Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339
Abstract
:1. Introduction
2. Results
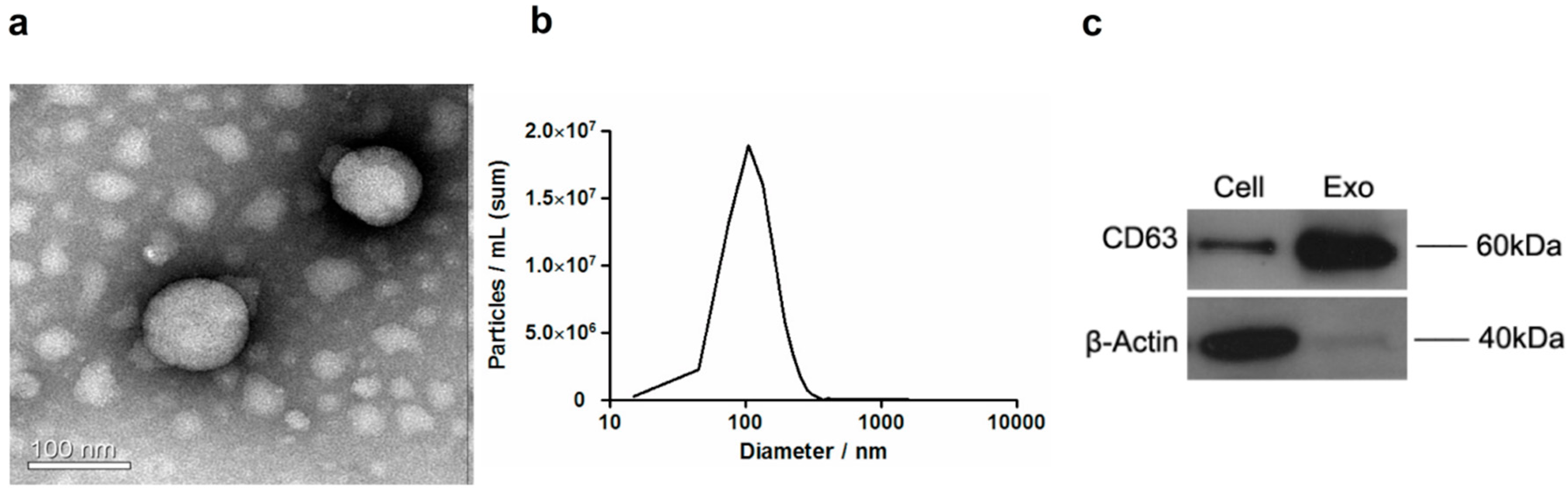
2.1. Extraction and Morphological Examination of Hepatocellular Carcinoma Cell (HCC)-Derived Exosomes
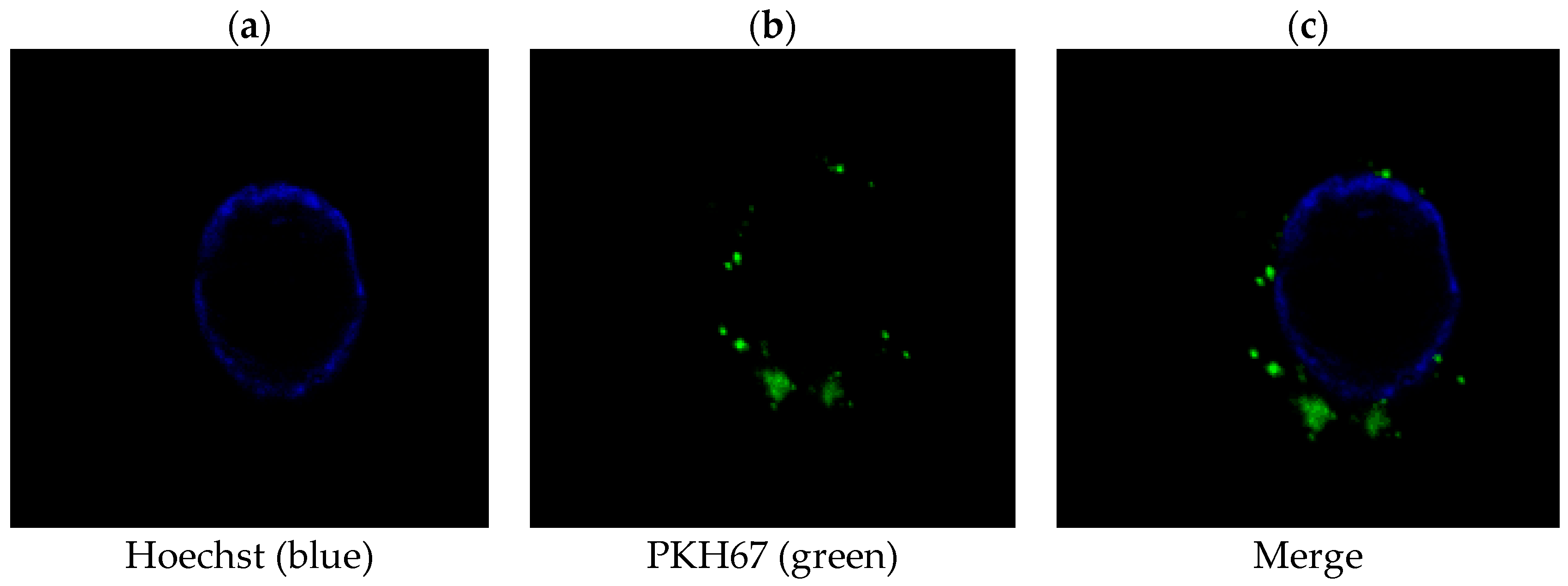
2.2. PLC/PRF/5-Derived Exosomes Can Be Internalized by THP-1 Cells
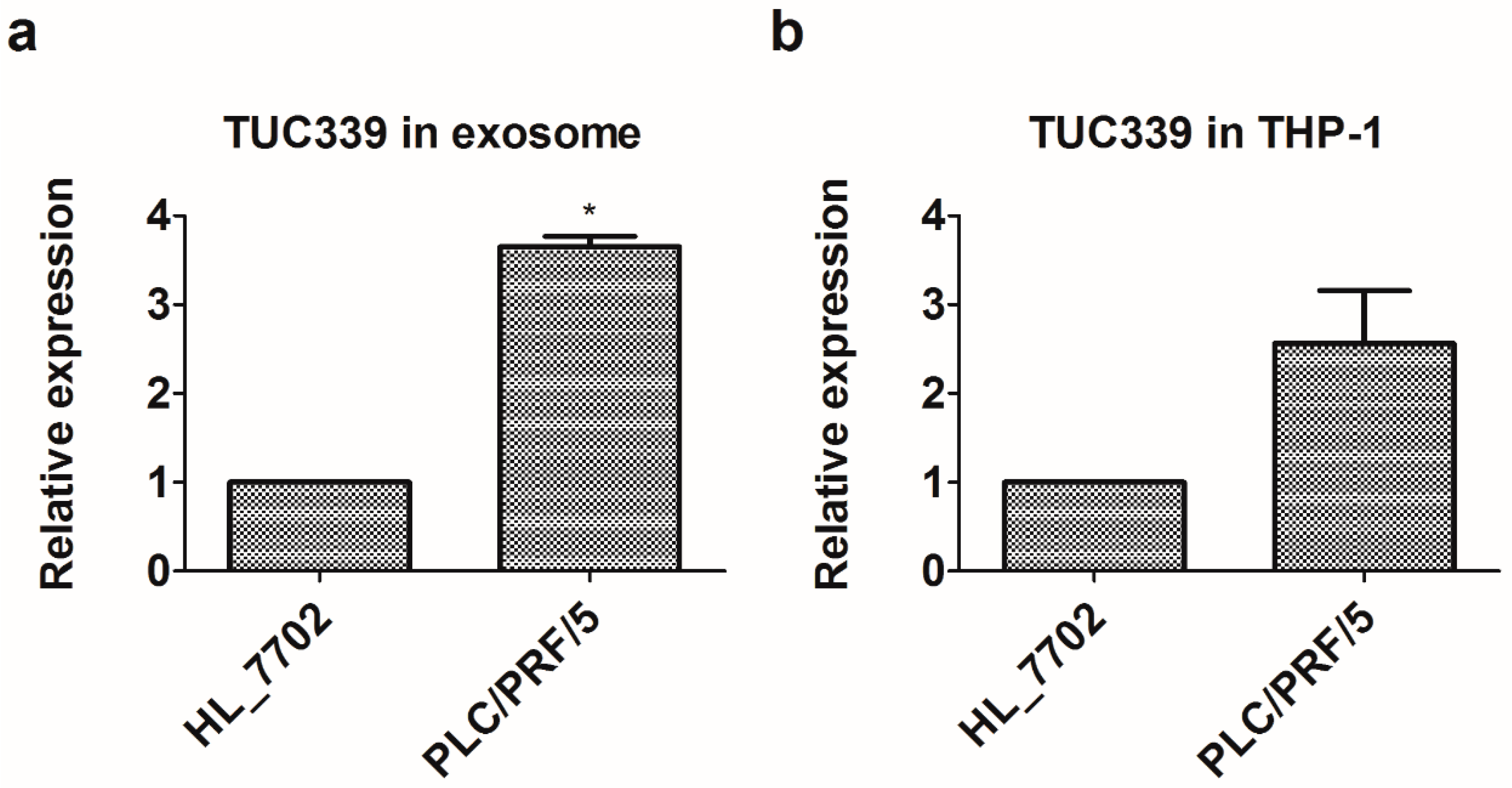
2.3. Long Non-Coding RNA TUC339 is Enriched in PLC/PRF/5-Derived Exosomes
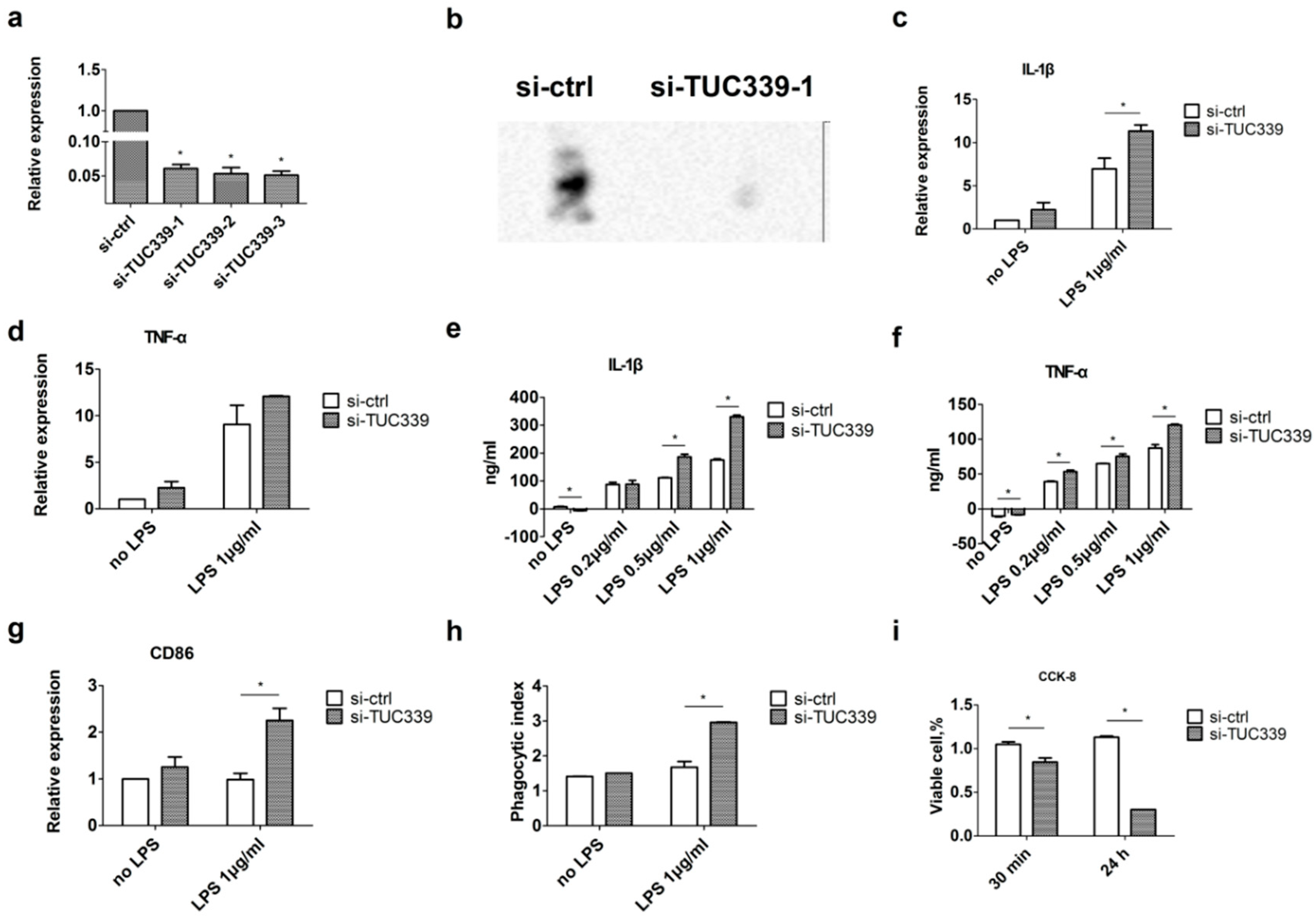
2.4. Knockdown of TUC339 in THP-1 Cells Leads to Increased Pro-Inflammatory Cytokine Production, Increased co-Stimulatory Molecule Expression, Enhanced Phagocytosis, and Reduced Viability
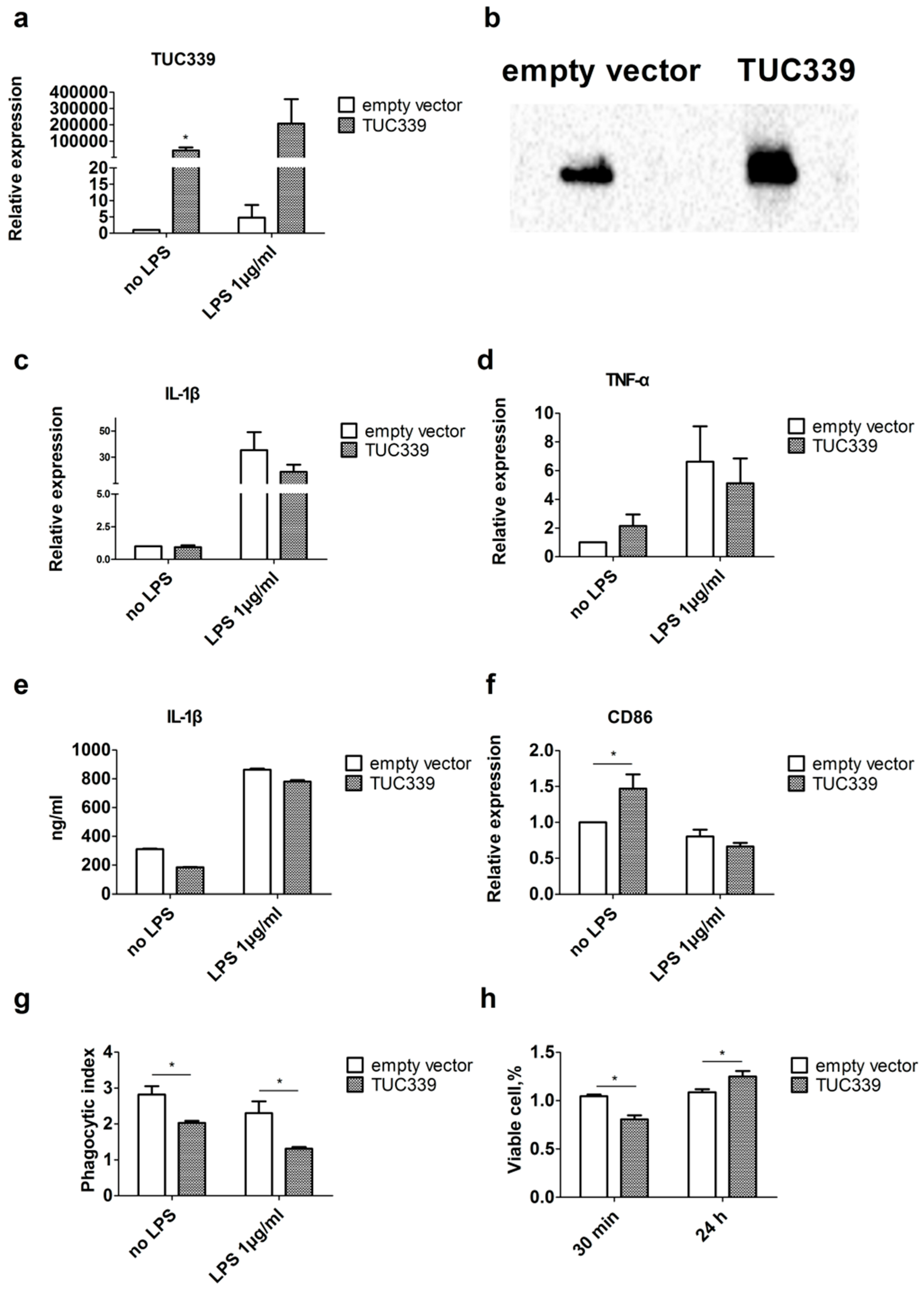
2.5. Over-Expression of TUC339 in THP-1 Cells Leads to Reduced Pro-Inflammatory Cytokine Production, Decreased co-Stimulatory Molecule Expression, and Compromised Phagocytosis
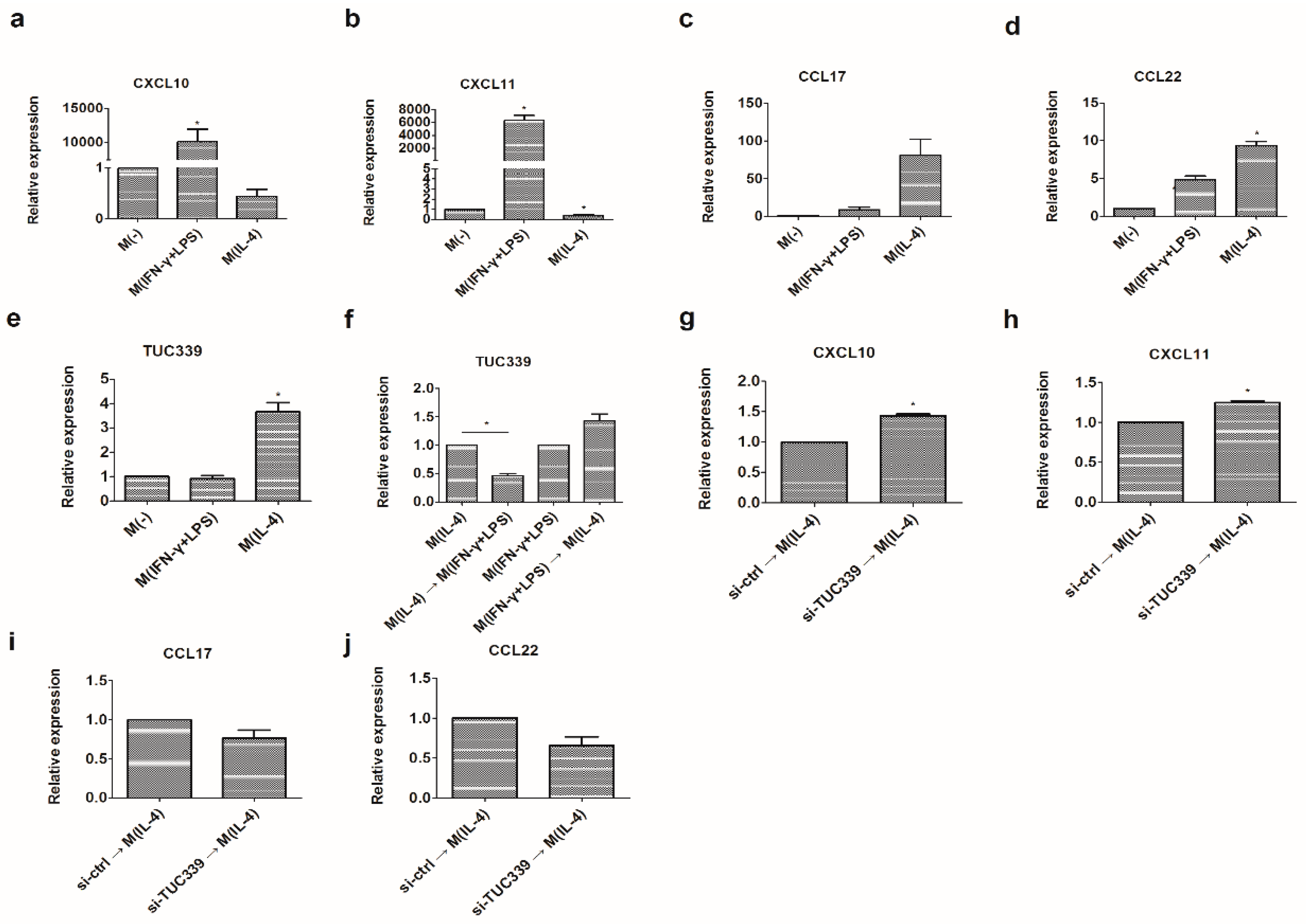
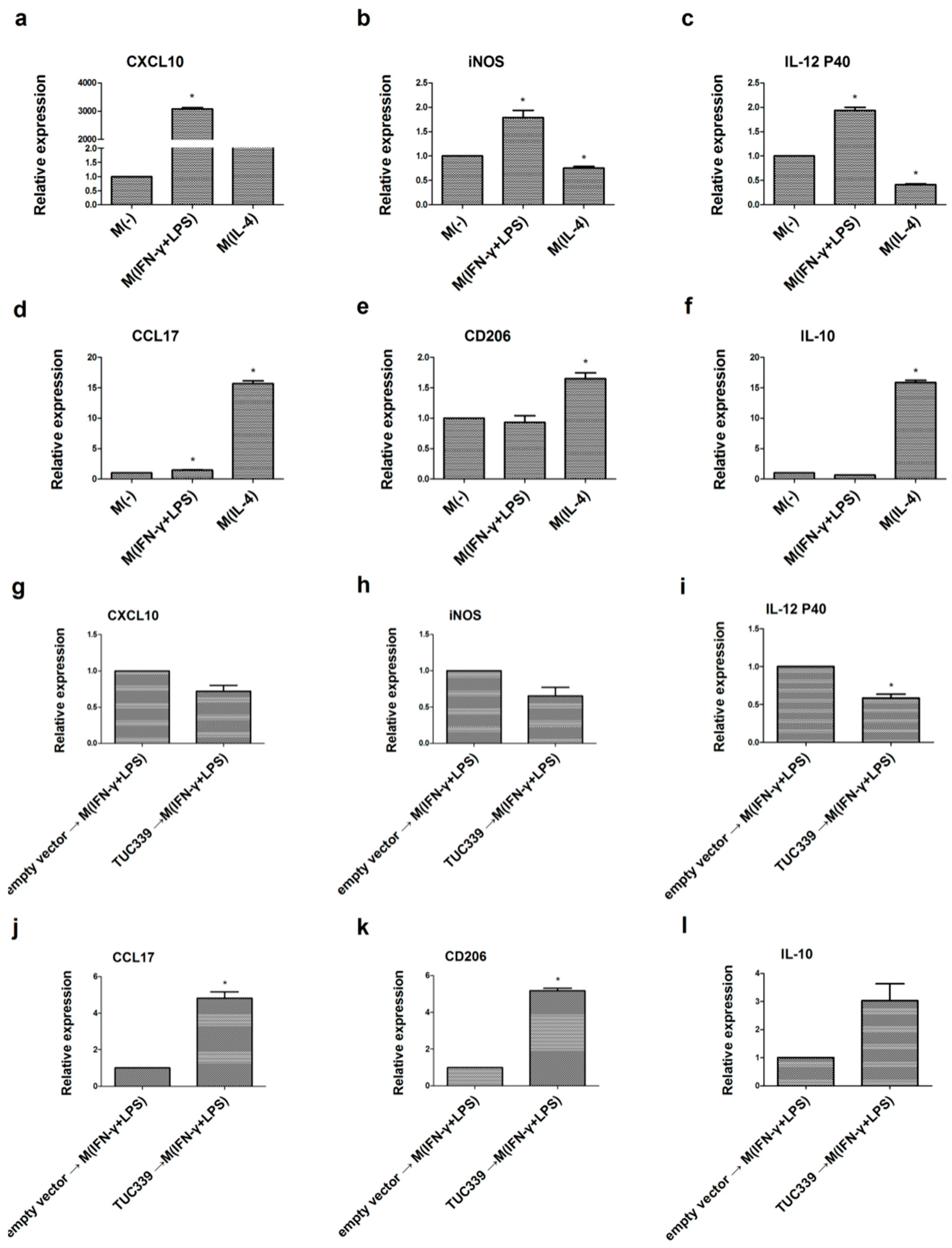
2.6. TUC339 is Required for M(IL-4) Macrophages Polarization
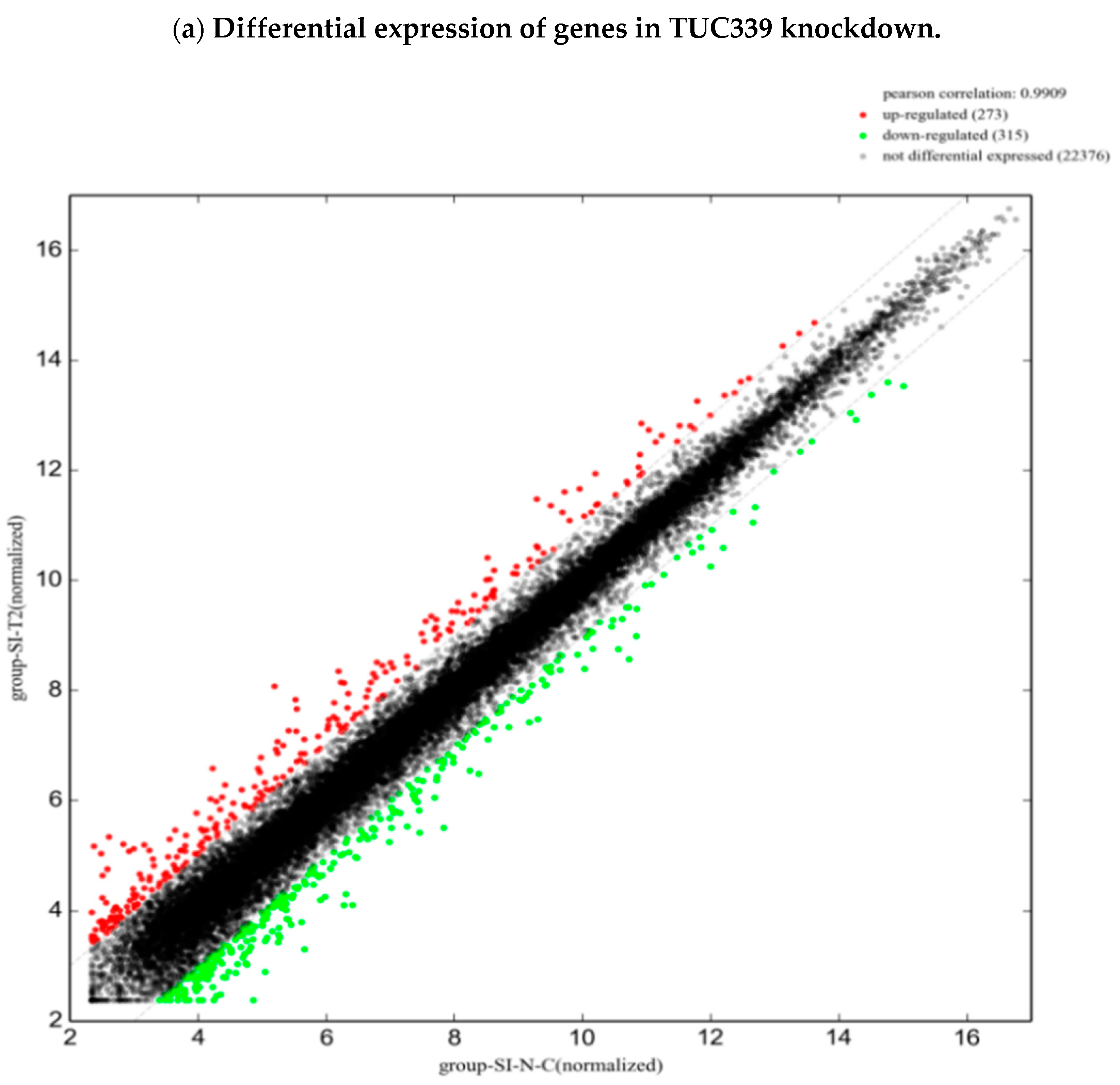
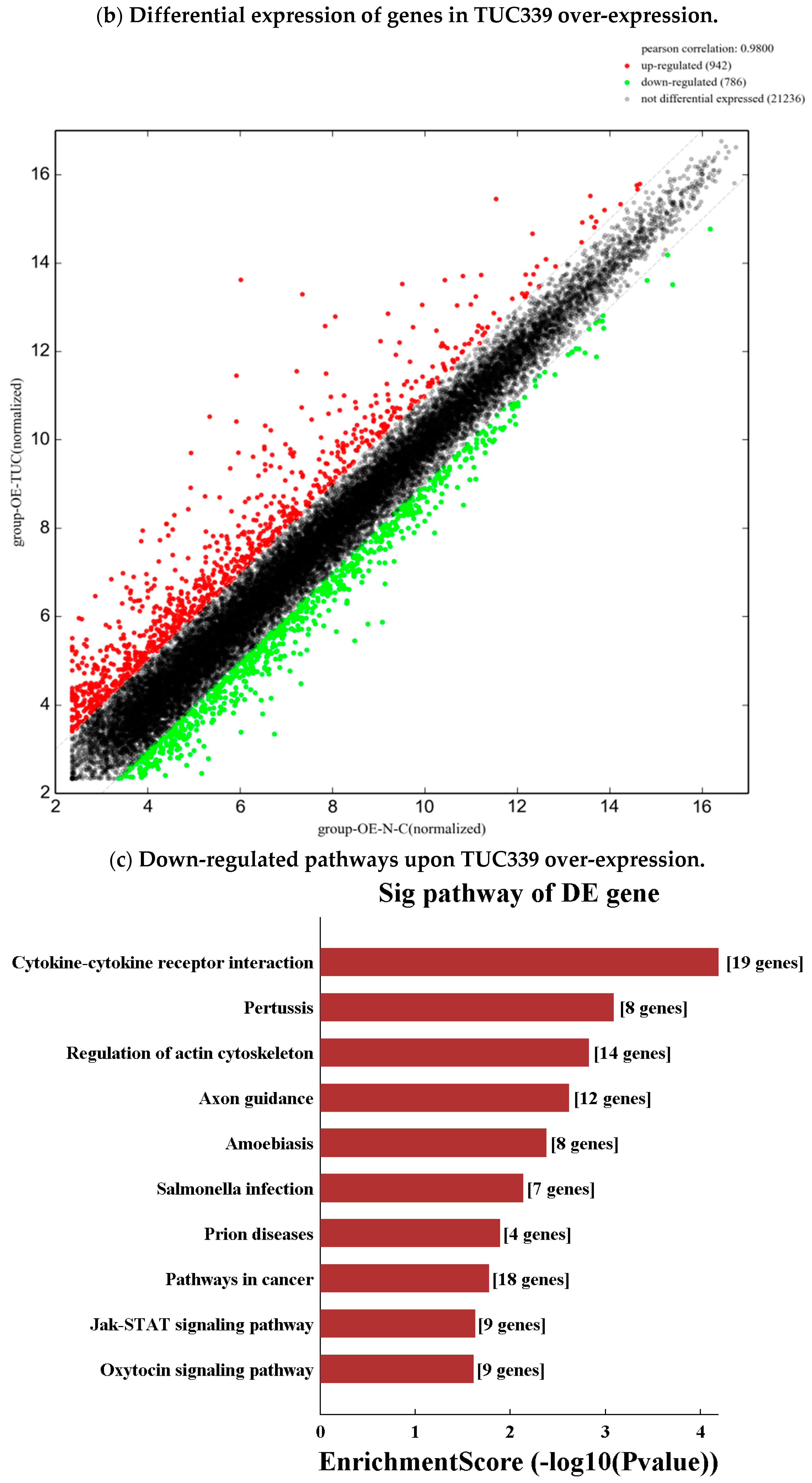
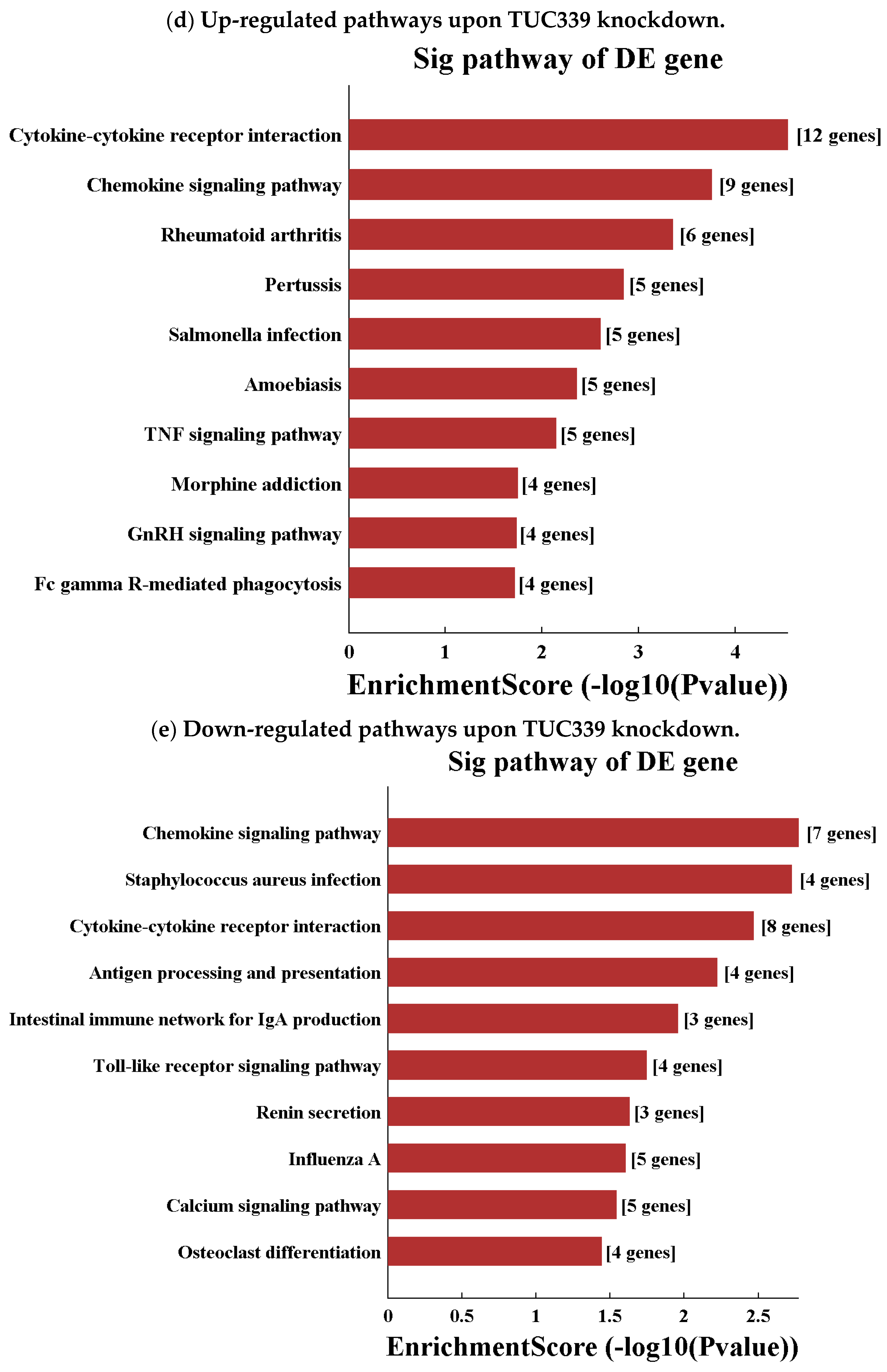
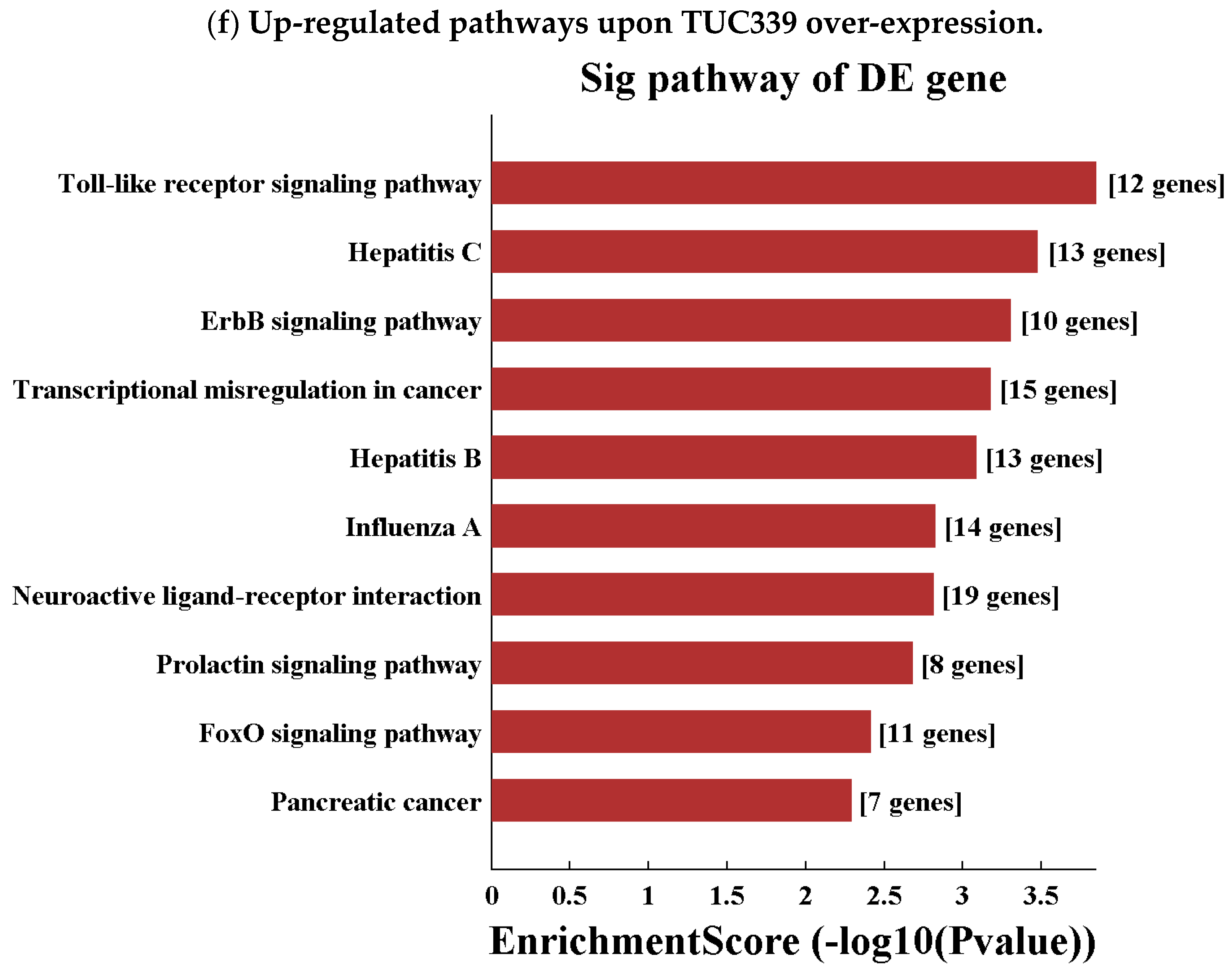
2.7. Transcriptome-Wide Analysis Reveals Pathways Downstream of TUC339
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Isolation of Exosomes Derived from PLC/PRF/5 Cells
4.3. Transmission Electron Microscopy
4.4. Nanoparticle Tracking Analysis
4.5. Western Blot
4.6. Cellular Internalization of PLC/PRF/5 Cells-Derived Exosomes
4.7. Macrophage-Differentiation and Polarization Conditions
4.8. Phagocytosis Assay
4.9. CCK-8 Assay
4.10. Real-Time PCR
4.11. Transfection of siRNA
4.12. ELISA
4.13. Northern Blot
4.14. Gene Over-Expression
4.15. Genome-Wide Gene Expression Analysis
4.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Bartolomei, M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013, 152, 1308–1323. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Meng, X.M.; Huang, C.; Wu, B.M.; Zhang, L.; Lv, X.W.; Li, J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014, 344, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Aune, T.M.; Spurlock, C.R. Long non-coding RNAs in innate and adaptive immunity. Virus Res. 2016, 212, 146–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.; O’Brien, K.; Kelleher, F.C.; Corcoran, C.; Germano, S.; Radomski, M.W.; Crown, J.; O’Driscoll, L. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods Mol. Biol. 2011, 784, 181–195. [Google Scholar] [PubMed]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.P.; et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Shenoda, B.B.; Ajit, S.K. Modulation of Immune Responses by Exosomes Derived from Antigen-Presenting Cells. Clin. Med. Insights Pathol. 2016, 9 (Suppl. 1), 1–8. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol. Cancer Res. 2014, 12, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Kogure, T.; Haga, H.; Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014, 4, 458–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, M.; Chen, W.; Xiang, A.; Wang, R.; Chen, H.; Pan, J.; Pang, H.; An, H.; Wang, X.; Hou, H.; et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol. Cancer 2017, 16, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Huang, C.; Lin, Z.; Zhan, S.; Kong, L.; Fang, C.; Li, J. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal 2014, 26, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013, 34, 216–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 3, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Kogure, T.; Yan, I.K.; Lin, W.L.; Patel, T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer 2013, 7–8, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D. Isolation and molecular characterization of extracellular vesicles. Methods 2015, 87, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Li, S.; Li, G.; Zhang, S.; Tang, X.; Ni, S.; Jian, X.; Xu, C.; Zhu, J.; Lu, M. The role of extracellular vesicles in mediating progression, metastasis and potential treatment of hepatocellular carcinoma. Oncotarget 2017, 2, 3683–3695. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 3, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Andreola, G.; Rivoltini, L.; Castelli, C.; Huber, V.; Perego, P.; Deho, P.; Squarcina, P.; Accornero, P.; Lozupone, F.; Lugini, L.; et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J. Exp. Med. 2002, 10, 1303–1316. [Google Scholar] [CrossRef]
- Huber, V.; Fais, S.; Iero, M.; Lugini, L.; Canese, P.; Squarcina, P.; Zaccheddu, A.; Colone, M.; Arancia, G.; Gentile, M.; et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: Role in immune escape. Gastroenterology 2005, 7, 1796–1804. [Google Scholar] [CrossRef]
- Aderem, A.; Underhill, D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999, 17, 593–623. [Google Scholar] [CrossRef] [PubMed]
- Rougerie, P.; Miskolci, V.; Cox, D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: Role and regulation of the actin cytoskeleton. Immunol. Rev. 2013, 256, 222–239. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.E.; Manfredi, A.A. How macrophages ring the inflammation alarm. Proc. Natl. Acad. Sci. USA 2014, 111, 2866–2867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Luo, Q.; Yao, F.; Qing, C.; Ye, J.; Deng, Y.; Li, J. Identification of Differentially Expressed Long Non-coding RNAs in Polarized Macrophages. Sci. Rep. 2016, 6, 19705. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Xu, Y.; Lai, Y.; He, W.; Li, Y.; Wang, R.; Luo, X.; Chen, R.; Chen, T. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J. Cell. Biochem. 2018, 119, 2951–2963. [Google Scholar] [CrossRef] [PubMed]
- Ti, D.; Hao, H.; Tong, C.; Liu, J.; Dong, L.; Zheng, J.; Zhao, Y.; Liu, H.; Fu, X.; Han, W. LPS-preconditioned mesenchymal stromal cells modify macrophage polarization for resolution of chronic inflammation via exosome-shuttled let-7b. J. Transl. Med. 2015, 13, 308. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 19, 2958. https://doi.org/10.3390/ijms19102958
Li X, Lei Y, Wu M, Li N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. International Journal of Molecular Sciences. 2018; 19(10):2958. https://doi.org/10.3390/ijms19102958
Chicago/Turabian StyleLi, Xue, Yi Lei, Miao Wu, and Nan Li. 2018. "Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339" International Journal of Molecular Sciences 19, no. 10: 2958. https://doi.org/10.3390/ijms19102958




