Considerations of AOX Functionality Revealed by Critical Motifs and Unique Domains
Abstract
1. Introduction
2. Alternative Oxidase (AOX) Regulation in Monocots
3. Probing AOX Function: Is Past Performance an Indicator of Future Failure?
4. Beyond the Diiron Center: Can Functionality Be Defined by the Hydrophobic Cavity?
5. Exploration of the Dimer Interface in Monocot AOX Isoforms
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bolser, D.M.; Kerhornou, A.; Walts, B.; Kersey, P. Triticeae Resources in Ensembl Plants. Plant Cell Physiol. 2015, 56, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Richter, T.E.; Ronald, P.C. The evolution of disease resistance genes. Plant Mol. Biol. 2000, 42, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Fu, Y.B.; Pavlicek, T.; Khalifa, S.; Tavasi, M.; Beiles, A. Evolution of wild cereals during 28 years of global warming in Israel. Proc. Natl. Acad. Sci. USA 2012, 109, 3412–3415. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.X.; Waugh, R.; Langridge, P.; Close, T.J.; Wise, R.P.; Graner, A.; Matsumoto, T.; Sato, K.; Schulman, A.; Muehlbauer, G.J.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Gurel, F.; Ozturk, Z.N.; Ucarli, C.; Rosellini, D. Barley Genes as Tools to Confer Abiotic Stress Tolerance in Crops. Front. Plant Sci. 2016, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Garcia-Pereira, M.J.; Gracia, M.P.; Igartua, E.; Casas, A.M.; Contreras-Moreira, B. Large Differences in Gene Expression Responses to Drought and Heat Stress between Elite Barley Cultivar Scarlett and a Spanish Landrace. Front. Plant Sci. 2017, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Parida, S.K.; Raghuvanshi, S.; Kapoor, S.; Khurana, P.; Khurana, J.P.; Tyagi, A.K. Rice Improvement through Genome-Based Functional Analysis and Molecular Breeding in India. Rice 2016, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Wang, J.; Zeigler, R.S. The 3000 rice genomes project: New opportunities and challenges for future rice research. Gigascience 2014, 3, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Ahmadikhah, A.; Marufinia, A. Effect of reduced plant height on drought tolerance in rice. 3 Biotech 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.H.; McDonald, A.E.; Arnholdt-Schmitt, B.; Fernandes de Melo, D. A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion 2014, 19 (Pt B), 172–183. [Google Scholar] [CrossRef]
- Saha, B.; Borovskii, G.; Panda, S.K. Alternative oxidase and plant stress tolerance. Plant Signal. Behav. 2016, 11, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Brew-Appiah, R.A.T.; York, Z.B.; Krishnan, V.; Roalson, E.H.; Sanguinet, K.A. Genome-wide identification and analysis of the ALTERNATIVE OXIDASE gene family in diploid and hexaploid wheat. PLoS ONE 2018, 13, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Abu-Romman, S.; Shatnawi, M.; Hasan, M.; Qrunfleh, I.; Omar, S.; Salem, N. cDNA cloning and expression analysis of a putative alternative oxidase HsAOX1 from wild barley (Hordeum spontaneum). Genes Genomics 2012, 34, 59–66. [Google Scholar] [CrossRef]
- Wanniarachchi, V.R.; Dametto, L.; Sweetman, C.; Shavrukov, Y.; Day, D.A.; Jenkins, C.L.D.; Soole, K.L. Alternative Respiratory Pathway Component Genes (AOX and ND) in Rice and Barley and Their Response to Stress. Int. J. Mol. Sci. 2018, 19, 915. [Google Scholar] [CrossRef] [PubMed]
- Selinski, J.; Scheibe, R.; Day, D.A.; Whelan, J. Alternative Oxidase Is Positive for Plant Performance. Trends Plant Sci. 2018, 23, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Van Aken, O.; Elsässer, M.; Schwarzländer, M. Mitochondrial Energy Signaling and Its Role in the Low Oxygen Stress Response of Plants. Plant Physiol. 2018, 176, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, I.; Vermeirssen, V.; Van Aken, O.; Vandepoele, K.; Murcha, M.W.; Law, S.R.; Inze, A.; Ng, S.; Ivanova, A.; Rombaut, D.; et al. The Membrane-Bound NAC Transcription Factor ANAC013 Functions in Mitochondrial Retrograde Regulation of the Oxidative Stress Response in Arabidopsis. Plant Cell 2013, 25, 3472–3490. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.N.; Zheng, H.Q.; Wu, N.Y.; Chien, C.H.; Huang, H.D.; Lee, T.Y.; Chiang-Hsieh, Y.F.; Hou, P.F.; Yang, T.Y.; Chang, W.C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef] [PubMed]
- Butow, R.A.; Avadhani, N.G. Mitochondrial signaling: The Retrograde Response. Mol. Cell 2004, 14, 1–15. [Google Scholar] [CrossRef]
- Yurina, N.P.; Odintsova, M.S. Signal Transduction Pathways of Plant Mitochondria: Retrograde Regulation. Russ. J. Plant Physiol. 2010, 57, 7–19. [Google Scholar] [CrossRef]
- Crawford, T.; Lehotai, N.; Strand, A. The role of retrograde signals during plant stress responses. J. Exp. Bot. 2018, 69, 2783–2795. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; De Clercq, I.; Van Aken, O.; Law, S.R.; Ivanova, A.; Willems, P.; Giraud, E.; Van Breusegem, F.; Whelan, J. Anterograde and Retrograde Regulation of Nuclear Genes Encoding Mitochondrial Proteins during Growth, Development, and Stress. Mol. Plant 2014, 7, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.L.; Shiba, T.; Young, L.; Harada, S.; Kita, K.; Ito, K. Unraveling the Heater: New Insights into the Structure of the Alternative Oxidase. Annu. Rev. Plant Biol. 2013, 64, 637–663. [Google Scholar] [CrossRef] [PubMed]
- May, B.; Young, L.; Moore, A.L. Structural insights into the alternative oxidases: Are all oxidases made equal? Biochem. Soc. Trans. 2017, 45, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Nobre, T.; Campos, M.D.; Lucic-Mercy, E.; Arnholdt-Schmitt, B. Misannotation Awareness: A Tale of Two Gene-Groups. Front. Plant Sci. 2016, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shiba, T.; Kido, Y.; Sakamoto, K.; Inaoka, D.K.; Tsuge, C.; Tatsumi, R.; Takahashi, G.; Balogun, E.O.; Nara, T.; Aoki, T.; et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc. Natl. Acad. Sci. USA 2013, 110, 4580–4585. [Google Scholar] [CrossRef] [PubMed]
- Young, L.; May, B.; Pendlebury-Watt, A.; Shearman, J.; Elliott, C.; Albury, M.S.; Shiba, T.; Inaoka, D.K.; Harada, S.; Kita, K.; et al. Probing the ubiquinol-binding site of recombinant Sauromatum guttatum alternative oxidase expressed in E. coli membranes through site-directed mutagenesis. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Crichton, P.G.; Albury, M.S.; Affourtit, C.; Moore, A.L. Mutagenesis of the Sauromatum guttatum alternative oxidase reveals features important for oxygen binding and catalysis. Biochim. Biophys. Acta Bioenerg. 2010, 1797, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Selinski, J.; Hartmann, A.; Kordes, A.; Deckers-Hebestreit, G.; Whelan, J.; Scheibe, R. Analysis of Posttranslational Activation of Alternative Oxidase Isoforms. Plant Physiol. 2017, 174, 2113–2127. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Finkemeier, I. Plant mitochondrial retrograde signaling: Post-translational modifications enter the stage. Front. Plant Sci. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dawson, N.L.; Orengo, C.A. Diversity in protein domain superfamilies. Curr. Opin. Genet. Dev. 2015, 35, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.; Browse, J. Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 2011, 66, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Waschburger, E.; Kulcheski, F.R.; Veto, N.M.; Margis, R.; Margis-Pinheiro, M.; Turchetto-Zolet, A.C. Genome-wide analysis of the Glycerol-3-Phosphate Acyltransferase (GPAT) gene family reveals the evolution and diversification of plant GPATs. Genet. Mol. Biol. 2018, 41, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Schwerdt, J.G.; MacKenzie, K.; Wright, F.; Oehme, D.; Wagner, J.M.; Harvey, A.J.; Shirley, N.J.; Burton, R.A.; Schreiber, M.; Halpin, C.; et al. Evolutionary Dynamics of the Cellulose Synthase Gene Superfamily in Grasses. Plant Physiol. 2015, 168, 968–983. [Google Scholar] [CrossRef] [PubMed]
- Little, A.; Schwerdt, J.G.; Shirley, N.J.; Khor, S.F.; Neumann, K.; O’Donovan, L.A.; Lahnstein, J.; Collins, H.M.; Henderson, M.; Fincher, G.B.; et al. Revised Phylogeny of the Cellulose Synthase Gene Superfamily: Insights into Cell Wall Evolution. Plant Physiol. 2018, 177, 1124–1141. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, R.; Salvi, D.; Brandi, V.; Angelini, R.; Ascenzi, P.; Polticelli, F. Molecular Evolution of Alternative Oxidase Proteins: A Phylogenetic and Structure Modeling Approach. J. Mol. Evol. 2016, 82, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Arpino, J.A.J.; Reddington, S.C.; Halliwell, L.M.; Rizkallah, P.J.; Jones, D.D. Random Single Amino Acid Deletion Sampling Unveils Structural Tolerance and the Benefits of Helical Registry Shift on GFP Folding and Structure. Structure 2014, 22, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.S.; Wei, X.; Dong, X.; Xu, L.; Liu, J.; Jiang, B. Structural plasticity of green fluorescent protein to amino acid deletions and fluorescence rescue by folding-enhancing mutations. BMC Biochem. 2015, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Toth-Petroczy, A.; Tawfik, D.S. Hopeful (Protein InDel) Monsters? Structure 2014, 22, 803–804. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, L.V.; Mokshina, N.E.; Nazipova, A.R.; Gorshkova, T.A. Systemic Use of “Limping” Enzymes in Plant Cell Walls. Russ. J. Plant Physiol. 2017, 64, 808–821. [Google Scholar] [CrossRef]
- Routledge, S.J.; Mikaliunaite, L.; Patel, A.; Clare, M.; Cartwright, S.P.; Bawa, Z.; Wilks, M.D.B.; Low, F.; Hardy, D.; Rothnie, A.J.; et al. The synthesis of recombinant membrane proteins in yeast for structural studies. Methods 2016, 95, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Umbach, A.L.; Ng, V.S.; Siedow, J.N. Regulation of plant alternative oxidase activity: A tale of two cysteines. Biochim. Biophys. Acta Bioenerg. 2006, 1757, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.P.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. Techniques for the Analysis of Protein-Protein Interactions in Vivo. Plant Physiol. 2016, 171, 727–758. [Google Scholar] [CrossRef] [PubMed]
- Bontinck, M.; Van Leene, J.; Gadeyne, A.; De Rybel, B.; Eeckhout, D.; Nelissen, H.; De Jaeger, G. Recent Trends in Plant Protein Complex Analysis in a Developmental Context. Front. Plant Sci. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Y.; Remmelzwaal, S.; Heck, A.J.R.; Akhmanova, A.; Liu, F. Facilitating identification of minimal protein binding domains by cross-linking mass spectrometry. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Seymour, R.S. Expression of uncoupling protein and alternative oxidase depends on lipid or carbohydrate substrates in thermogenic plants. Biol. Lett. 2005, 1, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Karpova, O.V.; Kuzmin, E.V.; Elthon, T.E.; Newton, K.J. Differential Expression of Alternative Oxidase Genes in Maize Mitochondrial Mutants. Plant Cell 2002, 14, 3271–3284. [Google Scholar] [CrossRef] [PubMed]
- Polidoros, A.N.; Mylona, P.V.; Pasentsis, K.; Scandalios, J.G.; Tsaftaris, A.S. The maize alternative oxidase 1a (Aox1a) gene is regulated by signals related to oxidative stress. Redox Rep. 2005, 10, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Camacho, A.; Moreno-Sanchez, R.; Bernal-Lugo, I. Control of superoxide production in mitochondria from maize mesocotyls. FEBS Lett. 2004, 570, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Erdal, S.; Genisel, M. The property of progesterone to mitigate cold stress in maize is linked to a modulation of the mitochondrial respiratory pathway. Theor. Exp. Plant Physiol. 2016, 28, 385–393. [Google Scholar] [CrossRef]
- Silva-Neta, I.C.; Pinho, E.V.; Veiga, A.D.; Pinho, R.G.; Guimaraes, R.M.; Caixeta, F.; Santos, H.O.; Marques, T.L. Expression of genes related to tolerance to low temperature for maize seed germination. Genet. Mol Res. 2015, 14, 2674–2690. [Google Scholar] [CrossRef] [PubMed]
- Dutra, S.M.F.; Von Pinho, E.V.R.; Santos, H.O.; Lima, A.C.; Von Pinho, R.G.; Carvalho, M.L.M. Genes related to high temperature tolerance during maize seed germination. Genet. Mol Res. 2015, 14, 18047–18058. [Google Scholar] [CrossRef] [PubMed]
- Dahal, K.; Vanlerberghe, G.C. Improved chloroplast energy balance during water deficit enhances plant growth: More crop per drop. J. Exp. Bot. 2018, 69, 1183–1197. [Google Scholar] [CrossRef] [PubMed]
- Allwright, M.R.; Taylors, G. Molecular Breeding for Improved Second Generation Bioenergy Crops. Trends Plant. Sci. 2016, 21, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Udvardi, M. Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. J. Exp. Bot. 2018, 69, 855–865. [Google Scholar] [CrossRef] [PubMed]
| HvAOX1a | HvAOX1c | HvAOX1d1 | HvAOX1d2 | OsAOX1a | OsAOX1c | OsAOX1e | OsAOX1d | |
|---|---|---|---|---|---|---|---|---|
| ANAC013 | 3 | 1 | - | - | 3 | - | - | - |
| ANAC017 | 2 | 1 | - | - | 2 | - | - | - |
| AtWRKY63 | - | - | - | 1 | - | - | - | - |
| ANAC053 | 2 | - | - | - | 2 | - | - | - |
| ANAC078 | 1 | - | - | - | - | - | 1 | - |
| ABI4 | 3 | - | 4 | 3 | - | - | - | - |
| CTTGNNNNNCAMG | 2 | 2 | - | - | 2 | - | - | - |
| YTTGNNNNNVAMV | 4 | 2 | 1 | 2 | 6 | 2 | 1 | 2 |
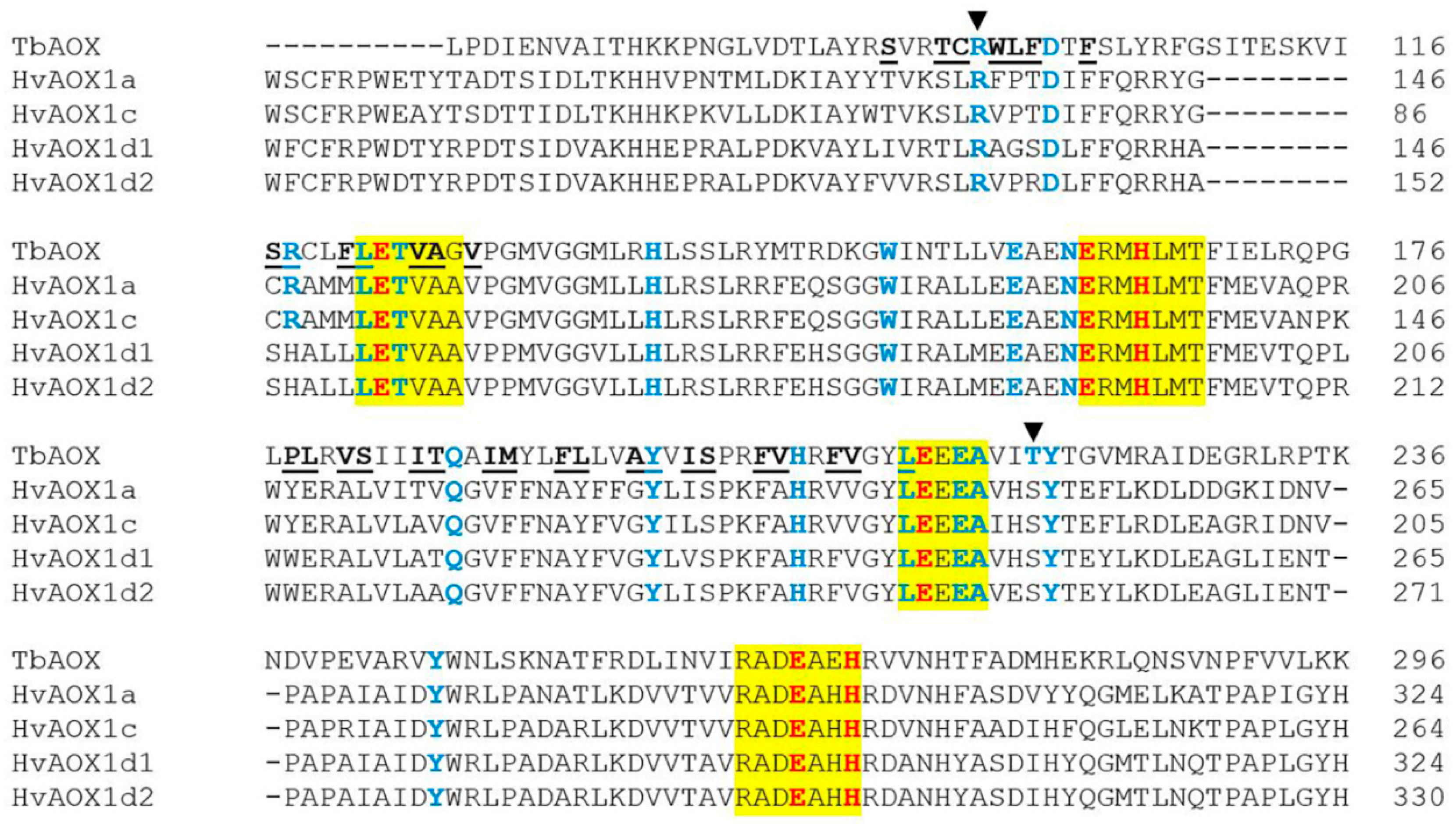
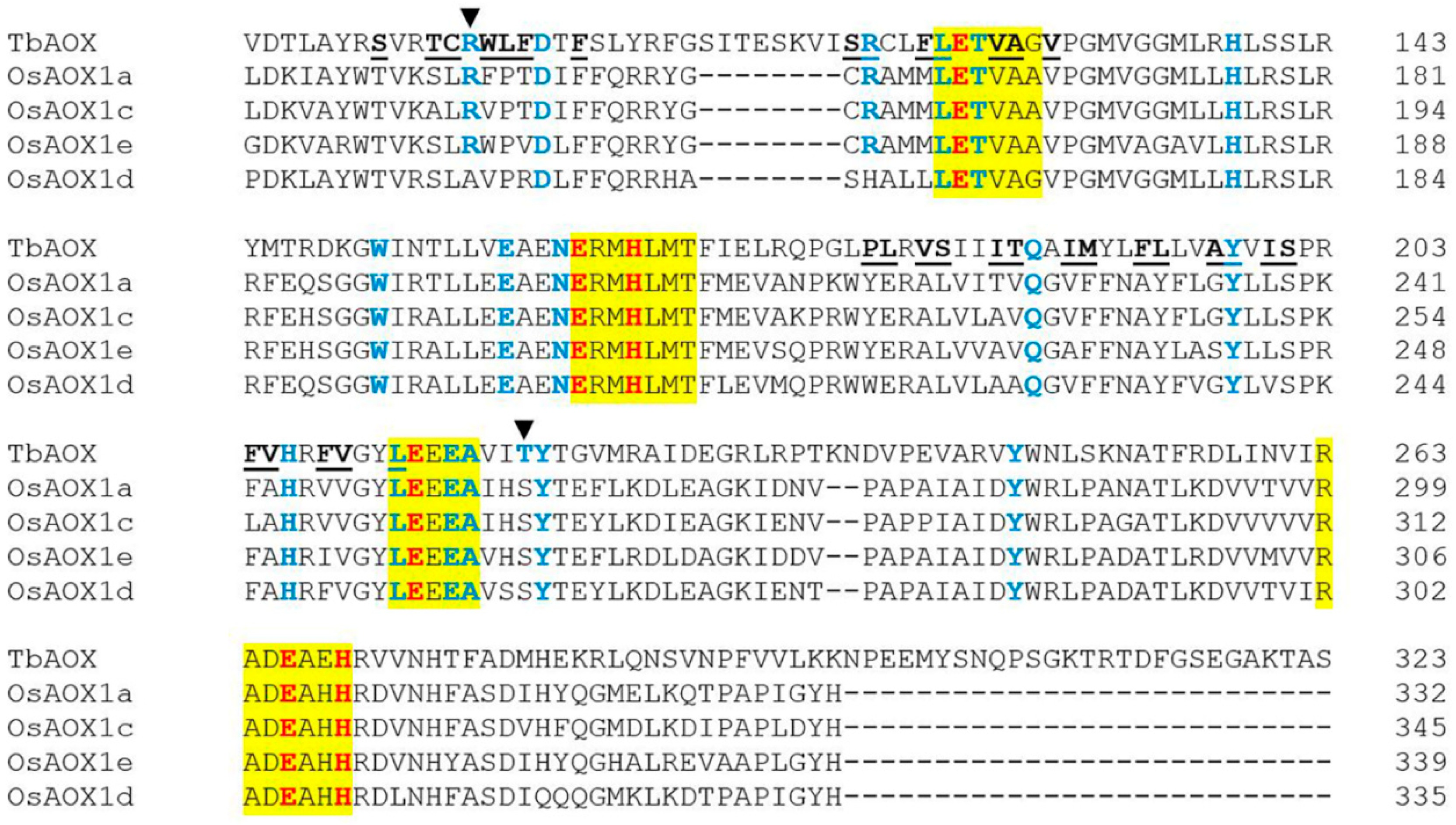
| AOX Isoforms | TbAOX Residues and Positions in the Hydrophobic Cavity | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | T | C | W | L | F | S | R | F | P | L | V | S | I | T | I | M | F | L | A | I | F | V | F | ||
| 91 | 94 | 95 | 97 | 98 | 99 | 117 | 118 | 121 | 178 | 179 | 181 | 182 | 185 | 186 | 189 | 190 | 193 | 194 | 197 | 200 | 204 | 205 | 208 | ||
| HvAOX1a | T | S | L | F | P | T | C | R | M | Y | E | A | L | T | V | V | F | A | Y | G | I | F | A | V | |
| TaAOX1a-2AL | T | S | L | F | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | I | F | A | V | |
| TaAOX1a-2BL | T | S | L | F | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | I | F | A | V | |
| TaAOX1a-2DL | T | S | L | F | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | I | F | A | V | |
| TaAOX1a-like-2DL | - | - | - | - | - | - | - | - | - | Y | E | A | L | A | V | V | F | A | Y | G | V | F | A | V | |
| TuAOX1a * | T | S | L | F | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | I | F | A | V | |
| AetAOX1a * | T | S | L | F | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | I | F | A | V | |
| OsAOX1a | T | S | L | F | P | T | C | R | M | Y | E | A | L | T | V | V | F | A | Y | G | L | F | A | V | |
| HvAOX1c | T | S | L | V | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | L | F | A | V | |
| TaAOX1c-6AL | T | S | L | V | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | I | F | A | V | |
| TaAOX1c-6BL | T | S | L | V | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | V | F | A | V | |
| TaAOX1c-6DL | T | S | L | V | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | V | F | A | V | |
| TuAOX1c * | T | S | L | V | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | V | F | A | V | |
| OsAOX1c | T | A | L | V | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | G | L | L | A | V | |
| Put.TaAOX1e-3DS | T | A | I | W | P | T | C | R | M | Y | E | A | L | V | V | V | F | A | Y | T | A | V | A | M | |
| AetAOX1e * | T | A | M | W | P | T | C | R | M | Y | E | A | L | A | V | V | F | A | Y | T | A | V | A | M | |
| OsAOX1e | T | S | L | W | P | V | C | R | M | Y | E | A | L | A | V | A | F | A | Y | S | L | F | A | I | |
| OsAOX1d | T | S | L | V | P | R | S | H | L | W | E | A | L | A | A | V | F | A | Y | G | V | F | A | F | |
| dG1 | HvAOX1d1 | I | T | L | A | G | S | S | H | L | W | E | A | L | A | T | V | F | A | Y | G | V | F | A | F |
| TaAOX1d-2AL.1 | I | T | L | A | G | S | S | H | L | W | E | A | L | A | T | V | F | A | Y | G | V | F | A | F | |
| put.TaAOX1d-like-4AS | - | - | - | - | - | - | S | H | L | C | E | A | L | P | T | V | F | A | Y | G | V | F | A | F | |
| TuAOX1d.2 * | I | T | L | K | G | S | S | H | L | W | E | A | L | A | T | V | F | A | Y | G | V | F | A | F | |
| AetAOX1d-like * | I | T | L | A | G | S | S | H | L | - | - | - | - | - | - | V | F | A | Y | G | I | L | - | - | |
| dG2 | HvAOX1d2 | V | S | L | V | P | R | S | H | L | W | E | A | L | A | A | V | F | A | Y | G | I | F | A | F |
| TaAOX1d-2AL.2 | V | S | L | V | P | R | S | H | L | W | E | A | L | A | A | V | F | A | Y | G | I | F | A | F | |
| TaAOX1d-2DL | V | S | L | V | P | R | S | H | L | W | E | A | L | A | A | V | F | A | Y | G | I | F | A | F | |
| TuAOX1d.1 * | V | S | L | V | P | R | S | H | L | W | E | A | L | A | A | V | F | A | Y | G | I | F | A | F | |
| AetAOX1d * | V | S | L | V | P | R | S | H | L | W | E | A | L | A | A | V | F | A | Y | G | I | F | A | F | |
| AOX Isoforms | Completely Conserved with TbAOX | Semi-Conserved with TbAOX | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H138 | L142 | R143 | R163 | L166 | Q187 | M131 | M135 | L139 | S141 | M145 | R147 | D148 | L156 | A159 | M167 | R180 | I183 | ||
| HvAOX1a | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TaAOX1a-2AL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TaAOX1a-2BL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TaAOX1a-2DL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TaAOX1a-like-2DL | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V | |
| TuAOX1a * | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| AetAOX1a * | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| OsAOX1a | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| HvAOX1c | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TaAOX1c-6AL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TaAOX1c-6BL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TaAOX1c-6DL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| TuAOX1c * | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| OsAOX1c | H | L | R | R | L | Q | M | M | L | S | F | H | S | L | A | M | R | V | |
| put.TaAOX1e-3DS | H | L | R | R | L | Q | M | A | L | S | F | Q | S | L | A | M | R | V | |
| AetAOX1e * | H | L | R | R | L | Q | M | A | L | S | F | Q | S | L | A | M | R | V | |
| OsAOX1e | H | L | R | R | L | Q | M | A | L | S | F | H | S | L | A | M | R | V | |
| OsAOX1d | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V | |
| dG1 | HvAOX1d1 | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| TaAOX1d-2AL.1 | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V | |
| put.TaAOX1d-like-4AS | H | L | R | R | L | Q | M | V | L | S | F | H | N | M | A | M | R | V | |
| TuAOX1d.2 * | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V | |
| AetAOX1d-like * | H | L | R | R | L | - | M | V | L | S | F | H | S | M | A | M | - | - | |
| dG2 | HvAOX1d2 | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| TaAOX1d-2AL.2 | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V | |
| TaAOX1d-2DL | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V | |
| TuAOX1d.1 * | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V | |
| AetAOX1d * | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V | |
| AOX Isoforms | Critical Cysteines | Putative Dimer Status In Vivo | |||
|---|---|---|---|---|---|
| CysI | CysII | CysIII | |||
| HvAOX1a | C | C | L | Inactive | |
| TaAOX1a-2AL | C | C | L | Inactive | |
| TaAOX1a-2BL | C | C | L | Inactive | |
| TaAOX1a-2DL | C | C | L | Inactive | |
| TaAOX1a-like-2DL | - | - | L | Active | |
| TuAOX1a * | - | C | L | Active | |
| AetAOX1a * | - | C | L | Active | |
| OsAOX1a | C | C | L | Inactive | |
| HvAOX1c | C | C | L | Inactive | |
| TaAOX1c-6AL | C | C | L | Inactive | |
| TaAOX1c-6BL | C | C | L | Inactive | |
| TaAOX1c-6DL | C | C | L | Inactive | |
| TuAOX1c * | E | C | L | Active | |
| OsAOX1c | C | C | L | Inactive | |
| put.TaAOX1e-3DS | C | C | L | Inactive | |
| AetAOX1e * | C | C | L | Inactive | |
| OsAOX1e | C | C | L | Inactive | |
| OsAOX1d | S | S | L | Active | |
| dG1 | HvAOX1d1 | C | S | L | Active |
| TaAOX1d-2AL.1 | S | S | L | Active | |
| put.TaAOX1d-like-4AS | S | S | L | Active | |
| TuAOX1d.2 * | S | S | L | Active | |
| AetAOX1d-like * | S | S | L | Active | |
| dG2 | HvAOX1d2 | C | S | L | Active |
| TaAOX1d-2AL.2 | C | S | L | Active | |
| TaAOX1d-2DL | C | S | L | Active | |
| TuAOX1d.1 * | C | S | L | Active | |
| AetAOX1d * | C | S | L | Active | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brew-Appiah, R.A.T.; Sanguinet, K.A. Considerations of AOX Functionality Revealed by Critical Motifs and Unique Domains. Int. J. Mol. Sci. 2018, 19, 2972. https://doi.org/10.3390/ijms19102972
Brew-Appiah RAT, Sanguinet KA. Considerations of AOX Functionality Revealed by Critical Motifs and Unique Domains. International Journal of Molecular Sciences. 2018; 19(10):2972. https://doi.org/10.3390/ijms19102972
Chicago/Turabian StyleBrew-Appiah, Rhoda A. T., and Karen A. Sanguinet. 2018. "Considerations of AOX Functionality Revealed by Critical Motifs and Unique Domains" International Journal of Molecular Sciences 19, no. 10: 2972. https://doi.org/10.3390/ijms19102972
APA StyleBrew-Appiah, R. A. T., & Sanguinet, K. A. (2018). Considerations of AOX Functionality Revealed by Critical Motifs and Unique Domains. International Journal of Molecular Sciences, 19(10), 2972. https://doi.org/10.3390/ijms19102972





