Pathog. 2016, 12, e1005790. [Google Scholar] [CrossRef] [PubMed]
- Schwander, T.; Lo, N.; Beekman, M.; Oldroyd, B.P.; Keller, L. Nature versus nurture in social insect caste differentiation. Trends Ecol. Evol. 2010, 25, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Sagili, R.R.; Metz, B.N.; Lucas, H.M.; Chakrabarti, P.; Breece, C.R. Honey bees consider larval nutritional status rather than genetic relatedness when selecting larvae for emergency queen rearing. Sci. Rep. 2018, 8, 7679. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Liu, M.; Fu, Z.; Zhou, Z.; Kong, Y.; Liang, H.; Lin, Z.; Luo, J.; Zheng, H.; Wan, P.; et al. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet. 2017, 13, e1006946. [Google Scholar] [CrossRef] [PubMed]
- Masood, M.; Everett, C.P.; Chan, S.Y.; Snow, J.W. Negligible uptake and transfer of diet-derived pollen microRNAs in adult honey bees. RNA Biol. 2016, 13, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.P.; Raman, V.; Dhandapani, G.; Malhotra, E.V.; Sreevathsa, R.; Kumar, P.A.; Sharma, T.R.; Pattanayak, D. Silencing of HaAce1 gene by host-delivered artificial microRNA disrupts growth and development of Helicoverpa armigera. PLoS ONE 2018, 13, e0194150. [Google Scholar] [CrossRef] [PubMed]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.C. Climate Change Effects on Insects: Implications for Crop Protection and Food Security. J. Crop Improv. 2014, 28, 229–259. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.C.; Miao, X.X. Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2013, 20, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Zotti, M.J.; Smagghe, G. RNAi Technology for Insect Management and Protection of Beneficial Insects from Diseases: Lessons, Challenges and Risk Assessments. Neotrop. Entomol. 2015, 44, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, Q.; Luan, J.; Chung, S.H.; Eck, J.V.; Turgeon, R.; Douglas, A.E. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia Tabaci. Insect Biochem. Mol. Biol. 2017, 88, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206 Pt 24, 4393–4412. [Google Scholar] [CrossRef]
- Agrawal, A.; Rajamani, V.; Reddy, V.S.; Mukherjee, S.K.; Bhatnagar, R.K. Transgenic plants over-expressing insect-specific miRNA acquire insecticidal activity against Helicoverpa armigera: An alternative to Bt-toxin technology. Transgenic Res. 2015, 24, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, G.; Soreq, H. Termination and beyond: Acetylcholinesterase as a modulator of synaptic transmission. Cell Tissue Res. 2006, 326, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Song, X.; Wang, G.; Yang, K.; Wang, Y.; Niu, L.; Chen, X.; Fang, R. Plant-generated artificial small RNAs mediated aphid resistance. PLoS ONE 2014, 9, e97410. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, N.; Rewitz, K.F.; O’Connor, M.B. Ecdysone control of developmental transitions: Lessons from Drosophila research. Annu. Rev. Entomol. 2013, 58, 497–516. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Xiao, H.; Sun, Y.; Ding, S.; Situ, G.; Li, F. Transgenic microRNA-14 rice shows high resistance to rice stem borer. Plant Biotechnol. J. 2018. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Wong, A.Y.P.; Wang, S.; Jia, Q.; Chuang, W.-P.; Bendena, W.G.; Tobe, S.S.; Yang, S.H.; Chung, G.; Chan, T.-F.; et al. miRNA-Mediated Interactions in and between Plants and Insects. Int. J. Mol. Sci. 2018, 19, 3239. https://doi.org/10.3390/ijms19103239
Li C, Wong AYP, Wang S, Jia Q, Chuang W-P, Bendena WG, Tobe SS, Yang SH, Chung G, Chan T-F, et al. miRNA-Mediated Interactions in and between Plants and Insects. International Journal of Molecular Sciences. 2018; 19(10):3239. https://doi.org/10.3390/ijms19103239
Chicago/Turabian StyleLi, Chade, Annette Y. P. Wong, Shuang Wang, Qi Jia, Wen-Po Chuang, William G. Bendena, Stephen S. Tobe, Seung Hwan Yang, Gyuhwa Chung, Ting-Fung Chan, and et al. 2018. "miRNA-Mediated Interactions in and between Plants and Insects" International Journal of Molecular Sciences 19, no. 10: 3239. https://doi.org/10.3390/ijms19103239
APA StyleLi, C., Wong, A. Y. P., Wang, S., Jia, Q., Chuang, W.-P., Bendena, W. G., Tobe, S. S., Yang, S. H., Chung, G., Chan, T.-F., Lam, H.-M., Bede, J. C., & Hui, J. H. L. (2018). miRNA-Mediated Interactions in and between Plants and Insects. International Journal of Molecular Sciences, 19(10), 3239. https://doi.org/10.3390/ijms19103239