ABD-Derived Protein Blockers of Human IL-17 Receptor A as Non-IgG Alternatives for Modulation of IL-17-Dependent Pro-Inflammatory Axis
Abstract
1. Introduction
2. Results
2.1. Molecular Assembly and Production of Recombinant IL-17RA
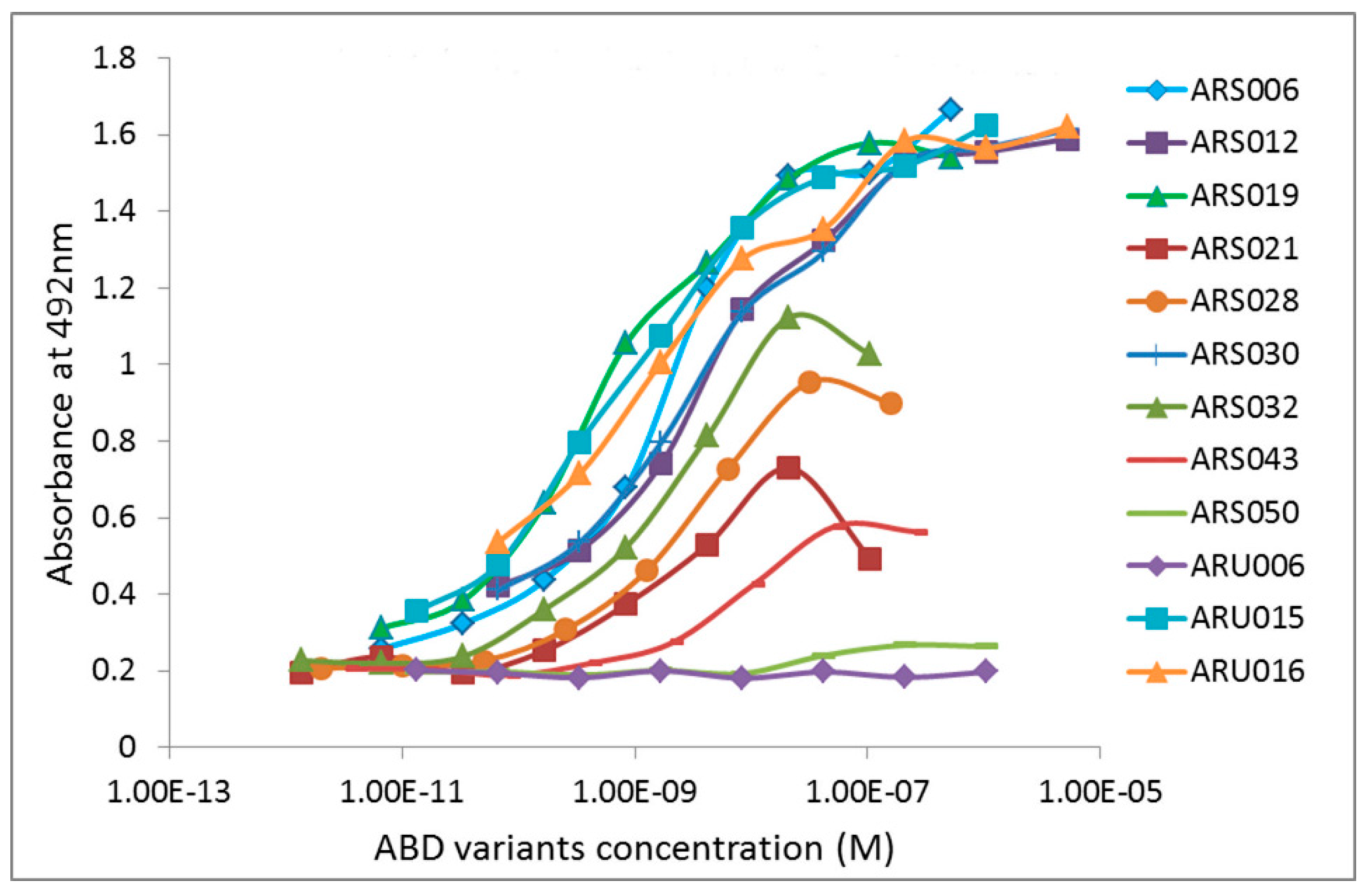
2.2. Selection of ABD-Derived Variants Producing IL-17RA-Targeted Proteins
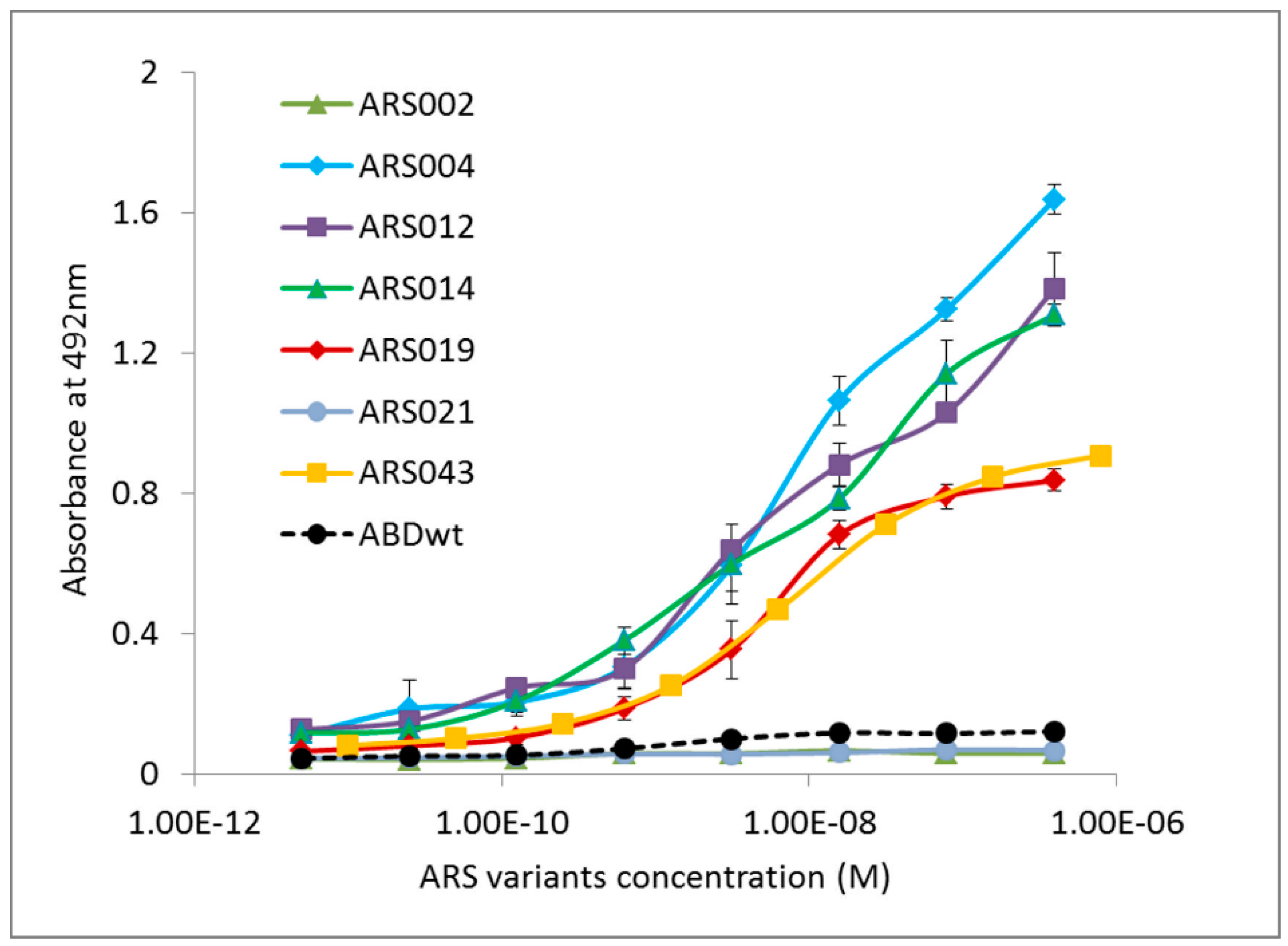
2.3. Characterization of Binding Properties of the Selected ARS Variants
2.4. Binding of the ARS Variants to Human Cells
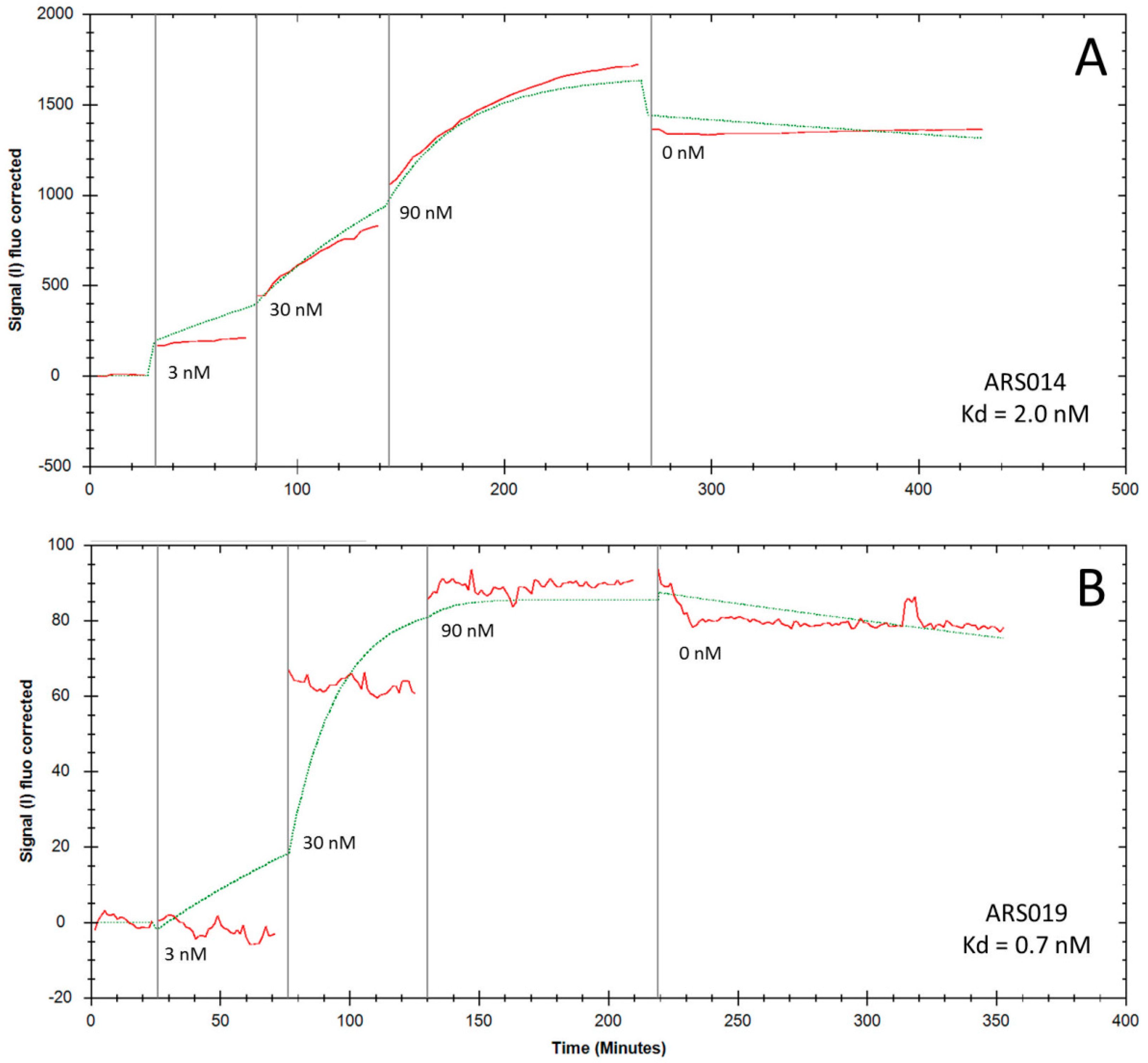
2.5. Determination of Binding Kinetics and Affinities of the ARS Variants Using Human Cells
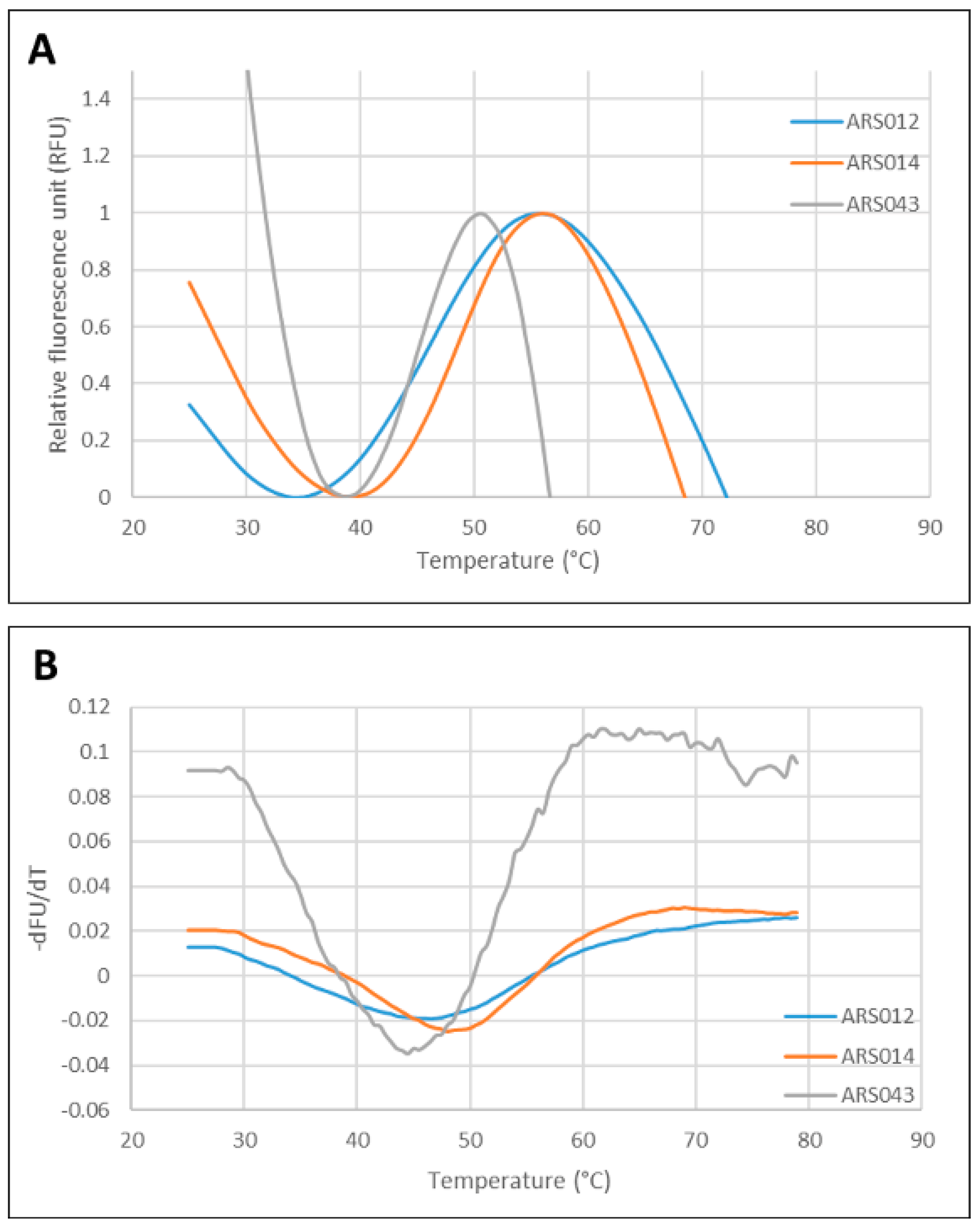
2.6. Thermal Stability of the ARS Binding Proteins
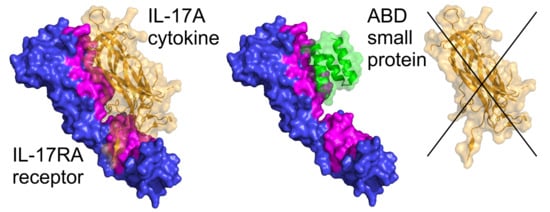
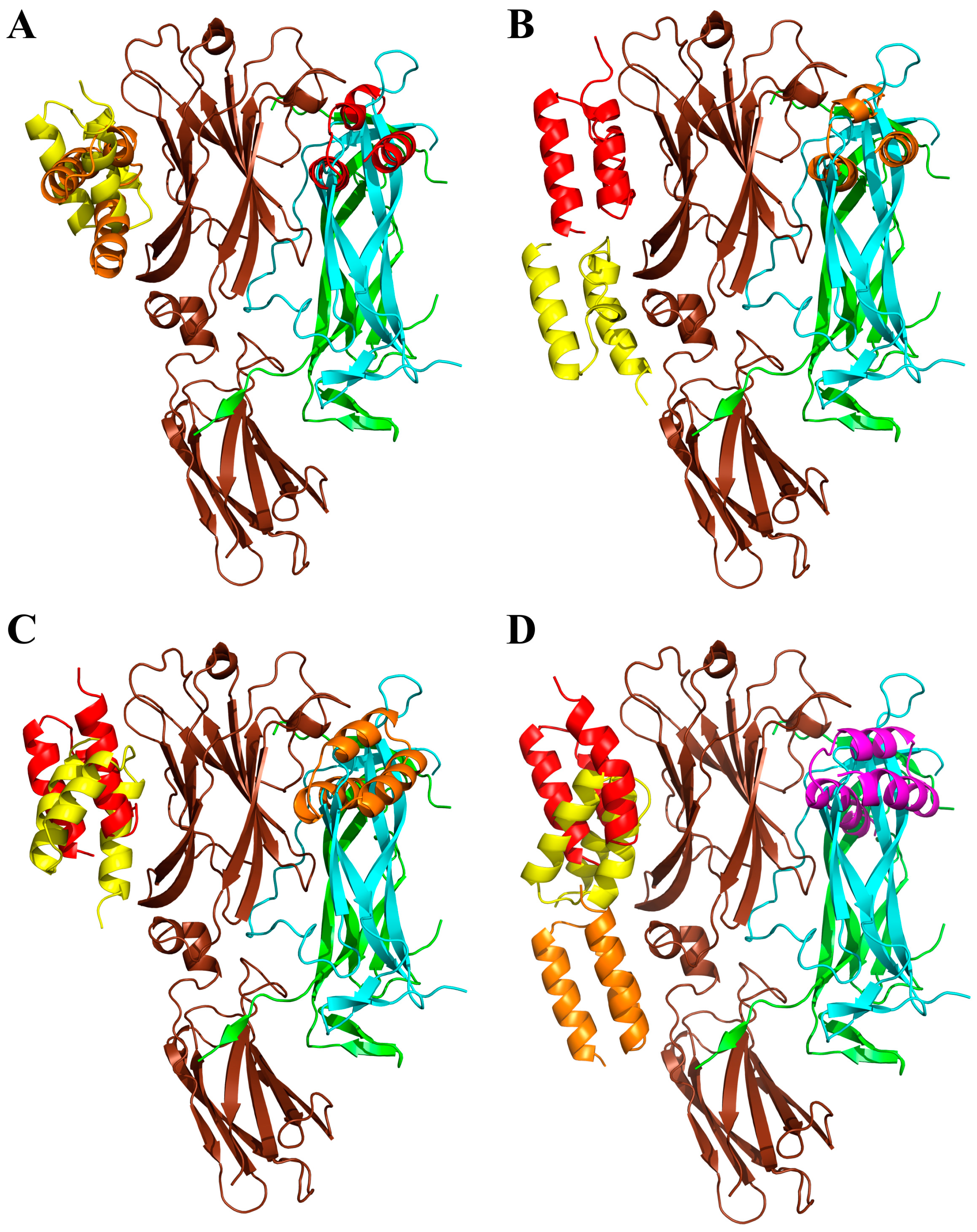
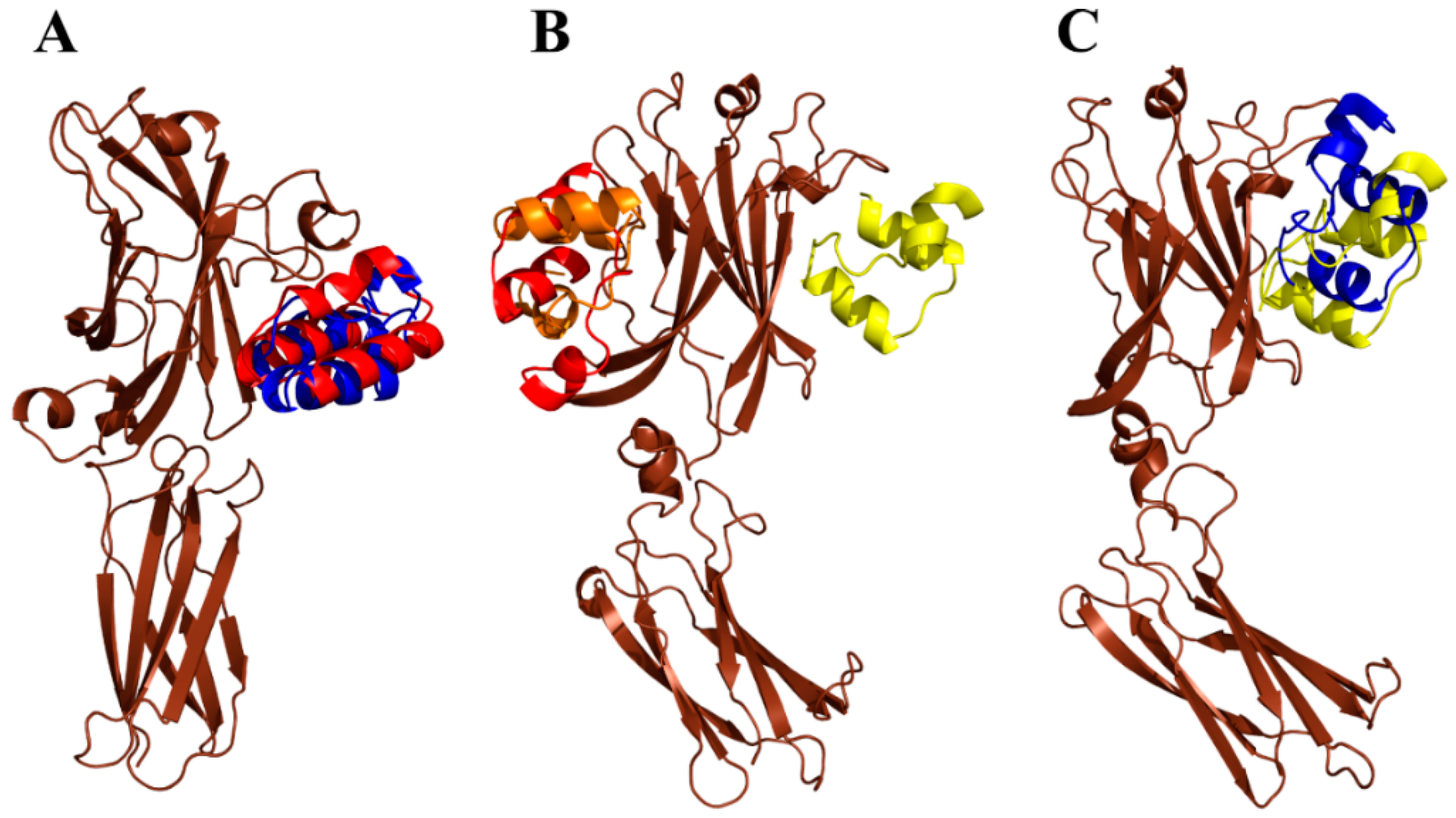
2.7. Modeling of ARS-IL17RA Interactions
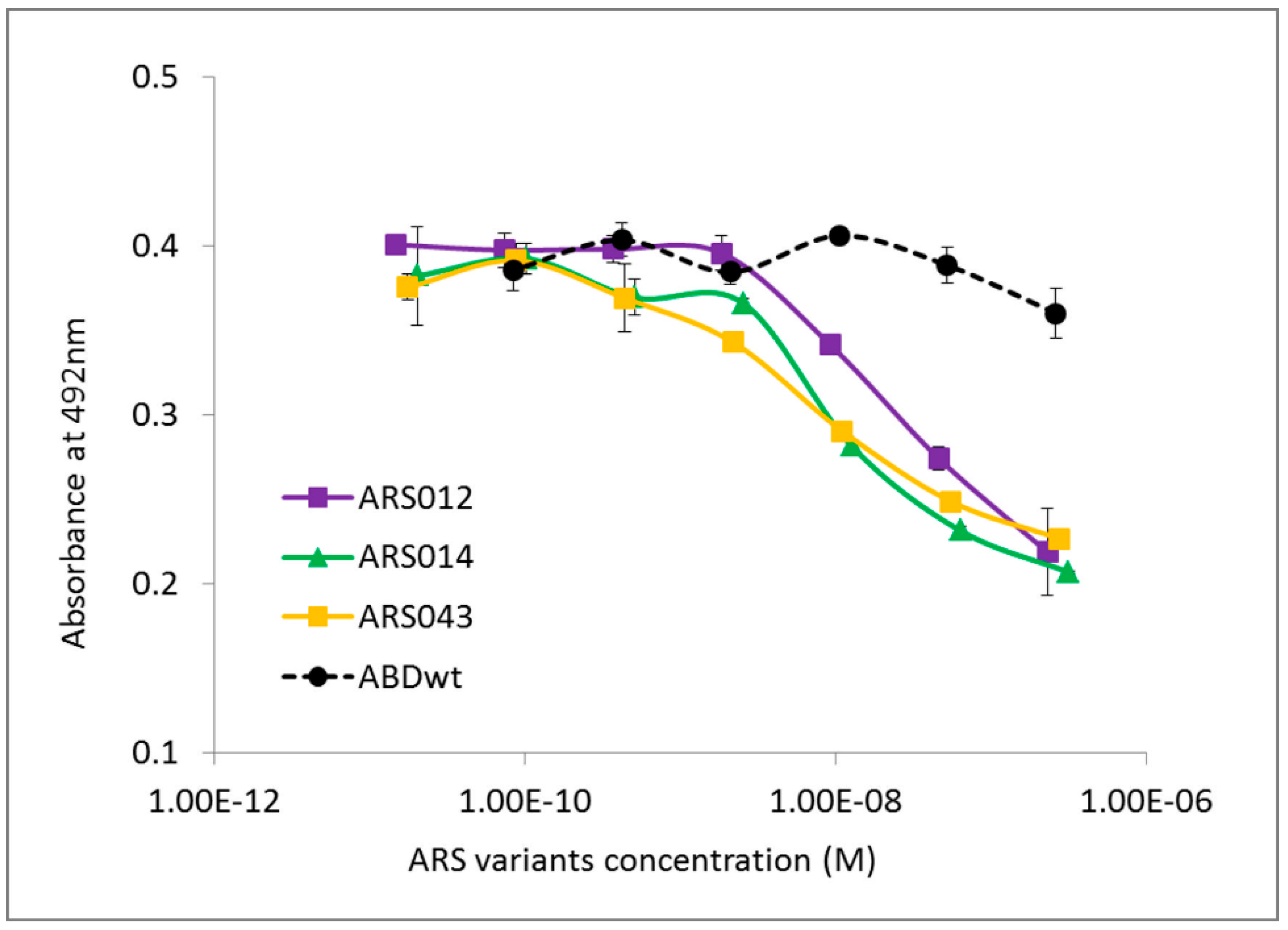
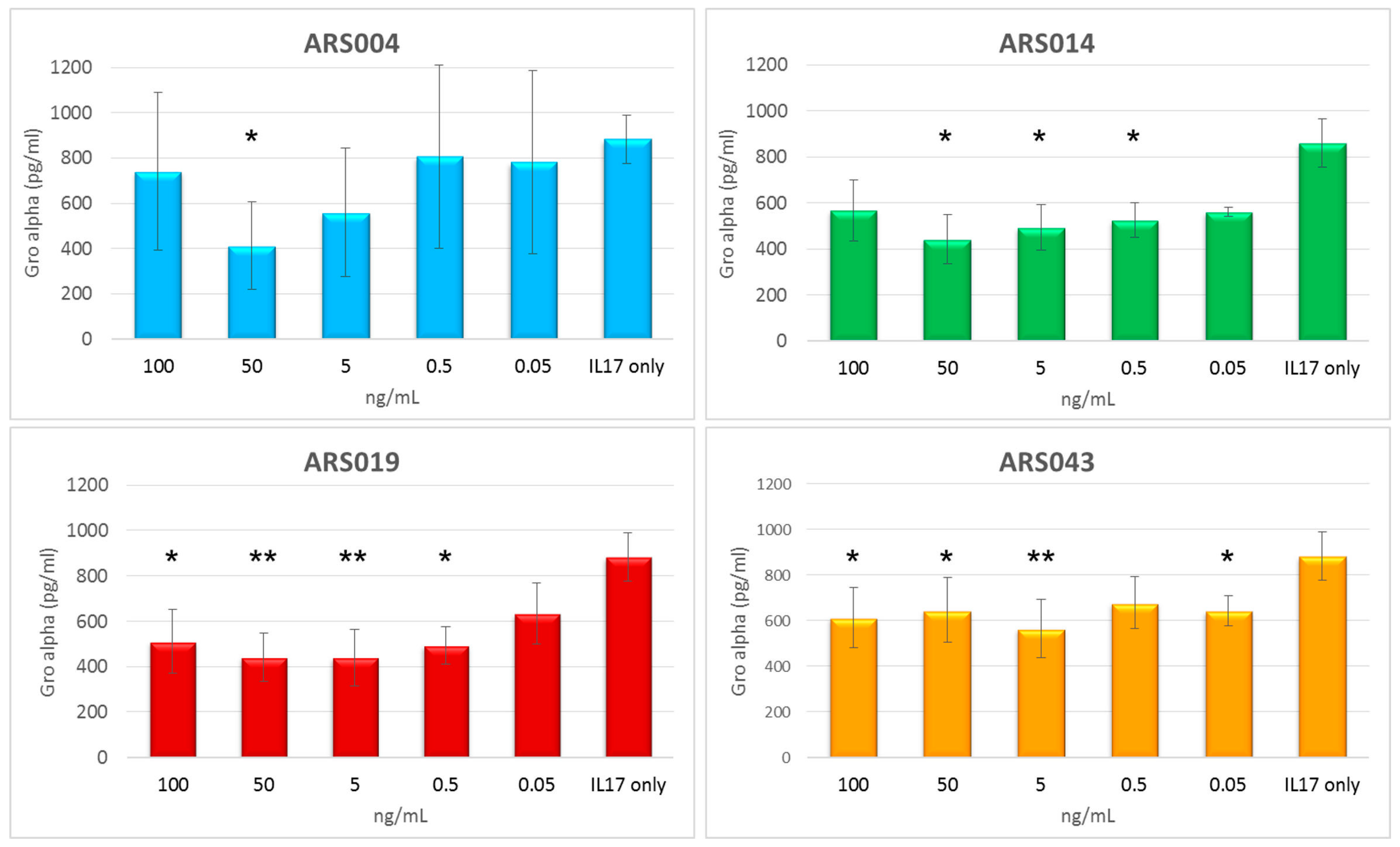
2.8. Immunomodulatory Potential of the ARS Binding Proteins
3. Discussion
4. Materials and Methods
4.1. Antibodies, Recombinant Proteins and Detection Agents
4.2. Cell Lines and Growth Conditions
4.3. Molecular Assembly and Production of Recombinant IL-17RA
4.4. ABD Library Construction and Ribosome Display Selection
4.5. Production of In Vivo Biotinylated Protein Binders
4.6. Screening of IL-17RA-Targeted Binders in ELISA
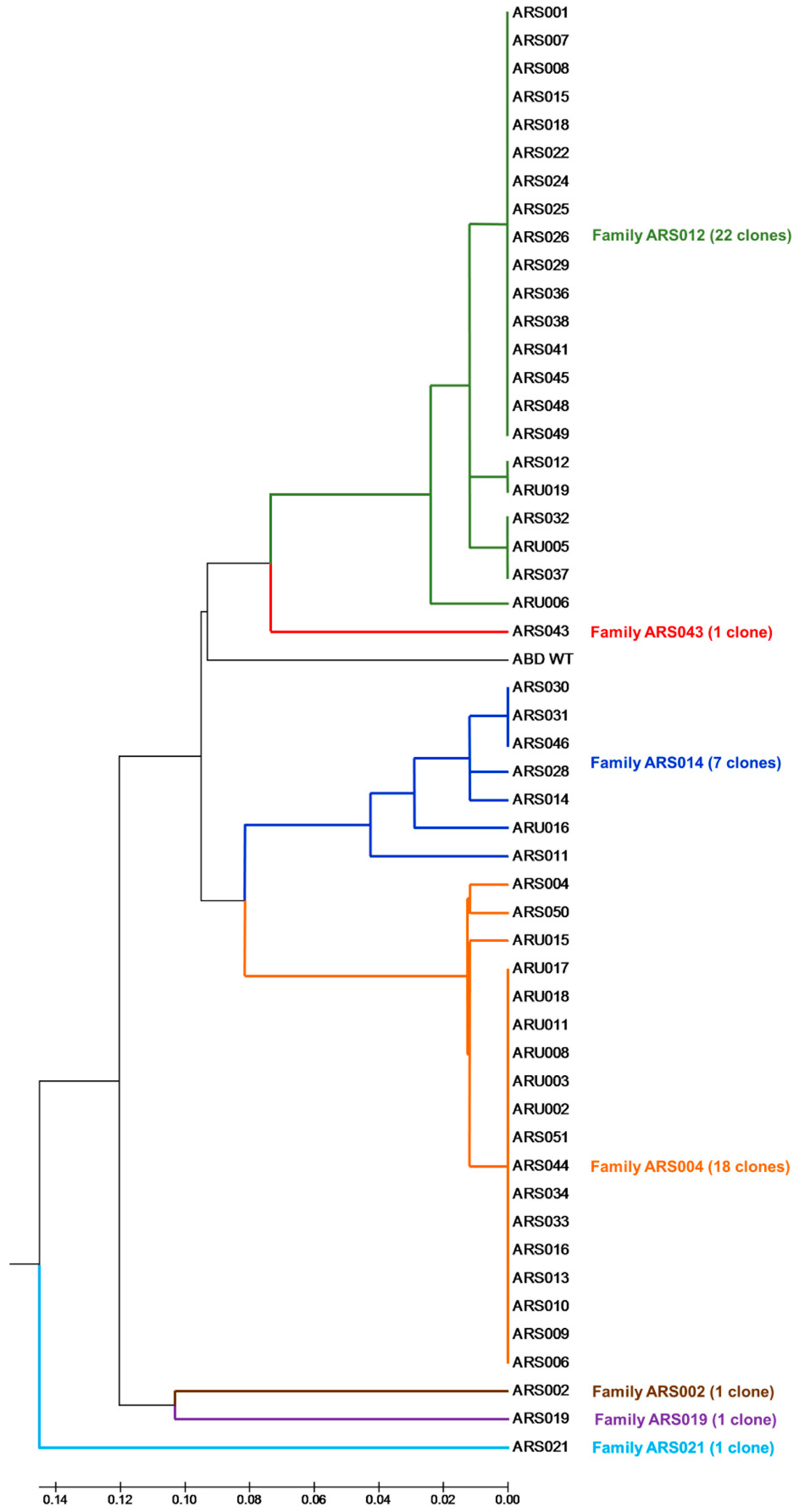
4.7. Sequence Analysis and Clustering of Selected ARS and ARU Variants
4.8. Modeling of ARS-IL17RA Interactions
4.9. Competition ELISA
4.10. Fluorescence-Based Thermal-Shift Assay
4.11. Flow Cytometry
4.12. Binding of ARS Proteins to Human Cells Analyzed by LigandTracer Green Line System
4.13. Testing of the Immunomodulatory Potential of ARS Ligands
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cua, D.J.; Sherlock, J.; Chen, Y.; Murphy, C.A.; Joyce, B.; Seymour, B.; Lucian, L.; To, W.; Kwan, S.; Churakova, T.; et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 2003, 421, 744–748. [Google Scholar] [CrossRef] [PubMed]
- Langrish, C.L.; Chen, Y.; Blumenschein, W.M.; Mattson, J.; Basham, B.; Sedgwick, J.D.; McClanahan, T.; Kastelein, R.A.; Cua, D.J. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005, 201, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Duerr, R.H.; Taylor, K.D.; Brant, S.R.; Rioux, J.D.; Silverberg, M.S.; Daly, M.J.; Steinhart, A.H.; Abraham, C.; Regueiro, M.; Griffiths, A.; et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006, 314, 1461–1463. [Google Scholar] [CrossRef] [PubMed]
- Oppmann, B.; Lesley, R.; Blom, B.; Timans, J.C.; Xu, Y.; Hunte, B.; Vega, F.; Yu, N.; Wang, J.; Singh, K.; et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 2000, 13, 715–725. [Google Scholar] [CrossRef]
- Parham, C.; Chirica, M.; Timans, J.; Vaisberg, E.; Travis, M.; Cheung, J.; Pflanz, S.; Zhang, R.; Singh, K.P.; Vega, F.; et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002, 168, 5699–5708. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Danilenko, D.M.; Valdez, P.; Kasman, I.; Eastham-Anderson, J.; Wu, J.; Ouyang, W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 2007, 445, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Chen, S.; Qian, H.; Huang, W. Interleukin-23: As a drug target for autoimmune inflammatory diseases. Immunology 2012, 135, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Dungan, L.S.; Mills, K.H. Caspase-1-processed IL-1 family cytokines play a vital role in driving innate IL-17. Cytokine 2011, 56, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Qian, Y. IL-17 family cytokines mediated signaling in the pathogenesis of inflammatory diseases. Cell. Signal. 2013, 25, 2335–2347. [Google Scholar] [CrossRef] [PubMed]
- Iwakura, Y.; Ishigame, H.; Saijo, S.; Nakae, S. Functional specialization of interleukin-17 family members. Immunity 2011, 34, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Restifo, N.P. T(H)17 cells in tumour immunity and immunotherapy. Nat. Rev. Immunol. 2010, 10, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Sutton, C.E.; Mielke, L.A.; Mills, K.H. IL-17-producing gammadelta T cells and innate lymphoid cells. Eur. J. Immunol. 2012, 42, 2221–2231. [Google Scholar] [CrossRef] [PubMed]
- Tonini, A.; Gualtieri, B.; Panduri, S.; Romanelli, M.; Chiricozzi, A. A new class of biologic agents facing the therapeutic paradigm in psoriasis: Anti-IL-23 agents. Expert Opin. Biol. Ther. 2018, 18, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Papp, K.A.; Griffiths, C.E.; Gordon, K.; Lebwohl, M.; Szapary, P.O.; Wasfi, Y.; Chan, D.; Hsu, M.C.; Ho, V.; Ghislain, P.D.; et al. Long-term safety of ustekinumab in patients with moderate-to-severe psoriasis: Final results from 5 years of follow-up. Br. J. Dermatol. 2013, 168, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, C.L.; Kimball, A.B.; Papp, K.A.; Yeilding, N.; Guzzo, C.; Wang, Y.; Li, S.; Dooley, L.T.; Gordon, K.B. PHOENIX 1 Study Investigators. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1). Lancet 2008, 371, 1665–1674. [Google Scholar] [CrossRef]
- Traczewski, P.; Rudnicka, L. Briakinumab for the treatment of plaque psoriasis. BioDrugs 2012, 26, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.B.; Langley, R.G.; Gottlieb, A.B.; Papp, K.A.; Krueger, G.G.; Strober, B.E.; Williams, D.A.; Gu, Y.; Valdes, J.M. A phase III, randomized, controlled trial of the fully human IL-12/23 mAb briakinumab in moderate-to-severe psoriasis. J. Investig. Dermatol. 2012, 132, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Reich, K.; Armstrong, A.W.; Foley, P.; Song, M.; Wasfi, Y.; Randazzo, B.; Li, S.; Shen, Y.K.; Gordon, K.B. Efficacy and safety of guselkumab, an anti-interleukin-23 monoclonal antibody, compared with adalimumab for the treatment of patients with moderate to severe psoriasis with randomized withdrawal and retreatment: Results from the phase III, double-blind, placebo- and active comparator-controlled VOYAGE 2 trial. J. Am. Acad. Dermatol. 2017, 76, 418–431. [Google Scholar] [PubMed]
- Langley, R.G.; Tsai, T.F.; Flavin, S.; Song, M.; Randazzo, B.; Wasfi, Y.; Jiang, J.; Li, S.; Puig, L. Efficacy and safety of guselkumab in patients with psoriasis who have an inadequate response to ustekinumab: Results of the randomized, double-blind, phase III NAVIGATE trial. Br. J. Dermatol. 2018, 178, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Papp, K.; Thaci, D.; Reich, K.; Riedl, E.; Langley, R.G.; Krueger, J.G.; Gottlieb, A.B.; Nakagawa, H.; Bowman, E.P.; Mehta, A.; et al. Tildrakizumab (MK-3222), an anti-interleukin-23p19 monoclonal antibody, improves psoriasis in a phase IIb randomized placebo-controlled trial. Br. J. Dermatol. 2015, 173, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Reich, K.; Papp, K.A.; Blauvelt, A.; Tyring, S.K.; Sinclair, R.; Thaci, D.; Nograles, K.; Mehta, A.; Cichanowitz, N.; Li, Q.; et al. Tildrakizumab versus placebo or etanercept for chronic plaque psoriasis (reSURFACE 1 and reSURFACE 2): Results from two randomised controlled, phase 3 trials. Lancet 2017, 390, 276–288. [Google Scholar] [CrossRef]
- Krueger, J.G.; Ferris, L.K.; Menter, A.; Wagner, F.; White, A.; Visvanathan, S.; Lalovic, B.; Aslanyan, S.; Wang, E.E.; Hall, D.; et al. Anti-IL-23A mAb BI 655066 for treatment of moderate-to-severe psoriasis: Safety, efficacy, pharmacokinetics, and biomarker results of a single-rising-dose, randomized, double-blind, placebo-controlled trial. J. Allergy Clin. Immunol. 2015, 136, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Hueber, W.; Patel, D.D.; Dryja, T.; Wright, A.M.; Koroleva, I.; Bruin, G.; Antoni, C.; Draelos, Z.; Gold, M.H.; Psoriasis Study, G.; et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci. Transl. Med. 2010, 2, 52ra72. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.U.; Vera, N.C.; Shealy, E.R.; Wetzel, M.; Feldman, S.R. A Review of the Use of Secukinumab for Psoriatic Arthritis. Rheumatol. Ther. 2017, 4, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Lubrano, E.; Perrotta, F.M. Secukinumab for ankylosing spondylitis and psoriatic arthritis. Ther. Clin. Risk Manag. 2016, 12, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Mease, P.J.; van der Heijde, D.; Ritchlin, C.T.; Okada, M.; Cuchacovich, R.S.; Shuler, C.L.; Lin, C.Y.; Braun, D.K.; Lee, C.H.; Gladman, D.D.; et al. Ixekizumab, an interleukin-17A specific monoclonal antibody, for the treatment of biologic-naive patients with active psoriatic arthritis: Results from the 24-week randomised, double-blind, placebo-controlled and active (adalimumab)-controlled period of the phase III trial SPIRIT-P1. Ann. Rheum. Dis. 2017, 76, 79–87. [Google Scholar] [PubMed]
- Blair, H.A. Brodalumab: A Review in Moderate to Severe Plaque Psoriasis. Drugs 2018, 78, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Papp, K.A.; Gordon, K.B.; Langley, R.G.; Lebwohl, M.G.; Gottlieb, A.B.; Rastogi, S.; Pillai, R.; Israel, R.J. Impact of previous biologic use on the efficacy and safety of brodalumab and ustekinumab in patients with moderate-to-severe plaque psoriasis: Integrated analysis of the randomized controlled trials AMAGINE-2 and AMAGINE-3. Br. J. Dermatol. 2018, 179, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Zaretsky, M.; Etzyoni, R.; Kaye, J.; Sklair-Tavron, L.; Aharoni, A. Directed evolution of a soluble human IL-17A receptor for the inhibition of psoriasis plaque formation in a mouse model. Chem. Biol. 2013, 20, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Skrlec, K.; Strukelj, B.; Berlec, A. Non-immunoglobulin scaffolds: A focus on their targets. Trends Biotechnol. 2015, 33, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.L.; Bedford, R.; Heseltine, S.J.; Tang, A.A.; Haza, K.Z.; Rao, A.; McPherson, M.J.; Tomlinson, D.C. Non-immunoglobulin scaffold proteins: Precision tools for studying protein-protein interactions in cancer. New Biotechnol. 2018, 45, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, J.N.; Li, J.; Biedermannova, L.; Kuchar, M.; Sipova, H.; Semeradtova, A.; Cerny, J.; Petrokova, H.; Mikulecky, P.; Polinek, J.; et al. Novel high-affinity binders of human interferon gamma derived from albumin-binding domain of protein G. Proteins 2012, 80, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Nilvebrant, J.; Astrand, M.; Lofblom, J.; Hober, S. Development and characterization of small bispecific albumin-binding domains with high affinity for ErbB3. Cell. Mol. Life Sci. 2013, 70, 3973–3985. [Google Scholar] [CrossRef] [PubMed]
- Nilvebrant, J.; Hober, S. The albumin-binding domain as a scaffold for protein engineering. Comput. Struct. Biotechnol. J. 2013, 6, e201303009. [Google Scholar] [CrossRef] [PubMed]
- Kuchar, M.; Vankova, L.; Petrokova, H.; Cerny, J.; Osicka, R.; Pelak, O.; Sipova, H.; Schneider, B.; Homola, J.; Sebo, P.; et al. Human interleukin-23 receptor antagonists derived from an albumin-binding domain scaffold inhibit IL-23-dependent ex vivo expansion of IL-17-producing T-cells. Proteins 2014, 82, 975–989. [Google Scholar] [CrossRef] [PubMed]
- Mareckova, L.; Petrokova, H.; Osicka, R.; Kuchar, M.; Maly, P. Novel binders derived from an albumin-binding domain scaffold targeting human prostate secretory protein 94 (PSP94). Protein Cell 2015, 6, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Zadravec, P.; Mareckova, L.; Petrokova, H.; Hodnik, V.; Perisic Nanut, M.; Anderluh, G.; Strukelj, B.; Maly, P.; Berlec, A. Development of Recombinant Lactococcus lactis Displaying Albumin-Binding Domain Variants against Shiga Toxin 1 B Subunit. PLoS ONE 2016, 11, e0162625. [Google Scholar] [CrossRef] [PubMed]
- Krizova, L.; Kuchar, M.; Petrokova, H.; Osicka, R.; Hlavnickova, M.; Pelak, O.; Cerny, J.; Kalina, T.; Maly, P. p19-targeted ABD-derived protein variants inhibit IL-23 binding and exert suppressive control over IL-23-stimulated expansion of primary human IL-17+ T-cells. Autoimmunity 2017, 50, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Spriggs, M.K.; Derry, J.M.; Strockbine, L.; Park, L.S.; VandenBos, T.; Zappone, J.D.; Painter, S.L.; Armitage, R.J. Molecular characterization of the human interleukin (IL)-17 receptor. Cytokine 1997, 9, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.A.; Suh, J.W.; Lee, K.H.; Kang, J.L.; Woo, S.Y. IL-17 and IL-22 enhance skin inflammation by stimulating the secretion of IL-1beta by keratinocytes via the ROS-NLRP3-caspase-1 pathway. Int. Immunol. 2012, 24, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Goetsch, L.; Haeuw, J.F.; Champion, T.; Lacheny, C.; N’Guyen, T.; Beck, A.; Corvaia, N. Identification of B- and T-cell epitopes of BB, a carrier protein derived from the G protein of Streptococcus strain G148. Clin. Diagn. Lab. Immunol. 2003, 10, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Song, X.; Chrunyk, B.A.; Shanker, S.; Hoth, L.R.; Marr, E.S.; Griffor, M.C. Crystal structures of interleukin 17A and its complex with IL-17 receptor A. Nat. Commun. 2013, 4, 1888. [Google Scholar] [CrossRef] [PubMed]
- Sarra, M.; Pallone, F.; Macdonald, T.T.; Monteleone, G. IL-23/IL-17 axis in IBD. Inflamm. Bowel Dis. 2010, 16, 1808–1813. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; McCuaig, S.; Franchini, F.; Powrie, F. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 2015, 15, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Skrlec, K.; Zadravec, P.; Hlavnickova, M.; Kuchar, M.; Vankova, L.; Petrokova, H.; Krizova, L.; Cerny, J.; Berlec, A.; Maly, P. p19-Targeting ILP Protein Blockers of IL-23/Th-17 Pro-Inflammatory Axis Displayed on Engineered Bacteria of Food Origin. Int. J. Mol. Sci. 2018, 19, 1933. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Nguyen, G.H.; Bowman, E.D.; Mathe, E.A.; Yuen, S.T.; Hawkes, J.E.; Croce, C.M.; Leung, S.Y.; Harris, C.C. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin. Cancer Res. 2009, 15, 5878–5887. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yi, T.; Zhang, W.; Pardoll, D.M.; Yu, H. IL-17 enhances tumor development in carcinogen-induced skin cancer. Cancer Res. 2010, 70, 10112–10120. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Li, H.; Yusuf, N.; Elmets, C.A.; Athar, M.; Katiyar, S.K.; Xu, H. IL-17 mediated inflammation promotes tumor growth and progression in the skin. PLoS ONE 2012, 7, e32126. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.U.; Frick, I.M.; Nilsson, H.; Kraulis, P.J.; Hober, S.; Jonasson, P.; Linhult, M.; Nygren, P.A.; Uhlen, M.; Bjorck, L.; et al. Structure, specificity, and mode of interaction for bacterial albumin-binding modules. J. Biol. Chem. 2002, 277, 8114–8120. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Brenke, R.; Comeau, S.R.; Vajda, S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins 2006, 65, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Eastman, P.; Pande, V.S. OpenMM: A Hardware Independent Framework for Molecular Simulations. Comput. Sci. Eng. 2015, 12, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]

| Protein | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABDwt | Y | Y | K | N | L | I | N | N | A | K | T | V | E | G | V | K | A | L | I | D | E | I | L | A | A | L | P |
| ARS004 | V | Y | K | - | L | I | N | Y | A | C | P | V | T | W | V | K | W | V | I | D | P | I | L | A | M | L | P |
| ARS012 | V | Y | K | - | L | I | N | M | A | L | Y | V | T | G | V | K | P | W | I | D | V | I | L | A | V | L | P |
| ARS014 | V | Y | K | N | L | I | N | Y | A | W | L | V | M | W | V | - | - | P | I | D | A | I | L | A | L | L | P |
| ARS019 | M | Y | K | N | V | I | N | I | A | W | W | V | S | I | V | K | Y | P | I | D | C | I | L | A | L | L | P |
| ARS043 | L | Y | K | N | M | I | N | M | A | L | W | V | T | G | V | K | W | L | I | D | P | I | L | A | T | L | P |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hlavničková, M.; Kuchař, M.; Osička, R.; Vaňková, L.; Petroková, H.; Malý, M.; Černý, J.; Arenberger, P.; Malý, P. ABD-Derived Protein Blockers of Human IL-17 Receptor A as Non-IgG Alternatives for Modulation of IL-17-Dependent Pro-Inflammatory Axis. Int. J. Mol. Sci. 2018, 19, 3089. https://doi.org/10.3390/ijms19103089
Hlavničková M, Kuchař M, Osička R, Vaňková L, Petroková H, Malý M, Černý J, Arenberger P, Malý P. ABD-Derived Protein Blockers of Human IL-17 Receptor A as Non-IgG Alternatives for Modulation of IL-17-Dependent Pro-Inflammatory Axis. International Journal of Molecular Sciences. 2018; 19(10):3089. https://doi.org/10.3390/ijms19103089
Chicago/Turabian StyleHlavničková, Marie, Milan Kuchař, Radim Osička, Lucie Vaňková, Hana Petroková, Michal Malý, Jiří Černý, Petr Arenberger, and Petr Malý. 2018. "ABD-Derived Protein Blockers of Human IL-17 Receptor A as Non-IgG Alternatives for Modulation of IL-17-Dependent Pro-Inflammatory Axis" International Journal of Molecular Sciences 19, no. 10: 3089. https://doi.org/10.3390/ijms19103089
APA StyleHlavničková, M., Kuchař, M., Osička, R., Vaňková, L., Petroková, H., Malý, M., Černý, J., Arenberger, P., & Malý, P. (2018). ABD-Derived Protein Blockers of Human IL-17 Receptor A as Non-IgG Alternatives for Modulation of IL-17-Dependent Pro-Inflammatory Axis. International Journal of Molecular Sciences, 19(10), 3089. https://doi.org/10.3390/ijms19103089