MET Activation by a Macrocyclic Peptide Agonist that Couples to Biological Responses Differently from HGF in a Context-Dependent Manner
Abstract
1. Introduction
2. Results
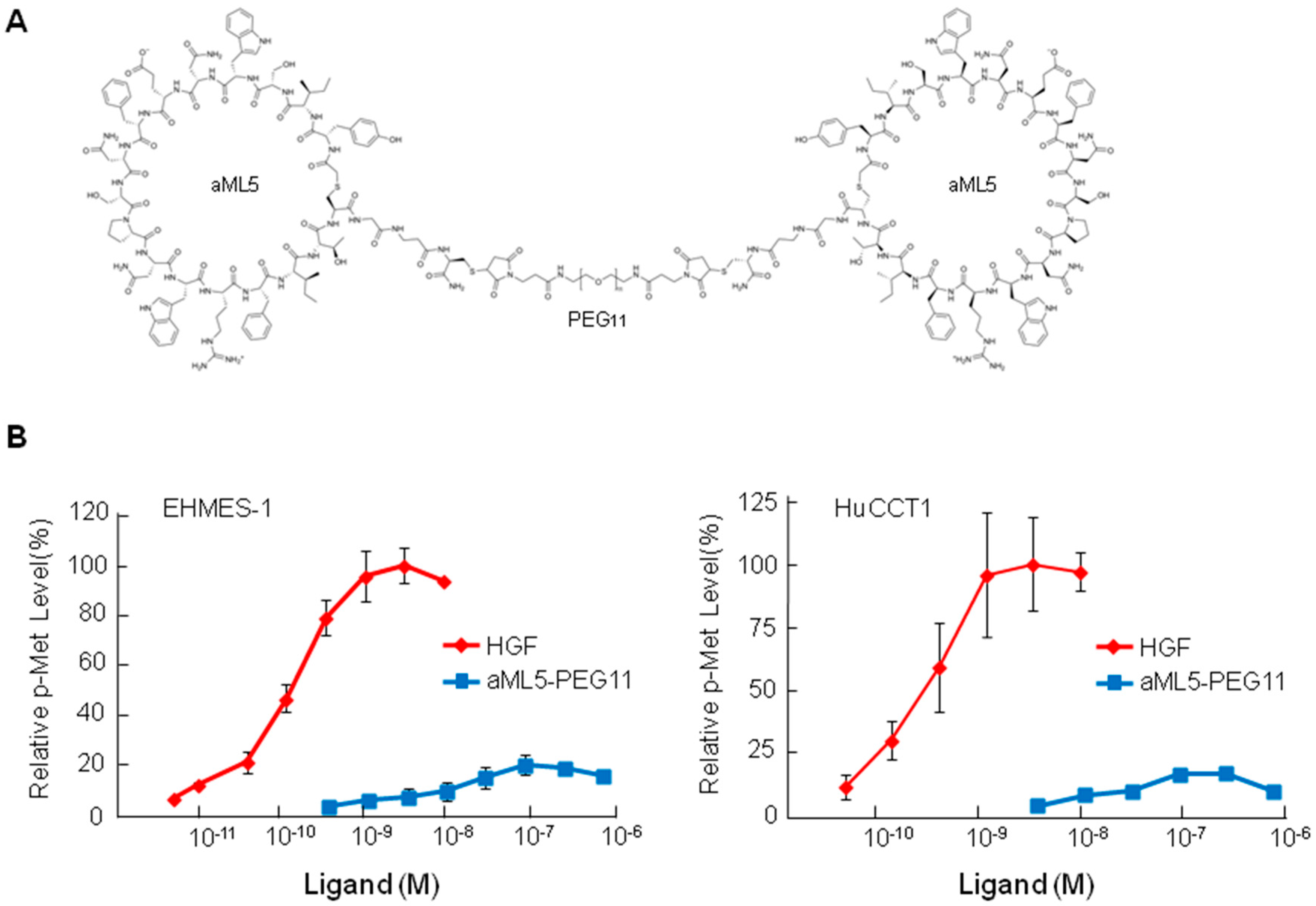
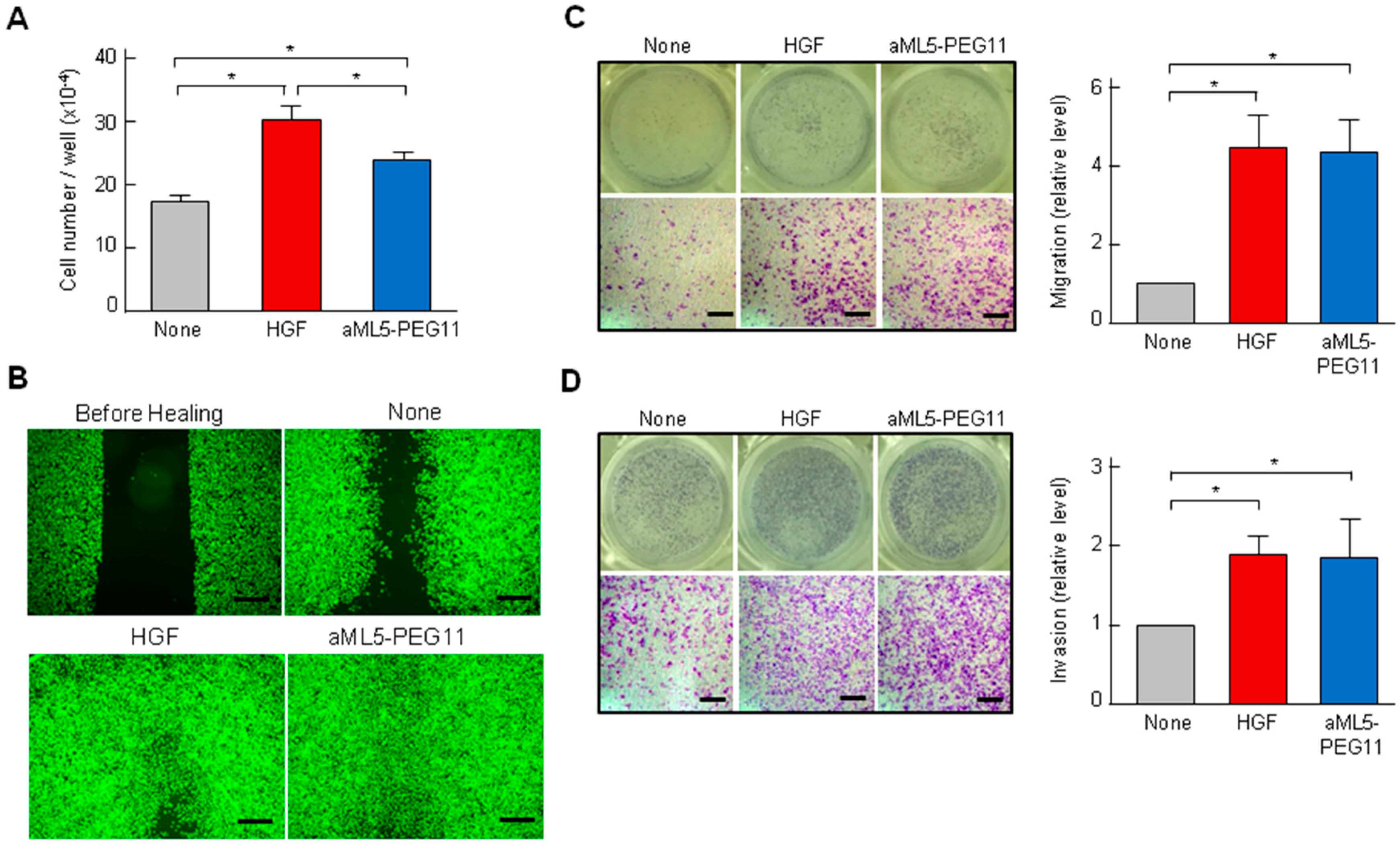
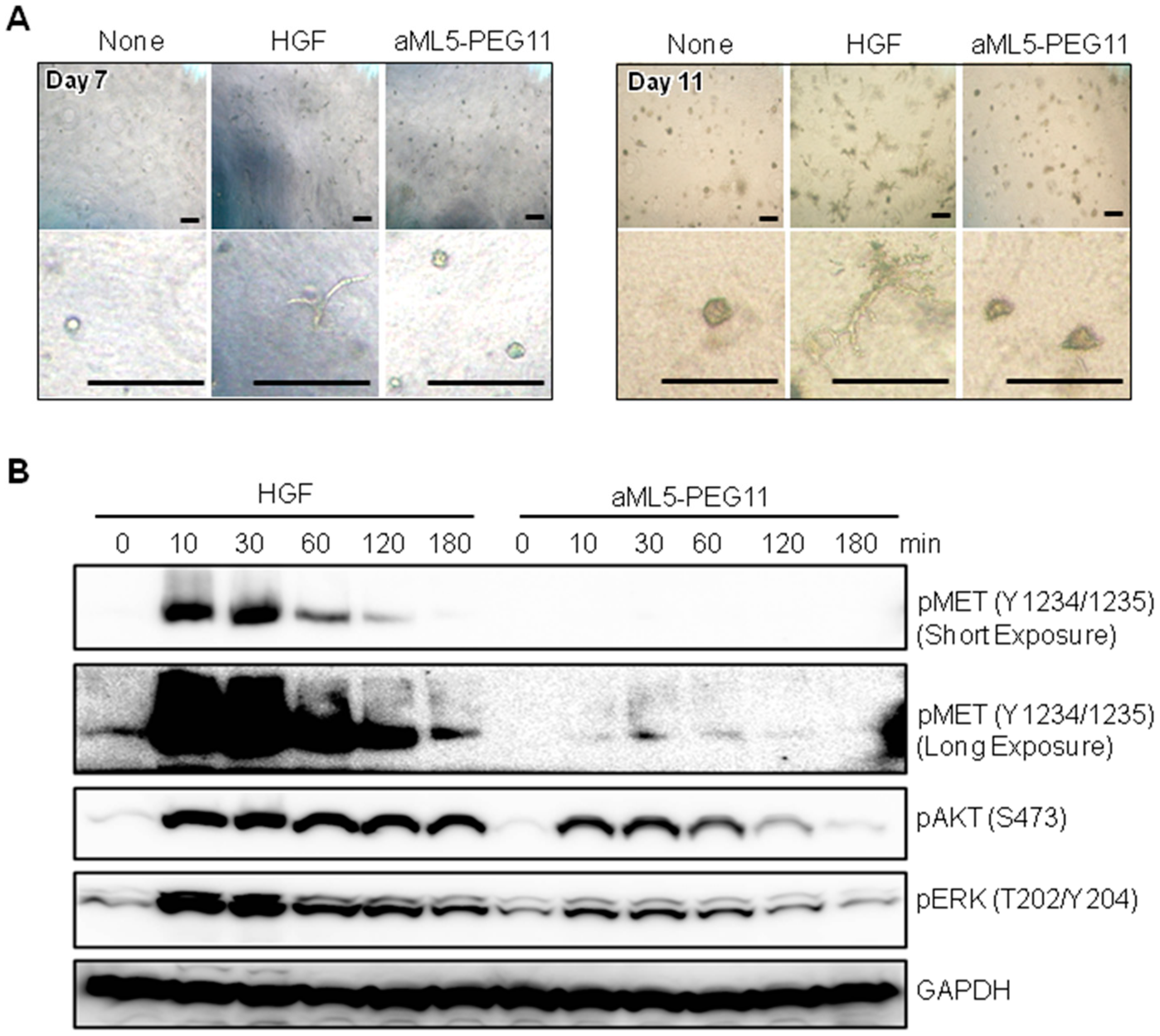
2.1. MET Receptor Activation and Biological Responses
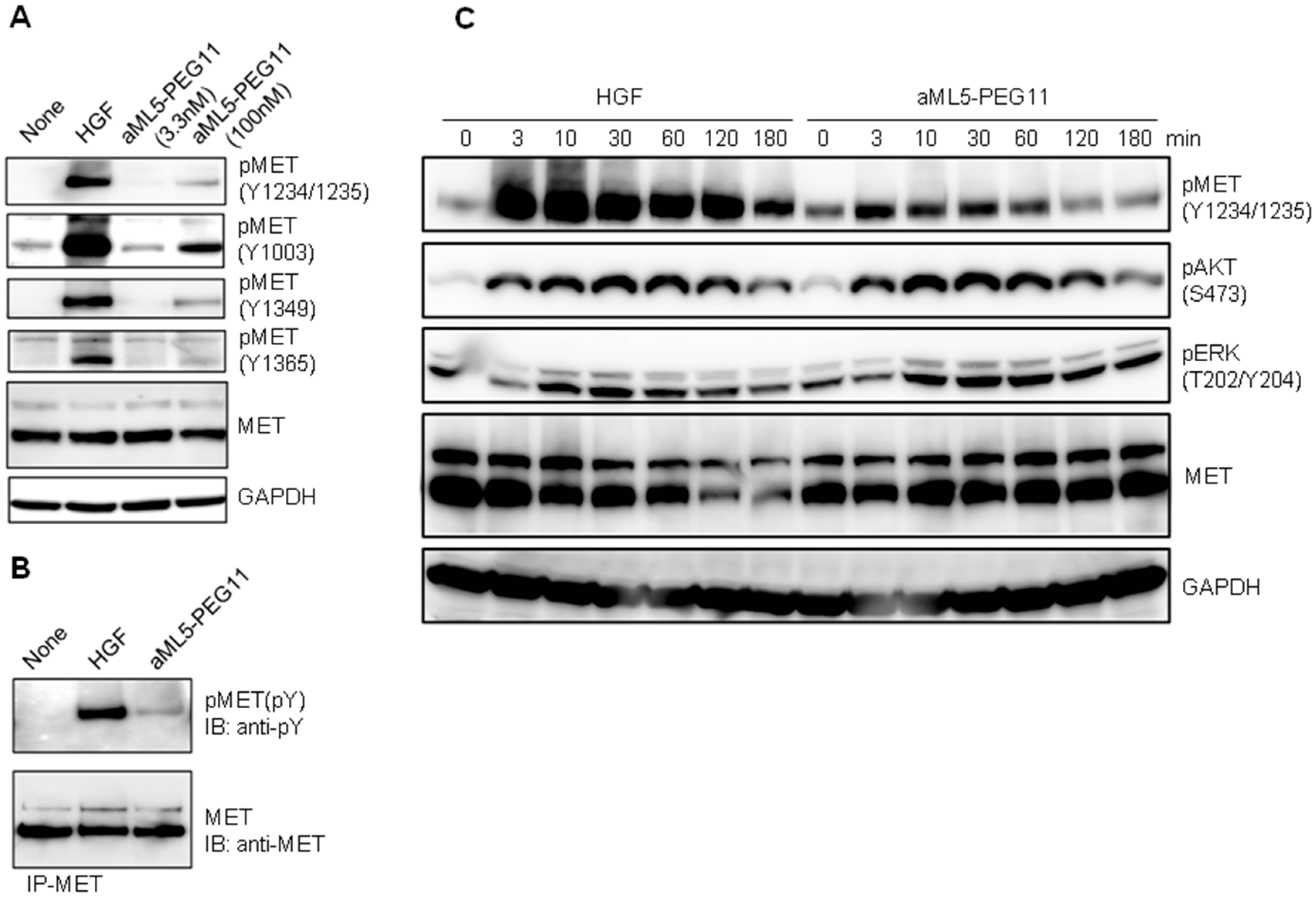
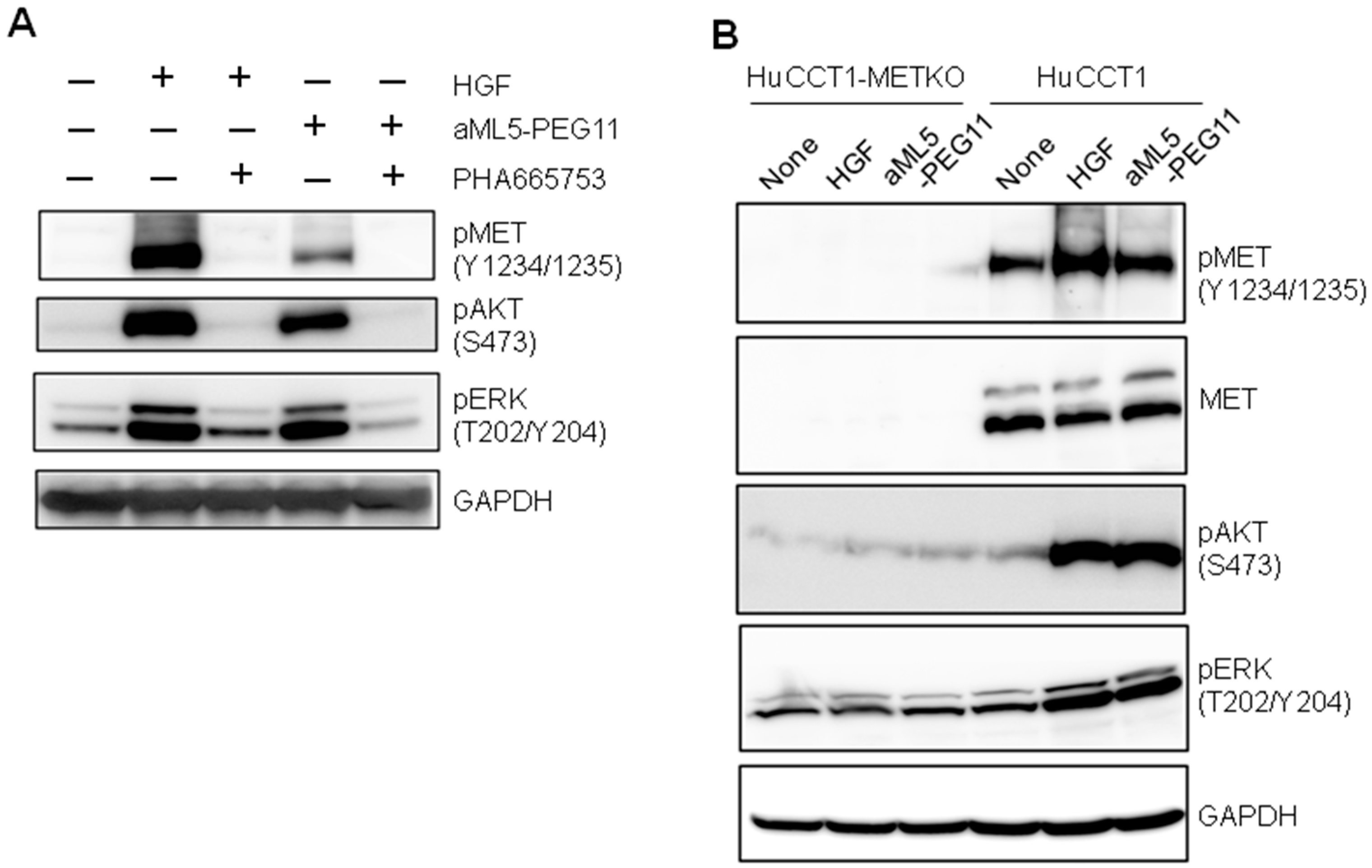
2.2. Signaling Molecule Activation Profiles
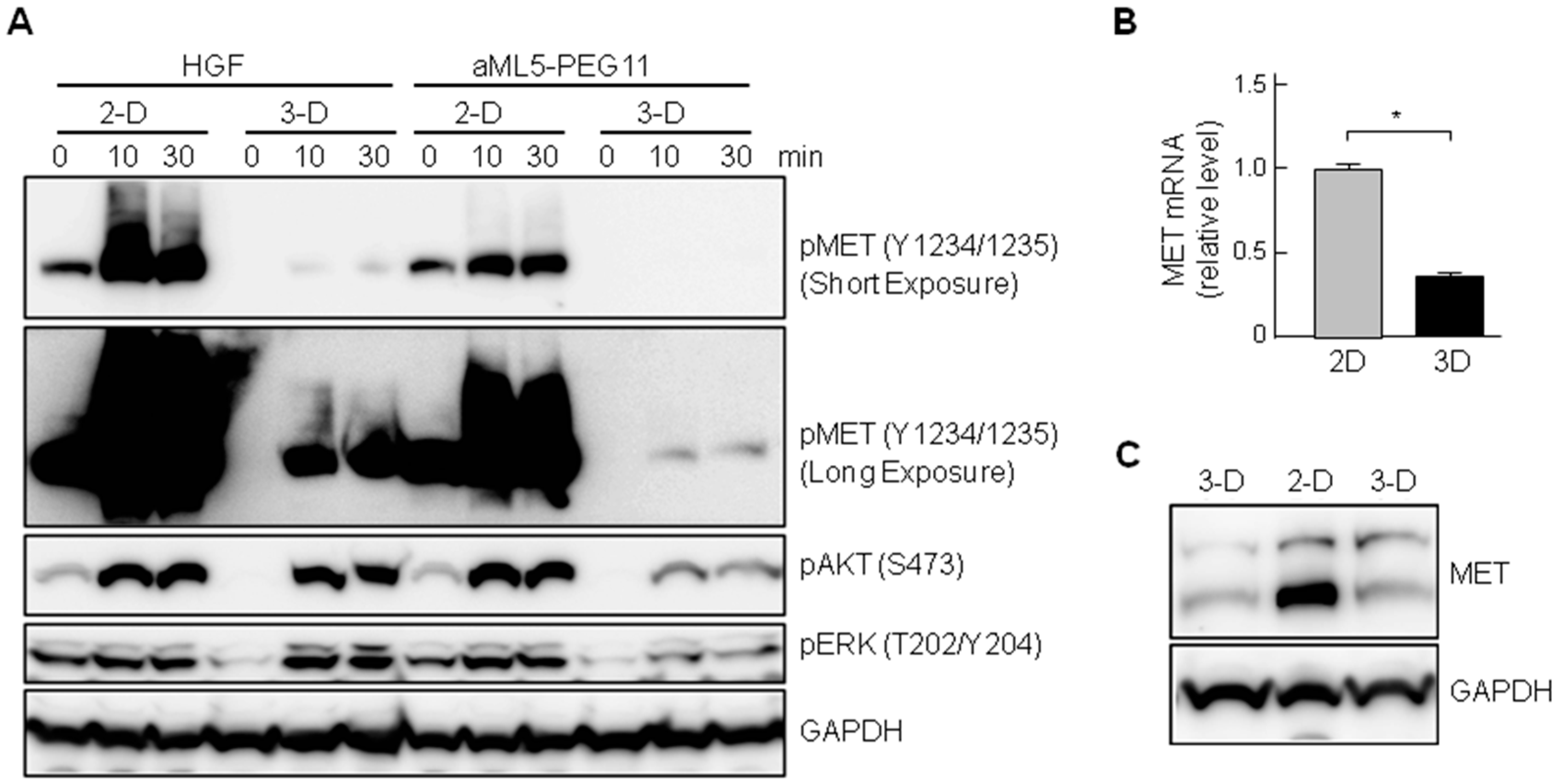
2.3. MET Activation Profile in 3-D Epithelial Morphogenesis
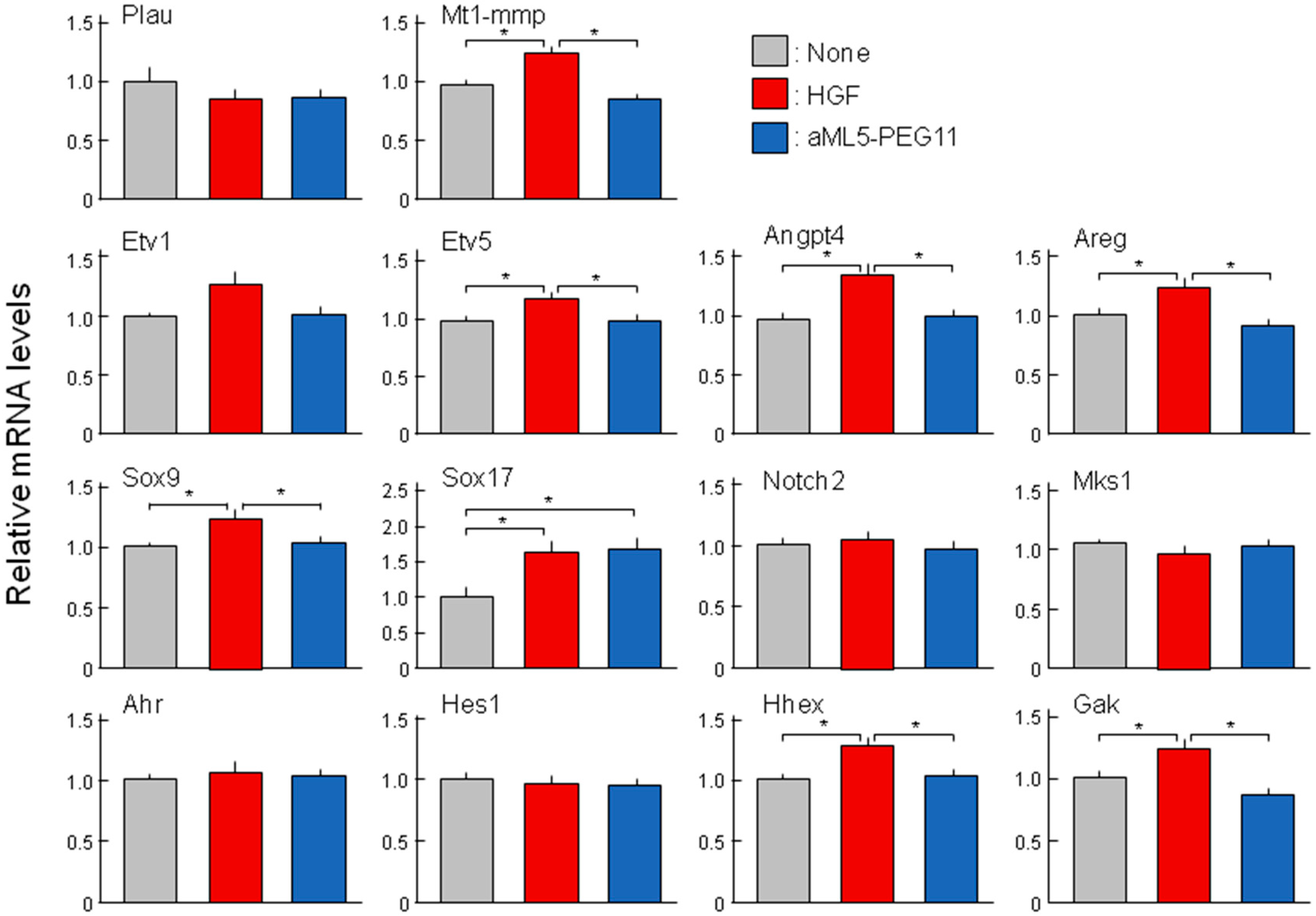
2.4. Analysis of Morphogenesis-Related Gene Expression Profiles
3. Discussion
4. Materials and Methods
4.1. Cells, HGF, Peptide, and Reagents
4.2. Establishment of MET Deficient Cells by Genome Editing
4.3. Cell-Based Phospho-MET Receptor Assay
4.4. Cell Growth Assay
4.5. Scratch Wound-Healing, Migration, and Invasion Assays
4.6. Branching Morphogenesis Assay
4.7. Western Blotting
4.8. Quantitative Real-Time PCR
4.9. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HGF | hepatocyte growth factor |
| RTKs | receptor tyrosine kinases |
| EpoR | erythropoietin receptor |
| EGFR | epidermal growth factor receptor |
| RaPID | random nonstandard peptides integrated discovery |
| ERK | extracellular signal-regulated kinase |
| AKT | protein kinase B |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| Plau | plasminogen activator urokinase |
| Mt1-mmp | Membrane type 1 metalloprotease |
| Etv | ETS translocation variant |
| Angptl4 | Angiopoietin Like 4 |
| Areg | Amphiregulin |
| Sox | SRY-Box |
| Mks1 | Meckel syndrome type 1 |
| Ahr | aryl-hydrocarbon receptor |
| Hes1 | hes family bHLH transcription factor 1 |
| Hhex | hematopoietically expressed homeobox |
| Gak | cyclin G associated kinase |
References
- Levin, D.; Schneider, W.M.; Hoffmann, H.H.; Yarden, G.; Busetto, A.G.; Manor, O.; Sharma, N.; Rice, C.M.; Schreiber, G. Multifaceted activities of type I interferon are revealed by a receptor antagonist. Sci. Signal. 2014, 7, ra50. [Google Scholar] [CrossRef] [PubMed]
- Macdonald-Obermann, J.L.; Pike, L.J. Different epidermal growth factor (EGF) receptor ligands show distinct kinetics and biased or partial agonism for homodimer and heterodimer formation. J. Biol. Chem. 2014, 289, 26178–26188. [Google Scholar] [CrossRef] [PubMed]
- Moraga, I.; Richter, D.; Wilmes, S.; Winkelmann, H.; Jude, K.; Thomas, C.; Suhoski, M.M.; Engleman, E.G.; Piehler, J.; Garcia, K.C. Instructive roles for cytokine-receptor binding parameters in determining signaling and functional potency. Sci. Signal. 2015, 8, ra114. [Google Scholar] [CrossRef] [PubMed]
- Moraga, I.; Wernig, G.; Wilmes, S.; Gryshkova, V.; Richter, C.P.; Hong, W.J.; Sinha, R.; Guo, F.; Fabionar, H.; Wehrman, T.S.; et al. Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell 2015, 160, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Riese, D.J. Ligand-based receptor tyrosine kinase partial agonists: New paradigm for cancer drug discovery? Expert Opin. Drug Discov. 2011, 6, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Yea, K.; Xie, J.; Zhang, H.; Zhang, W.; Lerner, R.A. Selection of multiple agonist antibodies from intracellular combinatorial libraries reveals that cellular receptors are functionally pleiotropic. Curr. Opin. Chem. Biol. 2015, 26, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Freed, D.M.; Bessman, N.J.; Kiyatkin, A.; Salazar-Cavazos, E.; Byrne, P.O.; Moore, J.O.; Valley, C.C.; Ferguson, K.M.; Leahy, D.J.; Lidke, D.S.; et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell 2017, 171, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Kaburaki, H.; Miyamoto, S.; Katayama, J.; Watanabe, Y. Discovery of a novel thrombopoietin mimic agonist peptide. J. Biochem. 1997, 122, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Tarasova, A.; Haylock, D.N.; Meagher, L.; Be, C.L.; White, J.; Nilsson, S.K.; Andrade, J.; Cartledge, K.; Winkler, D.A. Potent agonists of a hematopoietic stem cell cytokine receptor, c-Mpl. ChemMedChem 2013, 8, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Simonneau, C.; Leclercq, B.; Mougel, A.; Adriaenssens, E.; Paquet, C.; Raibaut, L.; Ollivier, N.; Drobecq, H.; Marcoux, J.; Cianférani, S.; et al. Semi-synthesis of a HGF/SF kringle one (K1) domain scaffold generates a potent in vivo MET receptor agonist. Chem. Sci. 2015, 6, 2110–2121. [Google Scholar] [CrossRef] [PubMed]
- Ueki, R.; Ueki, A.; Kanda, N.; Sando, S. Oligonucleotide-based mimetics of hepatocyte growth factor. Angew. Chem. Int. Ed. Engl. 2016, 55, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Sakai, K.; Suzuki, Y.; Ozawa, N.; Hatta, T.; Natsume, T.; Matsumoto, K.; Suga, H. Artificial human Met agonists based on macrocycle scaffolds. Nat. Commun. 2015, 6, 6373. [Google Scholar] [CrossRef] [PubMed]
- Furlan, A.; Kherrouche, Z.; Montagne, R.; Copin, M.C.; Tulasne, D. Thirty years of research on met receptor to move a biomarker from bench to bedside. Cancer Res. 2014, 74, 6737–6744. [Google Scholar] [CrossRef] [PubMed]
- Petrini, I. Biology of MET: A double life between normal tissue repair and tumor progression. Ann. Transl. Med. 2015, 3, 82. [Google Scholar] [CrossRef] [PubMed]
- Imamura, R.; Matsumoto, K. Hepatocyte growth factor in physiology and infectious diseases. Cytokine 2017, 98, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Abella, J.V.; Peschard, P.; Naujokas, M.A.; Lin, T.; Saucier, C.; Urbé, S.; Park, M. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol. Cell Biol. 2005, 25, 9632–9645. [Google Scholar] [CrossRef] [PubMed]
- Kong-Beltran, M.; Seshagiri, S.; Zha, J.; Zhu, W.; Bhawe, K.; Mendoza, N.; Holcomb, T.; Pujara, K.; Stinson, J.; Fu, L.; et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006, 66, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.C.; Maulik, G.; Christensen, J.; Salgia, R. c-Met: Structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003, 22, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Ponzetto, C.; Bardelli, A.; Zhen, Z.; Maina, F.; dalla Zonca, P.; Giordano, S.; Graziani, A.; Panayotou, G.; Comoglio, P.M. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 1994, 77, 261–271. [Google Scholar] [CrossRef]
- Weidner, K.M.; Sachs, M.; Riethmacher, D.; Birchmeier, W. Mutation of juxtamembrane tyrosine residue 1001 suppresses loss-of-function mutations of the met receptor in epithelial cells. Proc. Natl. Acad. Sci. USA 1995, 92, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Montesano, R.; Matsumoto, K.; Nakamura, T.; Orci, L. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell 1991, 67, 901–908. [Google Scholar] [CrossRef]
- Bissell, M.J.; Barcellos-Hoff, M.H. The influence of extracellular matrix on gene expression: Is structure the message? J. Cell Sci. Suppl. 1987, 8, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Delannoy-Courdent, A.; Fauquette, W.; Dong-Le Bourhis, X.F.; Boilly, B.; Vandenbunder, B.; Desbiens, X. Expression of c-ets-1 and uPA genes is associated with mammary epithelial cell tubulogenesis or neoplastic scattering. Int. J. Dev. Biol. 1996, 40, 1097–1108. [Google Scholar] [PubMed]
- Weaver, S.A.; Wolters, B.; Ito, N.; Woskowicz, A.M.; Kaneko, K.; Shitomi, Y.; Seiki, M.; Itoh, Y. Basal localization of MT1-MMP is essential for epithelial cell morphogenesis in 3D collagen matrix. J. Cell Sci. 2014, 127, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Chotteau-Lelievre, A.; Montesano, R.; Soriano, J.; Soulie, P.; Desbiens, X.; de Launoit, Y. PEA3 transcription factors are expressed in tissues undergoing branching morphogenesis and promote formation of duct-like structures by mammary epithelial cells in vitro. Dev. Biol. 2003, 259, 241–257. [Google Scholar] [CrossRef]
- Ciarloni, L.; Mallepell, S.; Brisken, C. Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc. Natl. Acad. Sci. USA 2007, 104, 5455–5460. [Google Scholar] [CrossRef] [PubMed]
- Raynaud, P.; Carpentier, R.; Antoniou, A.; Lemaigre, F.P. Biliary differentiation and bile duct morphogenesis in development and disease. Int. J. Biochem. Cell Biol. 2011, 43, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Stanger, B.Z. Molecular mechanisms of bile duct development. Int. J. Biochem. Cell Biol. 2011, 43, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Harstad, E.B.; Guite, C.A.; Thomae, T.L.; Bradfield, C.A. Liver deformation in Ahr-null mice: Evidence for aberrant hepatic perfusion in early development. Mol. Pharmacol. 2006, 69, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Zhao, X.; Yim, Y.I.; Eisenberg, E.; Greene, L.E. Essential role of cyclin-G-associated kinase (Auxilin-2) in developing and mature mice. Mol. Biol. Cell 2008, 19, 2766–2776. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Kataoka, H.; Date, K.; Nakamura, T. Cooperative interaction between α- and β-chains of hepatocyte growth factor on c-Met receptor confers ligand-induced receptor tyrosine phosphorylation and multiple biological responses. J. Biol. Chem. 1998, 273, 22913–22920. [Google Scholar] [CrossRef] [PubMed]
- Kirchhofer, D.; Yao, X.; Peek, M.; Eigenbrot, C.; Lipari, M.T.; Billeci, K.L.; Maun, H.R.; Moran, P.; Santell, L.; Wiesmann, C.; et al. Structural and functional basis of the serine protease-like hepatocyte growth factor beta-chain in Met binding and signaling. J. Biol. Chem. 2004, 279, 39915–39924. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.; Lazarus, R.A.; Yao, X.; Kirchhofer, D.; Wiesmann, C. Crystal structure of the HGF β-chain in complex with the Sema domain of the Met receptor. EMBO J. 2004, 23, 2325–2335. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, K.; Kitamura, A.; Naka, D.; Kitamura, N. An alternatively processed mRNA generated from human hepatocyte growth factor gene. Eur. J. Biochem. 1991, 197, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.M.; Rubin, J.S.; Bottaro, D.P.; Hirschfield, D.W.; Chedid, M.; Aaronson, S.A. Identification of a competitive HGF antagonist encoded by an alternative transcript. Science 1991, 254, 1382–1385. [Google Scholar] [CrossRef] [PubMed]
- Cioce, V.; Csaky, K.G.; Chan, A.M.; Bottaro, D.P.; Taylor, W.G.; Jensen, R.; Aaronson, S.A.; Rubin, J.S. Hepatocyte growth factor (HGF)/NK1 is a naturally occurring HGF/scatter factor variant with partial agonist/antagonist activity. J. Biol. Chem. 1996, 271, 13110–13115. [Google Scholar] [CrossRef] [PubMed]
- Day, R.M.; Cioce, V.; Breckenridge, D.; Castagnino, P.; Bottaro, D.P. Differential signaling by alternative HGF isoforms through c-Met: Activation of both MAP kinase and PI 3-kinase pathways is insufficient for mitogenesis. Oncogene 1999, 18, 3399–3406. [Google Scholar] [CrossRef] [PubMed]
- Tolbert, W.D.; Daugherty-Holtrop, J.; Gherardi, E.; Vande Woude, G.; Xu, H.E. Structural basis for agonism and antagonism of hepatocyte growth factor. Proc. Natl. Acad. Sci. USA 2010, 107, 13264–13269. [Google Scholar] [CrossRef] [PubMed]
- Mungunsukh, O.; Lee, Y.H.; Bottaro, D.P.; Day, R.M. The hepatocyte growth factor isoform NK2 activates motogenesis and survival but not proliferation due to lack of Akt activation. Cell Signal. 2016, 28, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, R.; Neel, B.G.; Rapoport, T.A. Mathematical models of protein kinase signal transduction. Mol. Cell 2002, 9, 957–970. [Google Scholar] [CrossRef]
- Ferrell, J.E., Jr.; Machleder, E.M. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 1998, 280, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Azeloglu, E.U.; Iyengar, R. Signaling networks: Information flow, computation, and decision making. Cold Spring Harb. Perspect. Biol. 2015, 7, a005934. [Google Scholar] [CrossRef] [PubMed]
- Rosário, M.; Birchmeier, W. How to make tubes: Signaling by the Met receptor tyrosine kinase. Trends Cell Biol. 2003, 13, 328–335. [Google Scholar] [CrossRef]
- Ho, C.C.M.; Chhabra, A.; Starkl, P.; Schnorr, P.J.; Wilmes, S.; Moraga, I.; Kwon, H.S.; Gaudenzio, N.; Sibilano, R.; Wehrman, T.S.; et al. Decoupling the functional pleiotropy of stem cell factor by tuning c-Kit signaling. Cell 2017, 168, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ge, H.; Lemon, B.; Vonderfecht, S.; Baribault, H.; Weiszmann, J.; Gupte, J.; Gardner, J.; Lindberg, R.; Wang, Z.; et al. Separating mitogenic and metabolic activities of fibroblast growth factor 19 (FGF19). Proc. Natl. Acad. Sci. USA 2010, 107, 14158–14163. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Rinella, M.E.; Abdelmalek, M.F.; Trotter, J.F.; Paredes, A.H.; Arnold, H.L.; Kugelmas, M.; Bashir, M.R.; Jaros, M.J.; Ling, L.; et al. NGM282 for treatment of non-alcoholic steatohepatitis: A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2018, 391, 1174–1185. [Google Scholar] [CrossRef]
- Chmielowiec, J.; Borowiak, M.; Morkel, M.; Stradal, T.; Munz, B.; Werner, S.; Wehland, J.; Birchmeier, C.; Birchmeier, W. c-Met is essential for wound healing in the skin. J. Cell Biol. 2007, 177, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Bevan, D.; Gherardi, E.; Fan, T.P.; Edwards, D.; Warn, R. Diverse and potent activities of HGF/SF in skin wound repair. J. Pathol. 2004, 203, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Matsumoto, K.; Tomioka, D.; Bessho, K.; Itami, S.; Yoshikawa, K.; Nakamura, T. Recombinant hepatocyte growth factor accelerates cutaneous wound healing in a diabetic mouse model. Growth Factors 2004, 22, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Buchstein, N.; Hoffmann, D.; Smola, H.; Lang, S.; Paulsson, M.; Niemann, C.; Krieg, T.; Eming, S.A. Alternative proteolytic processing of hepatocyte growth factor during wound repair. Am. J. Pathol. 2009, 174, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Ochiai, H.; Kaneko, T.; Mashimo, T.; Tokumasu, D.; Sakane, Y.; Suzuki, K.; Miyamoto, T.; Sakamoto, N.; Matsuura, S.; et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 2013, 3, 3379. [Google Scholar] [CrossRef] [PubMed]
- Ninagawa, S.; Okada, T.; Sumitomo, Y.; Kamiya, Y.; Kato, K.; Horimoto, S.; Ishikawa, T.; Takeda, S.; Sakuma, T.; Yamamoto, T.; et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 2014, 206, 347–356. [Google Scholar] [CrossRef] [PubMed]
| mRNAs | Sequences |
|---|---|
| Mt1-mmp | F: 5′-CACTGCCTACGAGAGGAAGG-3′ |
| R: 5′-TTGGGGTACTCGCTATCCAC-3′ | |
| Angptl4 | F: 5′-GTCCACCGACCTCCCGTTA-3′ |
| R: 5′-CCTCATGGTCTAGGTGCTTGT-3′ | |
| Areg | F: 5′-GAGCCGACTATGACTACTCAGA-3′ |
| R: 5′-TCACTTTCCGTCTTGTTTTGGG-3′ | |
| Etv1 | F: 5′-TGGCAGTTTTTGGTAGCTCTTC-3′ |
| R: 5′-CGGAGTGAACGGCTAAGTTTATC-3′ | |
| Etv5 | F: 5′-CAGTCAACTTCAAGAGGCTTGG-3′ |
| R: 5′-TGCTCATGGCTACAAGACGAC-3′ | |
| Sox9 | F: 5′-AGCGAACGCACATCAAGAC-3′ |
| R: 5′-CTGTAGGCGATCTGTTGGGG-3′ | |
| Sox17 | F: 5′-GTGGACCGCACGGAATTTG-3′ |
| R: 5′-GGAGATTCACACCGGAGTCA-3′ | |
| Notch2 | F: 5′-CCTTCCACTGTGAGTGTCTGA-3′ |
| R: 5′-AGGTAGCATCATTCTGGCAGG-3′ | |
| Hhex | F: 5′-TCAGAATCGACGCGCTAAATG-3′ |
| R: 5′-AGAGCTATCCAAAGAAGCACCT-3′ | |
| Gak | F: 5′-CCACCCGAACATTGTCCAGTT-3′ |
| R: 5′-AGAACCGTGTCGCACGAAA-3′ | |
| Mks1 | F: 5′-GCAAGAAAAACCGACGAATCTTT-3′ |
| R: 5′-TCGCTCGACCAAGAATGAAGG-3′ | |
| Ahr | F: 5′-ACATCACCTACGCCAGTCGC-3′ |
| R: 5′-TCTATGCCGCTTGGAAGGAT-3′ | |
| Hes1 | F: 5′-ACGTGCGAGGGCGTTAATAC-3′ |
| R: 5′-ATTGATCTGGGTCATGCAGTTG-3′ | |
| Met | F: 5′-AGATGAATGTGAATATGAAGTATC-3′ |
| R: 5′-CAGTCTTGTACTCAGCAACCT-3′ | |
| Gapdh | F: 5′-GAGTCAACGGATTTGGTCGT-3′ |
| R: 5′-GACAAGCTTCCCGTTCTCAG-3′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, W.; Sakai, K.; Imamura, R.; Ito, K.; Suga, H.; Sakuma, T.; Yamamoto, T.; Matsumoto, K. MET Activation by a Macrocyclic Peptide Agonist that Couples to Biological Responses Differently from HGF in a Context-Dependent Manner. Int. J. Mol. Sci. 2018, 19, 3141. https://doi.org/10.3390/ijms19103141
Miao W, Sakai K, Imamura R, Ito K, Suga H, Sakuma T, Yamamoto T, Matsumoto K. MET Activation by a Macrocyclic Peptide Agonist that Couples to Biological Responses Differently from HGF in a Context-Dependent Manner. International Journal of Molecular Sciences. 2018; 19(10):3141. https://doi.org/10.3390/ijms19103141
Chicago/Turabian StyleMiao, Wenyu, Katsuya Sakai, Ryu Imamura, Kenichiro Ito, Hiroaki Suga, Tetsushi Sakuma, Takashi Yamamoto, and Kunio Matsumoto. 2018. "MET Activation by a Macrocyclic Peptide Agonist that Couples to Biological Responses Differently from HGF in a Context-Dependent Manner" International Journal of Molecular Sciences 19, no. 10: 3141. https://doi.org/10.3390/ijms19103141
APA StyleMiao, W., Sakai, K., Imamura, R., Ito, K., Suga, H., Sakuma, T., Yamamoto, T., & Matsumoto, K. (2018). MET Activation by a Macrocyclic Peptide Agonist that Couples to Biological Responses Differently from HGF in a Context-Dependent Manner. International Journal of Molecular Sciences, 19(10), 3141. https://doi.org/10.3390/ijms19103141






