Peanut Stunt Virus and Its Satellite RNA Trigger Changes in Phosphorylation in N. benthamiana Infected Plants at the Early Stage of the Infection
Abstract
1. Introduction
2. Results
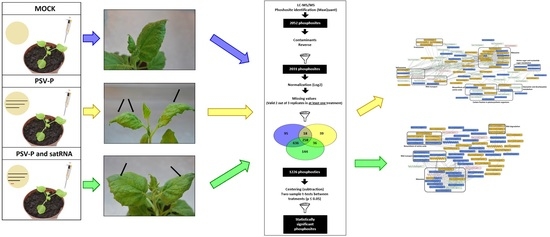
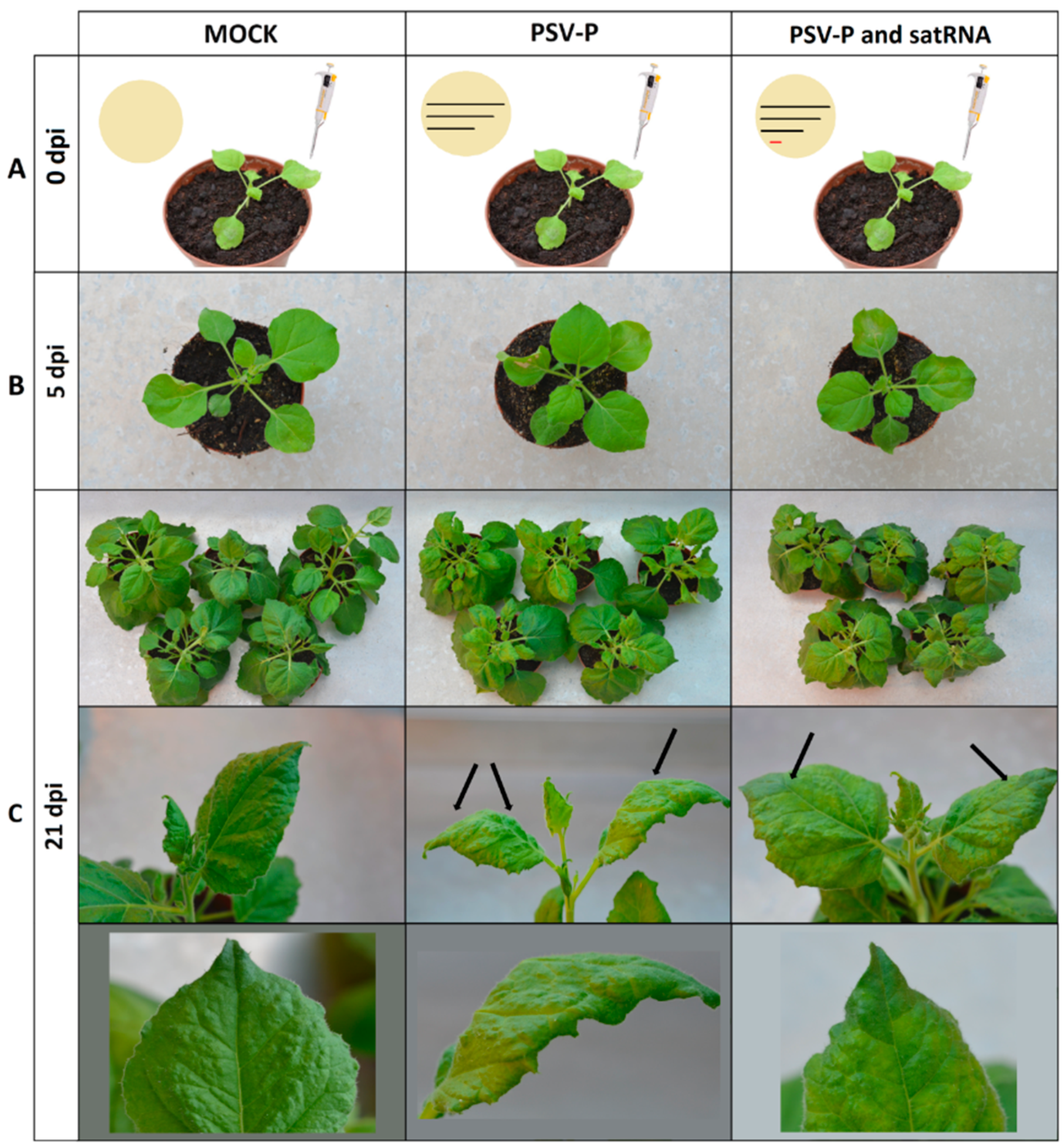
2.1. Experimental Set-Up for Proteome and Phosphoproteome Analyses of PSV-P- or PSV-P and satRNA-Infected N. benthamiana Plants
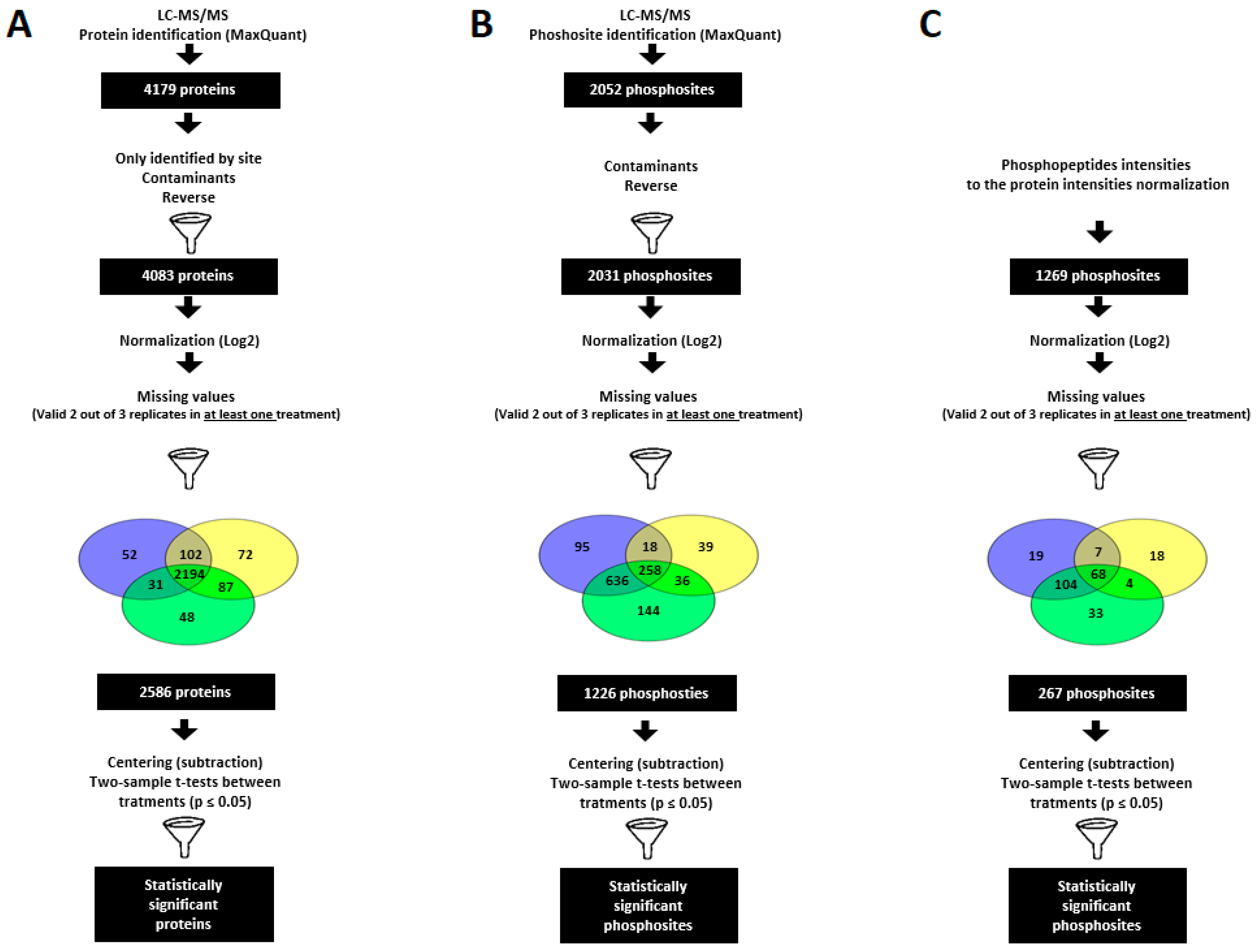
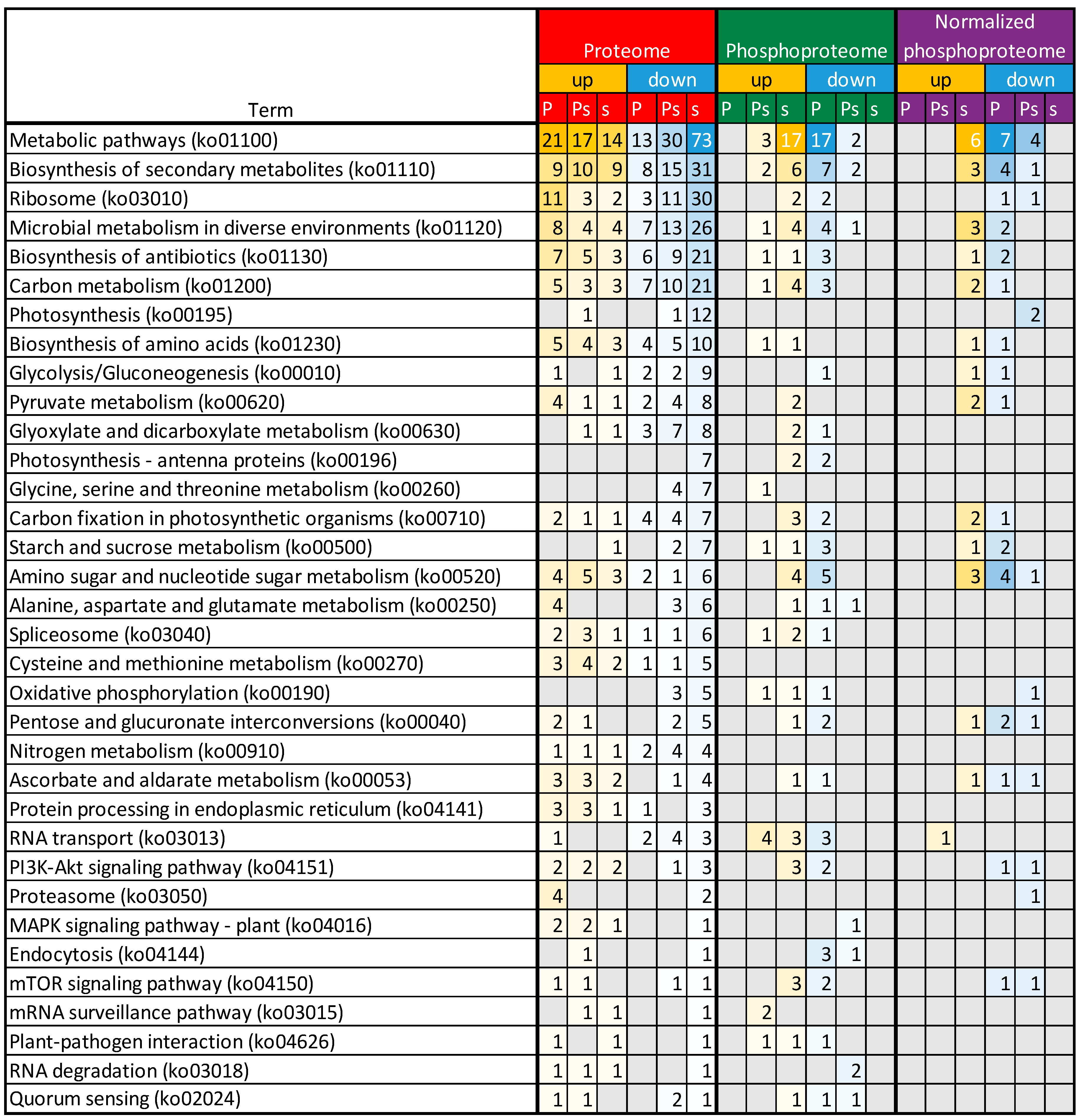
2.2. PSV-P and PSV-P and satRNA Infections Influence the N. benthamiana Proteome
2.3. PSV-P and PSV-P and satRNA Infections Influence the N. benthamiana Phosphoproteome
2.4. A Normalized N. benthamiana Phosphoproteome upon PSV-P and PSV-P and satRNA Infections
2.5. Evaluation of Correlation between Gene Expression and (Phospho)Protein Level
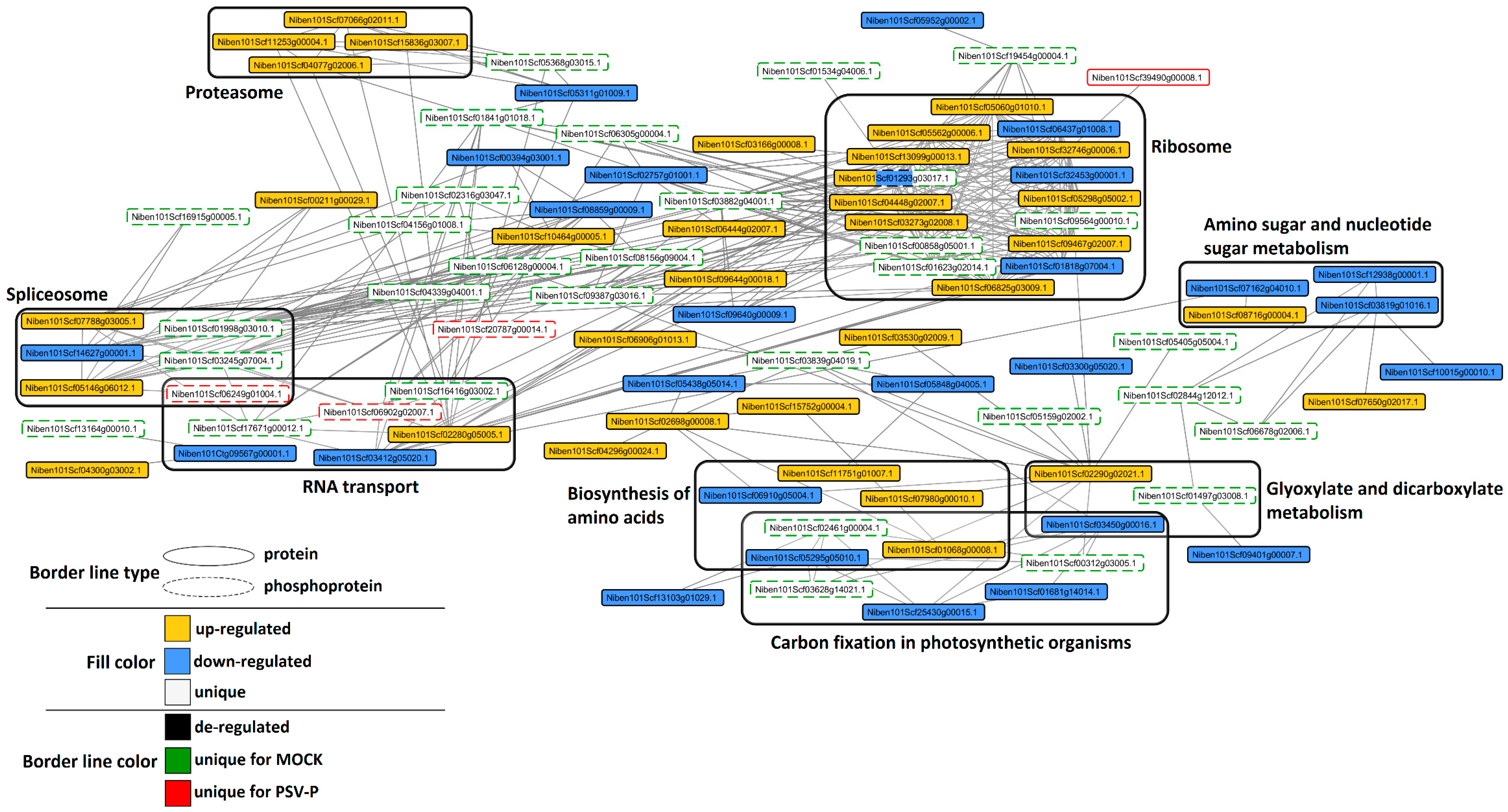
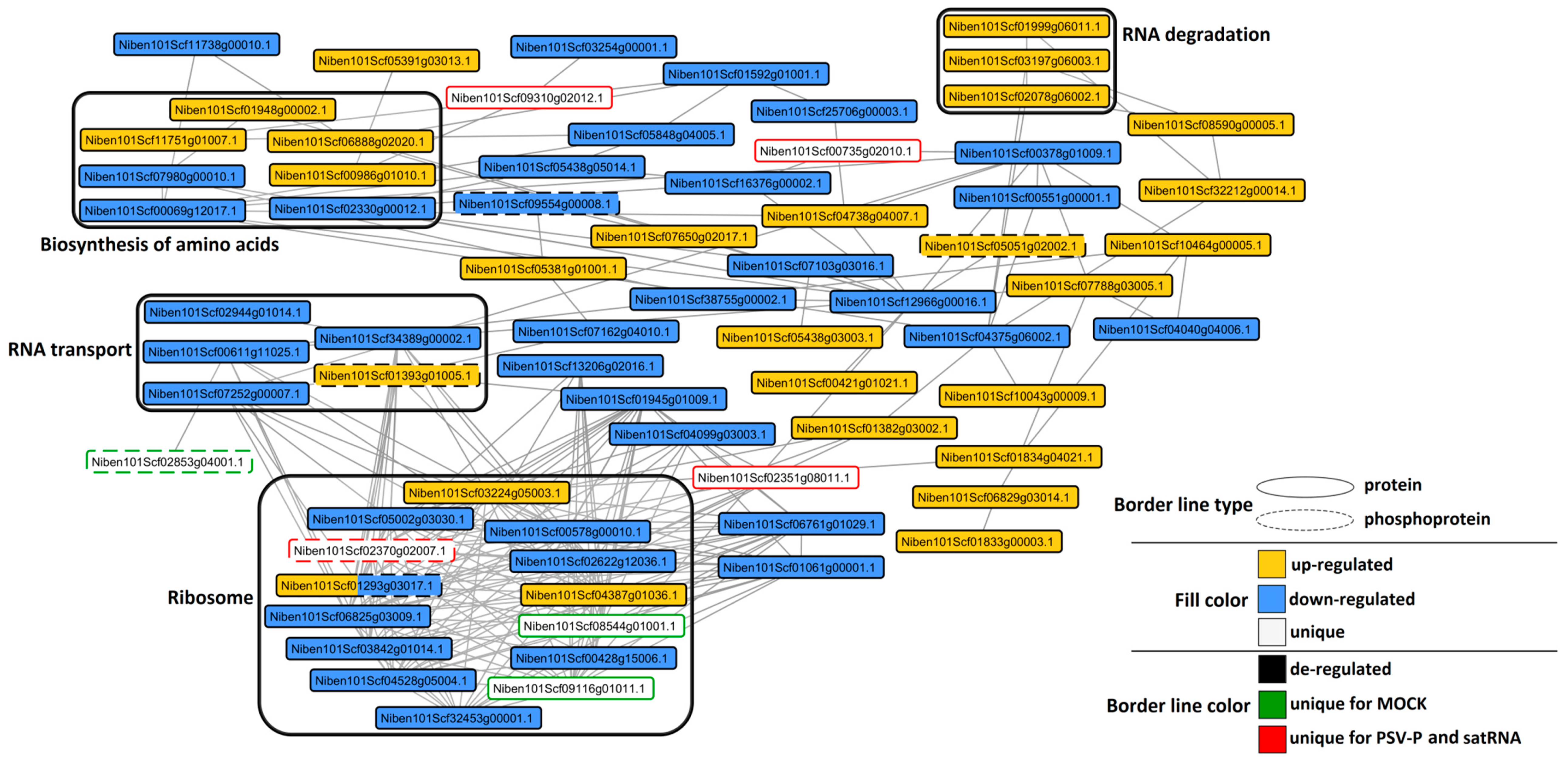
2.6. Analysis of Protein–Protein Interaction Networks in PSV-P and PSV-P and satRNA-Infected N. benthamiana
3. Discussion
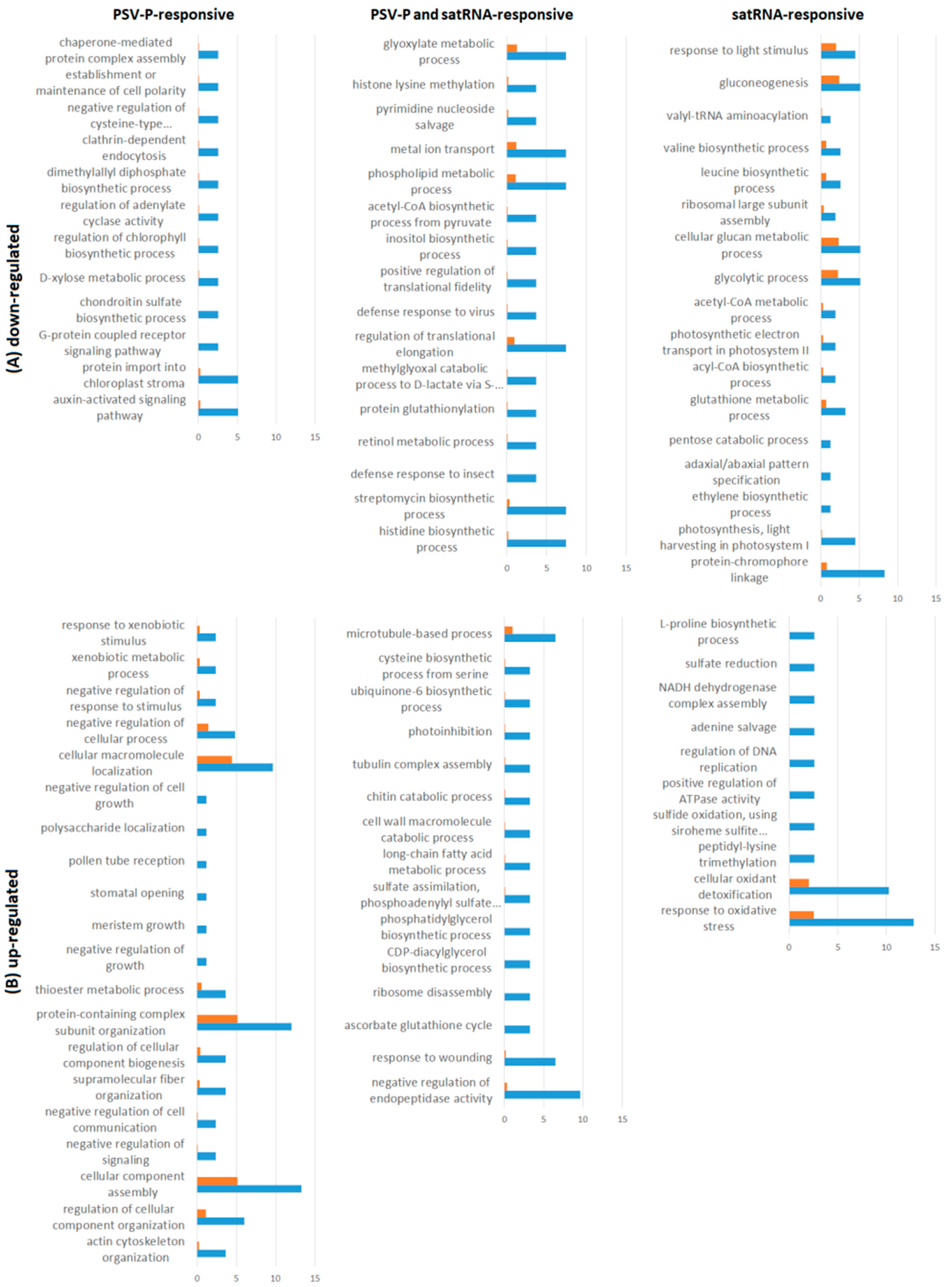
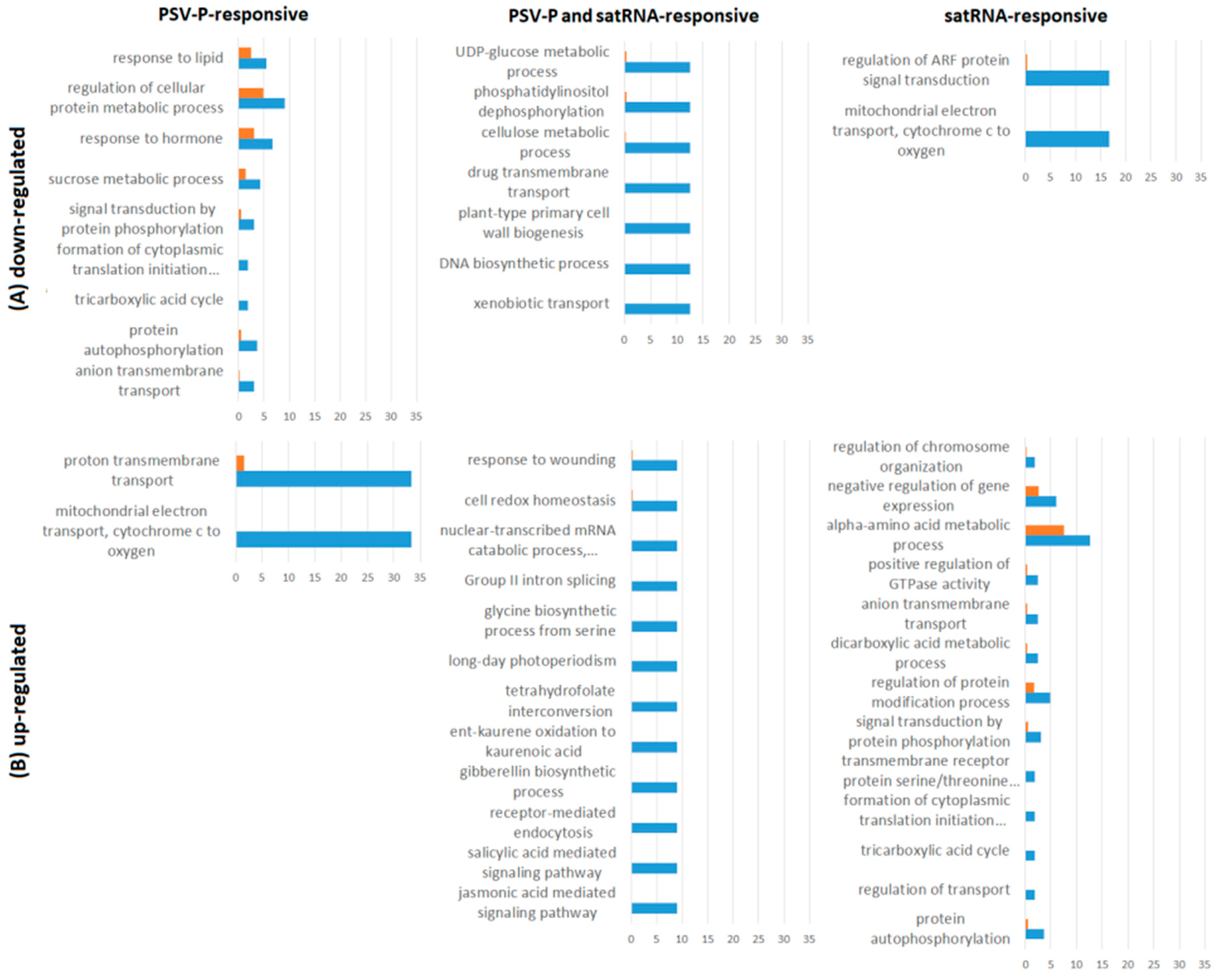
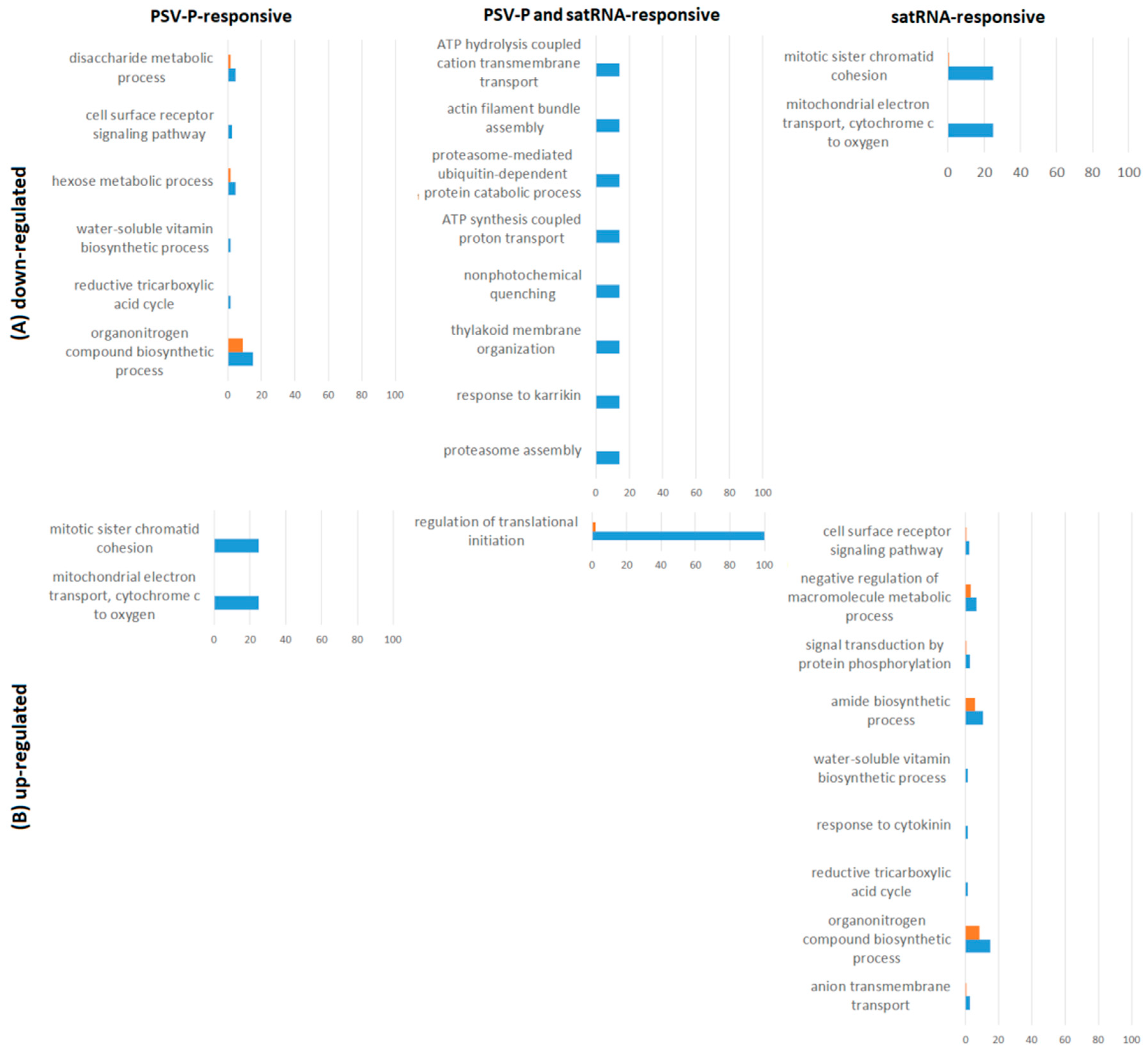
3.1. PSV-P and PSV-P and satRNA Infections Influenced N. benthamiana Signaling Pathways
3.2. PSV-P and PSV-P and satRNA Manipulate Primary Metabolism
3.3. N. benthamiana Plants Respond with Changes in the Level and Phosphorylation Status of Stress Response-Associated Proteins
3.4. PSV-P and PSV-P and satRNA Infection Results in Changes in RNA Processing
4. Materials and Methods
4.1. Plant and Virus Materials
4.2. RT-qPCR for Viral Genomic RNAs and N. benthamiana Transcripts Analyses
4.3. Protein Extraction
4.4. Mass Spectrometry
4.5. (Phospho)Proteomic Data Analysis
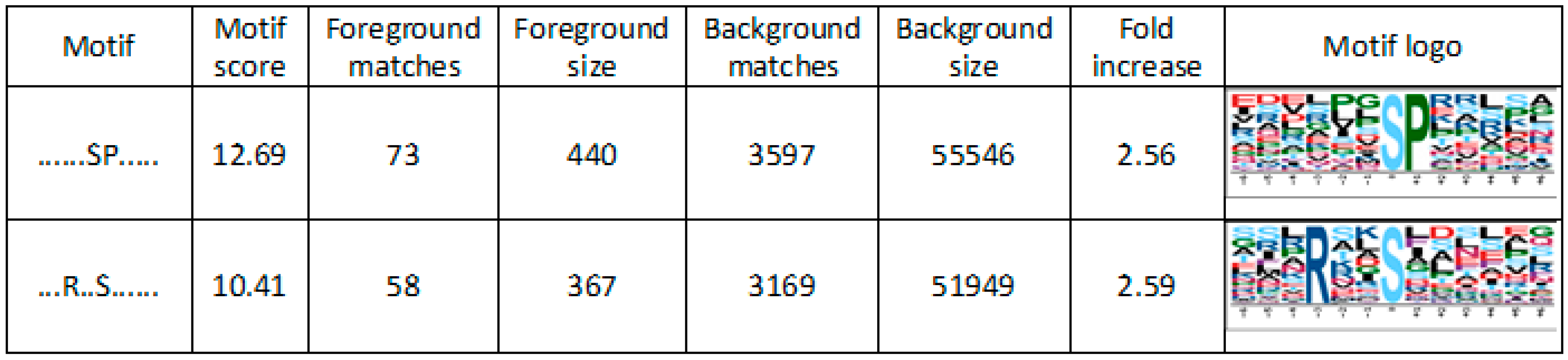
4.6. Motif-X Analysis
4.7. Functional Annotation of Differentially Expressed Genes and Pathway Analysis
4.8. STRING Analysis of Protein–Protein Interaction Networks
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hull, R. Plant Virology; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Gozdzicka-Jozefiak, A. Wirusologia Molekularna; Wydaw. Nauk. UAM Poznan: Poznań, Poland, 2004. [Google Scholar]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Sanford, J.; Johnston, S. The concept of parasite-derived resistance—Deriving resistance genes from the parasite’s own genome. J. Theor. Boil. 1985, 113, 395–405. [Google Scholar] [CrossRef]
- Obrępalska-Stęplowska, A.; Budziszewska, M.; Pospieszny, H. Complete nucleotide sequence of a Polish strain of Peanut stunt virus (PSV-P) that is related to but not a typical member of subgroup I. Acta Biochim. Pol. 2008, 55, 731–739. [Google Scholar] [PubMed]
- Netsu, O.; Hiratsuka, K.; Kuwata, S.; Hibi, T.; Ugaki, M.; Suzuki, M. Peanut stunt virus 2b cistron plays a role in viral local and systemic accumulation and virulence in Nicotiana benthamiana. Arch. Virol. 2008, 153, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-W.; Anderson, B.J.; Haase, H.R.; Symons, R.H. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 1994, 198, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Mushegian, A.; Koonin, E. Cell-to-cell movement of plant viruses. Arch. Virol. 1993, 133, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.E.; Roossinck, M.J.; Havelda, Z. Plant virus satellite and defective interfering RNAs: New paradigms for a new century. Annu. Rev. Phytopathol. 2004, 42, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Zhu, L.; Du, Z.; Zeng, R.; Feng, J.; Chen, J. Satellite RNA-mediated reduction of cucumber mosaic virus genomic RNAs accumulation in Nicotiana tabacum. Acta Biochim. Biophys. Sin. 2007, 39, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Obrępalska-Stęplowska, A.; Renaut, J.; Planchon, S.; Przybylska, A.; Wieczorek, P.; Barylski, J.; Palukaitis, P. Effect of temperature on the pathogenesis, accumulation of viral and satellite RNAs and on plant proteome in peanut stunt virus and satellite RNA-infected plants. Front. Plant Sci. 2015, 6, 903. [Google Scholar] [CrossRef] [PubMed]
- Obrępalska-Stęplowska, A.; Wieczorek, P.; Budziszewska, M.; Jeszke, A.; Renaut, J. How can plant virus satellite RNAs alter the effects of plant virus infection? A study of the changes in the Nicotiana benthamiana proteome after infection by Peanut stunt virus in the presence or absence of its satellite RNA. Proteomics 2013, 13, 2162–2175. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, S.; Wu, J.; Han, Z.; Wang, R.; Wu, L.; Zhang, H.; Chen, Y.; Hu, X. Phosphoproteomic analysis of the resistant and susceptible genotypes of maize infected with sugarcane mosaic virus. Amino Acids 2015, 47, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Choi, E.Y.; Choi, Y.J.; Ahn, J.H.; Park, O.K. Proteomics studies of post-translational modifications in plants. J. Exp. Bot. 2006, 57, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Kersten, B.; Agrawal, G.K.; Durek, P.; Neigenfind, J.; Schulze, W.; Walther, D.; Rakwal, R. Plant phosphoproteomics: An update. Proteomics 2009, 9, 964–988. [Google Scholar] [CrossRef] [PubMed]
- Friso, G.; van Wijk, K.J. Posttranslational protein modifications in plant metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [PubMed]
- Xing, T.; Laroche, A. Revealing plant defense signaling: Getting more sophisticated with phosphoproteomics. Plant Signal. Behav. 2011, 6, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ye, Z.; Nie, Y.; Zhang, J.; Wang, G.-L.; Wang, Z. Comparative phosphoproteome analysis of Magnaporthe oryzae-responsive proteins in susceptible and resistant rice cultivars. J. Proteom. 2015, 115, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Silva-Sanchez, C.; Zhang, T.; Chen, S.; Li, H. Phosphoproteomics technologies and applications in plant biology research. Front. Plant Sci. 2015, 6, 430. [Google Scholar] [CrossRef] [PubMed]
- Vijayapalani, P.; Chen, J.C.-F.; Liou, M.-R.; Chen, H.-C.; Hsu, Y.-H.; Lin, N.-S. Phosphorylation of bamboo mosaic virus satellite RNA (satBaMV)-encoded protein P20 downregulates the formation of satBaMV-P20 ribonucleoprotein complex. Nucleic Acids Res. 2011, 40, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Mills-Lujan, K.; Andrews, D.L.; Chou, C.-W.; Deom, C.M. The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. PLoS ONE 2015, 10, e0122356. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, Y.; Yoshioka, K.; Shigyo, T.; Takahashi, H.; Nyunoya, H. Phosphorylation of the movement protein of Cucumber mosaic virus in transgenic tobacco plants. Virus Genes 2002, 24, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Palukaitis, P.; Park, Y.I. Phosphorylation of cucumber mosaic virus RNA polymerase 2a protein inhibits formation of replicase complex. EMBO J. 2002, 21, 2292–2300. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.S.; Santos, A.A.; Machado, J.P.B.; Fontes, E.P. The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling. Virology 2008, 380, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Rajamäki, M.-L.; Xi, D.; Sikorskaite-Gudziuniene, S.; Valkonen, J.P.; Whitham, S.A. Differential Requirement of the Ribosomal Protein S6 and Ribosomal Protein S6 Kinase for Plant-Virus Accumulation and Interaction of S6 Kinase with Potyviral VPg. Mol. Plant Microbe Interact. 2017, 30, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.D.; Stes, E.; Van Bel, M.; Nelissen, H.; Maddelein, D.; Inzé, D.; Coppens, F.; Martens, L.; Gevaert, K.; De Smet, I. Up-to-Date Workflow for Plant (Phospho) proteomics Identifies Differential Drought-Responsive Phosphorylation Events in Maize Leaves. J. Proteome Res. 2016, 15, 4304–4317. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Melo-Braga, M.N.; Braga, T.V.; León, I.R.; Antonacci, D.; Nogueira, F.C.; Thelen, J.J.; Larsen, M.R.; Palmisano, G. Modulation of protein phosphorylation, glycosylation and acetylation in grape (Vitis vinifera) mesocarp and exocarp due to Lobesia botrana infection. Mol. Cell. Proteom. 2012, 11, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.H.; Moon, B.C.; Park, H.C.; Koo, S.C.; Jeon, J.M.; Cheong, Y.H.; Chung, W.S.; Lim, C.O.; Kim, J.-Y.; Yoon, B.-D. Rice OsERG3 encodes an unusual small C2-domain protein containing a Ca2+-binding module but lacking phospholipid-binding properties. Biochim. Biophys. Acta 2011, 1810, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, F.; Thomas, C.L.; Lederer, C.; Niu, Y.; Wang, D.; Maule, A.J. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 2005, 138, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, P.; Obrępalska-Stęplowska, A. Suppress to survive—Implication of plant viruses in PTGS. Plant Mol. Boil. Rep. 2015, 33, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Ryšlavá, H.; Müller, K.; Semorádová, Š.; Synková, H.; Čeřovská, N. Photosynthesis and activity of phosphoenolpyruvate carboxylase in Nicotiana tabacum L. leaves infected by Potato virus A and Potato virus Y. Photosynthetica 2003, 41, 357–363. [Google Scholar] [CrossRef]
- Obrępalska-Stęplowska, A.; Zmienko, A.; Wrzesińska, B.; Goralski, M.; Figlerowicz, M.; Zyprych-Walczak, J.; Siatkowski, I.; Pospieszny, H. The defense response of Nicotiana benthamiana to peanut stunt virus infection in the presence of symptom exacerbating satellite RNA. Viruses 2018, 10, 449. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Qin, Q.; Wang, Y.; Pu, Y.; Liu, L.; Wen, X.; Ji, S.; Wu, J.; Wei, C.; Ding, B. Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 2016, 12, e1005847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, M.; Wang, W.; Li, D.; Huang, Q.; Wang, Y.; Zheng, X.; Zhang, Z. Silencing of G proteins uncovers diversified plant responses when challenged by three elicitors in Nicotiana benthamiana. Plant Cell Environ. 2012, 35, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D. Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Boil. 1999, 50, 97–131. [Google Scholar] [CrossRef] [PubMed]
- Goff, K.E.; Ramonell, K.M. The role and regulation of receptor-like kinases in plant defense. Gene Regul. Syst. Boil. 2007, 1, 167–175. [Google Scholar] [CrossRef]
- Diehl, N.; Schaal, H. Make yourself at home: Viral hijacking of the PI3K/Akt signaling pathway. Viruses 2013, 5, 3192–3212. [Google Scholar] [CrossRef] [PubMed]
- Ouibrahim, L.; Rubio, A.G.; Moretti, A.; Montané, M.-H.; Menand, B.; Meyer, C.; Robaglia, C.; Caranta, C. Potyviruses differ in their requirement for TOR signalling. J. Gen. Virol. 2015, 96, 2898–2903. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, E.; del Toro, F.J.; Canto, T.; Tenllado, F. Identification of MAPKs as signal transduction components required for the cell death response during compatible infection by the synergistic pair Potato virus X-Potato virus Y. Virology 2017, 509, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Baldwin, I.T.; Wu, J. Arabidopsis Plants Having Defects in Nonsense-mediated mRNA Decay Factors UPF1, UPF2, and UPF3 Show Photoperiod-dependent Phenotypes in Development and Stress Responses. J. Integr. Plant Boil. 2012, 54, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Garcia, S.; Voinnet, O. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 2014, 16, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Balistreri, G.; Horvath, P.; Schweingruber, C.; Zünd, D.; McInerney, G.; Merits, A.; Mühlemann, O.; Azzalin, C.; Helenius, A. The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 2014, 16, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.-S.; Pedley, K.F.; Martin, G.B. Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 2010, 22, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, J.; Bang, B.; Kim, I.; Lee, H.-H.; Park, J.; Seo, Y.-S. Comparative analyses of Tomato yellow leaf curl virus C4 protein-interacting host proteins in healthy and infected tomato tissues. Plant Pathol. J. 2016, 32, 377. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zhang, Y.; Wang, C.; Lei, R.; Wu, Y.; Li, X.; Zhu, S. Cucumber mosaic virus coat protein induces the development of chlorotic symptoms through interacting with the chloroplast ferredoxin I protein. Sci. Rep. 2018, 8, 1205. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmová, N.; Procházková, D.; Sindelarova, M.; Sindelar, L. Photosynthesis in leaves of Nicotiana tabacum L. infected with tobacco mosaic virus. Photosynthetica 2005, 43, 597–602. [Google Scholar] [CrossRef]
- Magyarosy, A.C.; Buchanan, B.B.; Schürmann, P. Effect of a systemic virus infection on chloroplast function and structure. Virology 1973, 55, 426–438. [Google Scholar] [CrossRef]
- Fernández-Calvino, L.; Osorio, S.; Hernández, M.L.; Hamada, I.B.; Del Toro, F.J.; Donaire, L.; Yu, A.; Bustos, R.; Fernie, A.R.; Martínez-Rivas, J.M. Virus-induced alterations in primary metabolism modulate susceptibility to Tobacco rattle virus in Arabidopsis. Plant Physiol. 2014, 166, 1821–1838. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.P.; Ventura, J.A.; Aguilar, C.; Nakayasu, E.S.; Almeida, I.C.; Fernandes, P.; Zingali, R.B. Proteomic analysis of papaya (Carica papaya L.) displaying typical sticky disease symptoms. Proteomics 2011, 11, 2592–2602. [Google Scholar] [CrossRef] [PubMed]
- Larson, R.L.; Wintermantel, W.M.; Hill, A.; Fortis, L.; Nunez, A. Proteome changes in sugar beet in response to Beet necrotic yellow vein virus. Physiol. Mol. Plant Pathol. 2008, 72, 62–72. [Google Scholar] [CrossRef]
- Wu, L.; Han, Z.; Wang, S.; Wang, X.; Sun, A.; Zu, X.; Chen, Y. Comparative proteomic analysis of the plant–virus interaction in resistant and susceptible ecotypes of maize infected with sugarcane mosaic virus. J. Proteom. 2013, 89, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.A.; Berglund, P.; Beard, C.W.; Johnston, R.E. Ribosomal protein S6 associates with alphavirus nonstructural protein 2 and mediates expression from alphavirus messages. J. Virol. 2006, 80, 7729–7739. [Google Scholar] [CrossRef] [PubMed]
- Elvira, M.I.; Galdeano, M.M.; Gilardi, P.; García-Luque, I.; Serra, M.T. Proteomic analysis of pathogenesis-related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum chinense L 3 plants. J. Exp. Bot. 2008, 59, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhou, T.; Wu, X.; Hong, Y.; Fan, Z.; Li, H. Influence of cytoplasmic heat shock protein 70 on viral infection of Nicotiana benthamiana. Mol. Plant Pathol. 2008, 9, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Brizard, J.P.; Carapito, C.; Delalande, F.; Van Dorsselaer, A.; Brugidou, C. Proteome Analysis of Plant-Virus Interactome Comprehensive Data for Virus Multiplication Inside Their Hosts. Mol. Cell. Proteom. 2006, 5, 2279–2297. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Ni, H.; Chen, Q.; Sun, F.; Zhou, T.; Lan, Y.; Zhou, Y. Comparative proteomic analysis reveals the cross-talk between the responses induced by H2O2 and by long-term rice black-streaked dwarf virus infection in rice. PLoS ONE 2013, 8, e81640. [Google Scholar] [CrossRef] [PubMed]
- Di Carli, M.; Villani, M.E.; Bianco, L.; Lombardi, R.; Perrotta, G.; Benvenuto, E.; Donini, M. Proteomic analysis of the plant—Virus interaction in cucumber mosaic virus (CMV) resistant transgenic tomato. J. Proteome Res. 2010, 9, 5684–5697. [Google Scholar] [CrossRef] [PubMed]
- Casado-Vela, J.; Sellés, S.; Martínez, R.B. Proteomic analysis of tobacco mosaic virus-infected tomato (Lycopersicon esculentum M.) fruits and detection of viral coat protein. Proteomics 2006, 6, S196–S206. [Google Scholar] [CrossRef] [PubMed]
- Balistreri, G.; Bognanni, C.; Mühlemann, O. Virus escape and manipulation of cellular nonsense-mediated mRNA decay. Viruses 2017, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Dinesh-Kumar, S.; Baker, B.J. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 2000, 97, 1908–1913. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-C.; Gassmann, W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 2003, 15, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Piron, F.; Nicolaï, M.; Minoïa, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols; Springer: Berlin, Germany, 2000; pp. 365–386. [Google Scholar]
- Vu, L.D.; Zhu, T.; Verstraeten, I.; van de Cotte, B.; International Wheat Genome Sequencing Consortium; Gevaert, K.; De Smet, I. Temperature-induced changes in the wheat phosphoproteome reveal temperature-regulated interconversion of phosphoforms. J. Exp. Bot. 2018, 69, 4609–4624. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.D.; Verstraeten, I.; Stes, E.; Van Bel, M.; Coppens, F.; Gevaert, K.; De Smet, I. Proteome Profiling of Wheat Shoots from Different Cultivars. Front. Plant Sci. 2017, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2014, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Rios, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.F.; Schwartz, D. Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinform. 2011, 35, 24. [Google Scholar]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L. InterPro: The integrative protein signature database. Nucleic Acids Res. 2008, 37, D211–D215. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
| Treatment | Proteome | Phosphoproteome | Phosphoproteome after Normalization | |||
|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | |
| PSV-P-responsive | 111 | 54 | 4 | 87 | 2 | 20 |
| PSV-P and satRNA-responsive | 66 | 87 | 37 | 34 | 2 | 15 |
| satRNA-responsive | 56 | 218 | 84 | 4 | 19 | - |
| Validation | Proteomics | Phosphoproteomics | |||||
|---|---|---|---|---|---|---|---|
| Gene | Expression | P(H1) | Result | Protein Level | Result | Phosphorylation Level | Result |
| PSV-P-responsive | |||||||
| BIP | 1.131 | 0.085 | 1.384 | UP | |||
| ERG3 | 0.997 | 0.941 | 2.970 | UP | |||
| MCA | 0.949 | 0.133 | 1.552 | UP | |||
| BSL3-like | 1.108 | 0.015 | UP | 1.040 | 2.461 | UP | |
| FBP2like | 1.151 | 0.117 | 0.788 | 0.062 | DOWN | ||
| PMI1 | 1.284 | 0.005 | UP | 1.213 | 0.313 | DOWN | |
| PPC1 | 1.249 | 0.012 | UP | 0.975 | 0.152 | DOWN | |
| RS6 | 0.833 | 0.010 | DOWN | 3.340 | UP | 0.275 | DOWN |
| TPR-like1320 | 1.122 | 0.106 | 1.283 | UP | 0.250 | DOWN | |
| TSJT1 | 0.722 | 0 | DOWN | 1.073 | 0.282 | DOWN | |
| PSV-P and satRNA-responsive | |||||||
| BIP | 1.469 | 0 | UP | 1.279 | UP | ||
| GRP2 | 1.221 | 0 | UP | 1.204 | UP | ||
| PR2B | 0.580 | 0.018 | DOWN | 0.300 | DOWN | ||
| AGO1B | 1.166 | 0.004 | UP | 1.268 | 0.556 | DOWN | |
| EIF5 | 1.200 | 0 | UP | 1.300 | 2.069 | UP | |
| ECT5 | 1.109 | 0.012 | UP | 1.053 | 0.507 | DOWN | |
| RPN10 | 0.956 | 0.118 | 1.330 | 0.397 | DOWN | ||
| RS6 | 0.903 | 0.062 | 2.848 | UP | 0.312 | DOWN | |
| TPR-like1320 | 1.122 | 0.130 | 1.223 | 0.616 | DOWN | ||
| satRNA-responsive | |||||||
| AGO4 | 1.169 | 0.002 | UP | 0.757 | DOWN | ||
| ERG3 | 1.446 | 0 | UP | 0,359 | DOWN | ||
| GRP2 | 1.162 | 0.002 | UP | 1.299 | UP | ||
| PR2B | 1.202 | 0.345 | 0.201 | DOWN | |||
| PSB | 1.207 | 0 | UP | 0.839 | DOWN | ||
| FBP2like | 0.914 | 0.309 | 1.369 | 12.957 | UP | ||
| PGM1 | 0.980 | 0.682 | 0.850 | 3.042 | UP | ||
| PMI1 | 0.849 | 0.012 | DOWN | 0.771 | 3.285 | UP | |
| PPC1 | 0.897 | 0.222 | 0.835 | 5.959 | UP | ||
| TSJT1 | 1.148 | 0.001 | UP | 0.813 | 3.957 | UP | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wrzesińska, B.; Dai Vu, L.; Gevaert, K.; De Smet, I.; Obrępalska-Stęplowska, A. Peanut Stunt Virus and Its Satellite RNA Trigger Changes in Phosphorylation in N. benthamiana Infected Plants at the Early Stage of the Infection. Int. J. Mol. Sci. 2018, 19, 3223. https://doi.org/10.3390/ijms19103223
Wrzesińska B, Dai Vu L, Gevaert K, De Smet I, Obrępalska-Stęplowska A. Peanut Stunt Virus and Its Satellite RNA Trigger Changes in Phosphorylation in N. benthamiana Infected Plants at the Early Stage of the Infection. International Journal of Molecular Sciences. 2018; 19(10):3223. https://doi.org/10.3390/ijms19103223
Chicago/Turabian StyleWrzesińska, Barbara, Lam Dai Vu, Kris Gevaert, Ive De Smet, and Aleksandra Obrępalska-Stęplowska. 2018. "Peanut Stunt Virus and Its Satellite RNA Trigger Changes in Phosphorylation in N. benthamiana Infected Plants at the Early Stage of the Infection" International Journal of Molecular Sciences 19, no. 10: 3223. https://doi.org/10.3390/ijms19103223
APA StyleWrzesińska, B., Dai Vu, L., Gevaert, K., De Smet, I., & Obrępalska-Stęplowska, A. (2018). Peanut Stunt Virus and Its Satellite RNA Trigger Changes in Phosphorylation in N. benthamiana Infected Plants at the Early Stage of the Infection. International Journal of Molecular Sciences, 19(10), 3223. https://doi.org/10.3390/ijms19103223