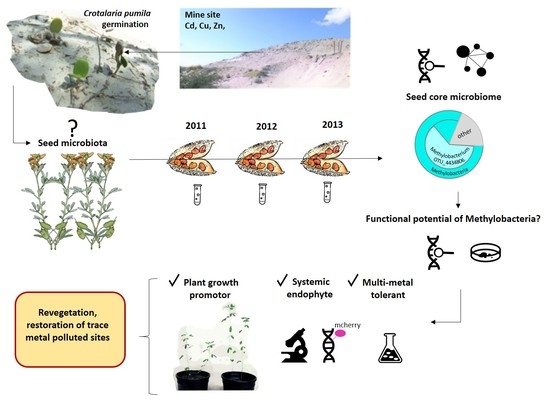
Seed Endophyte Microbiome of Crotalaria pumila Unpeeled: Identification of Plant-Beneficial Methylobacteria
Abstract
:1. Introduction
2. Results
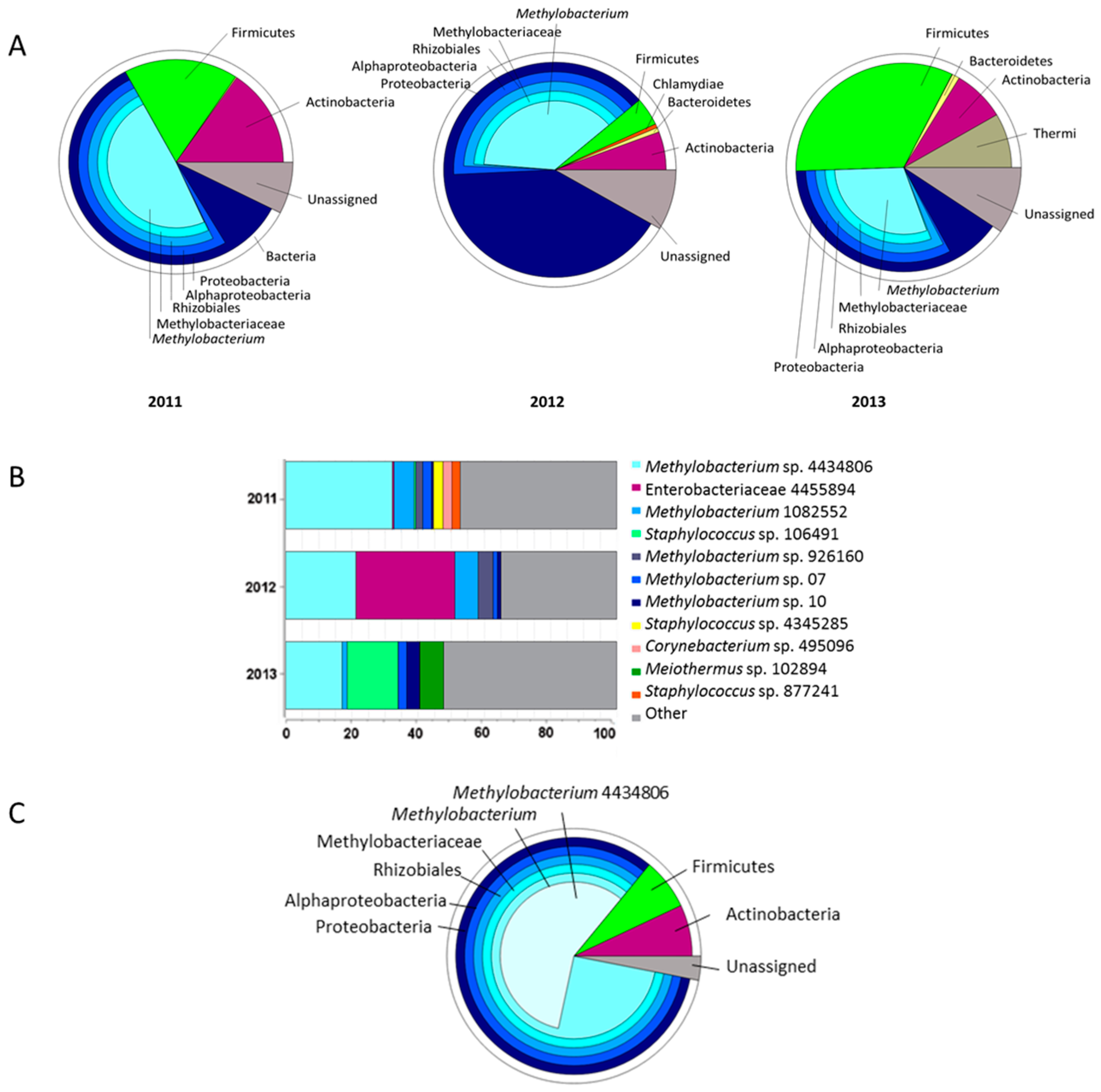
2.1. Seed Microbiome of C. pumila
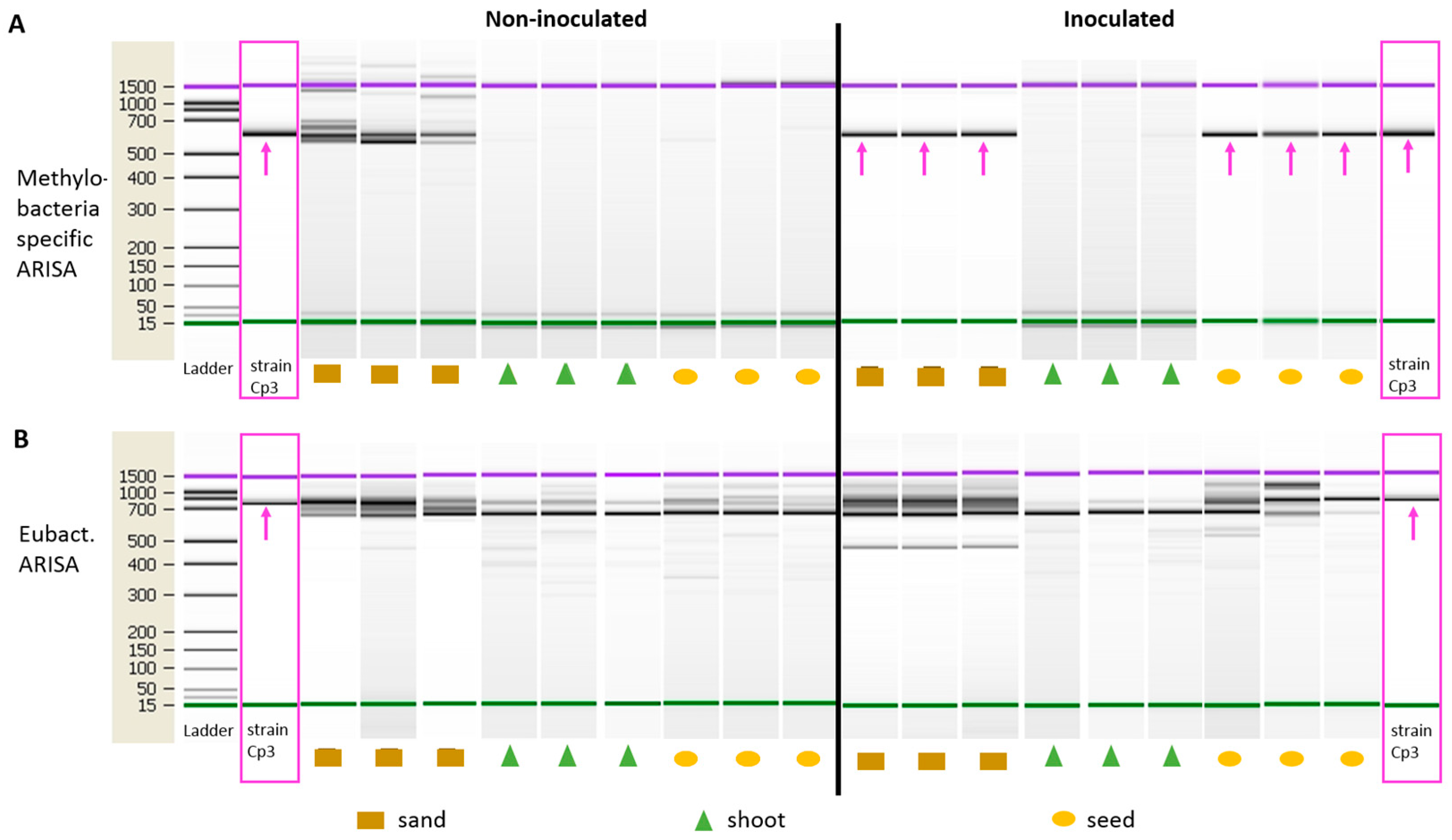
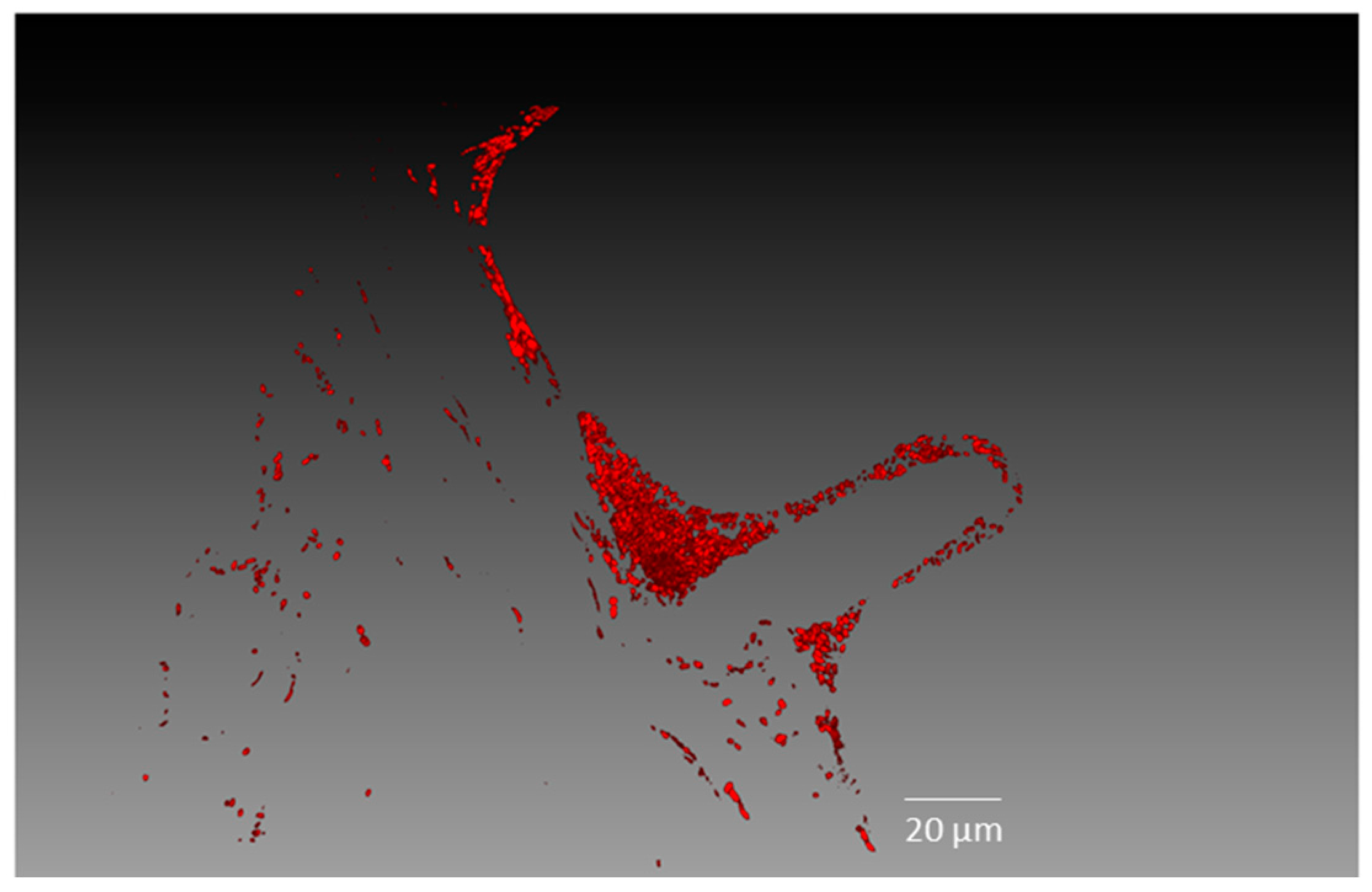
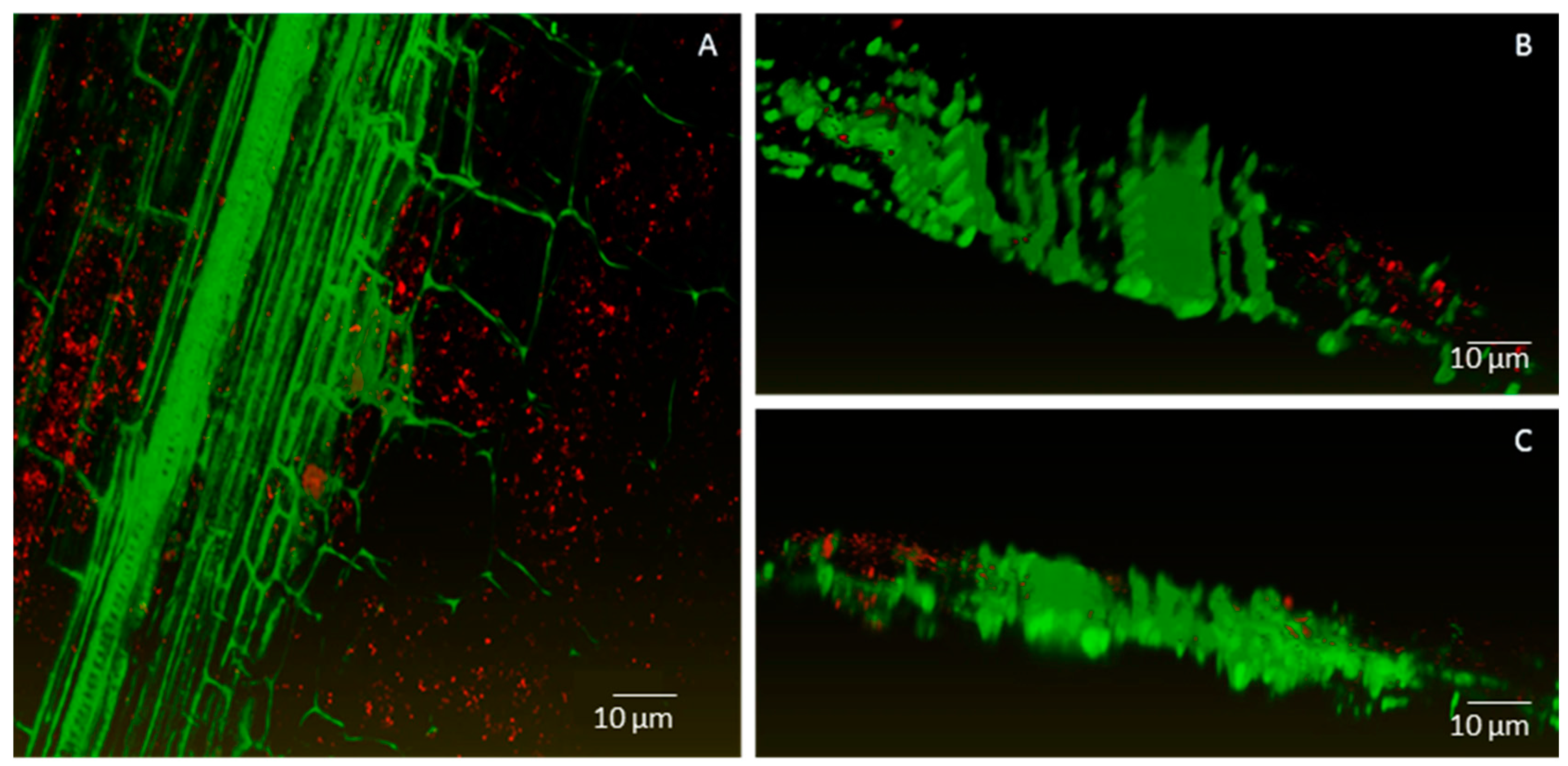
2.2. Endophytic Colonisation of Methylobacterium sp. Cp3 from Soil Substrate to Seed
2.3. Establishment of Methylobacterium sp. Cp3 in Plant Tissue in Presence of Trace Metals
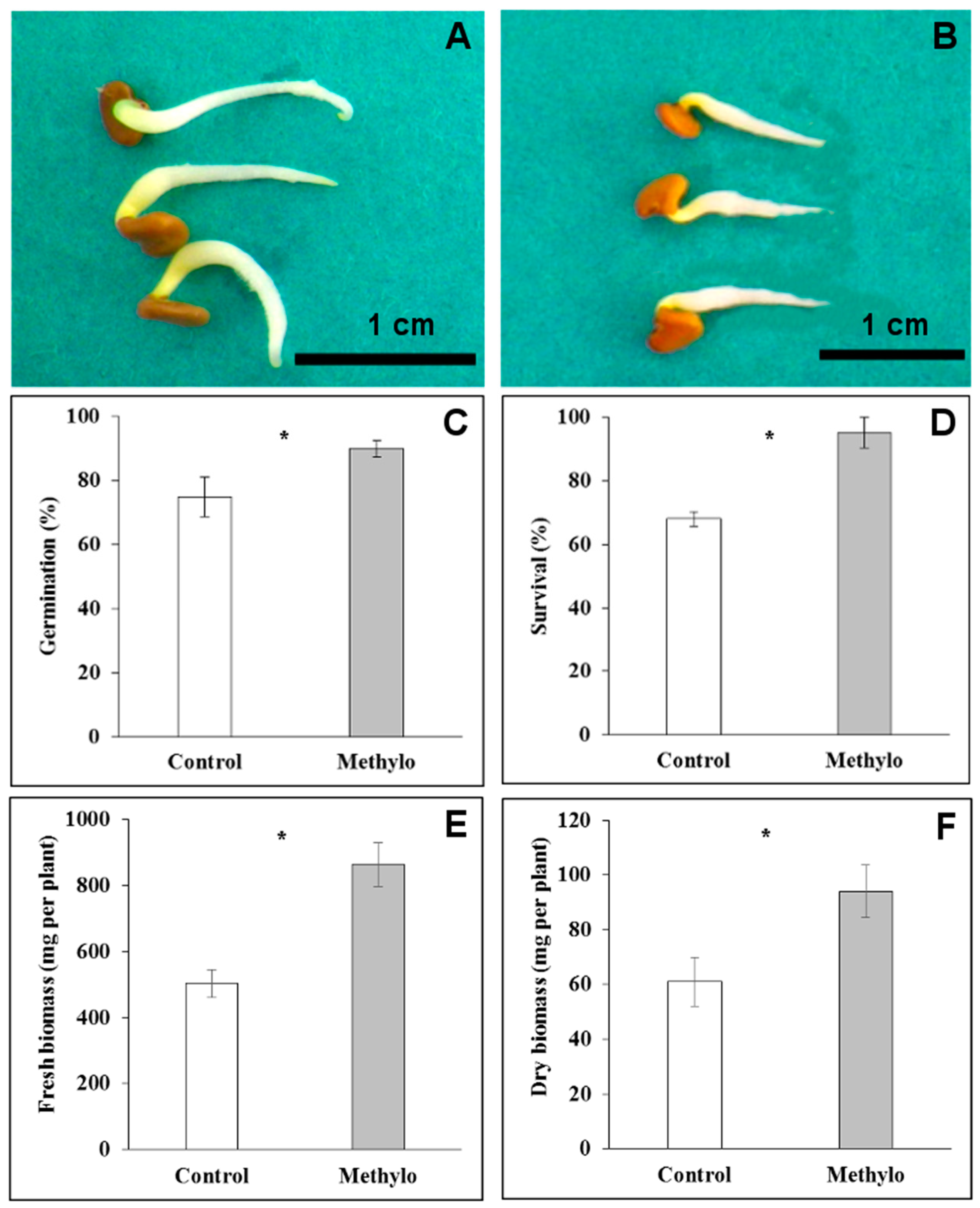
2.4. Strain Cp3 Inoculation Improves Seed Germination and Plantlet Survival under Cadmium Stress
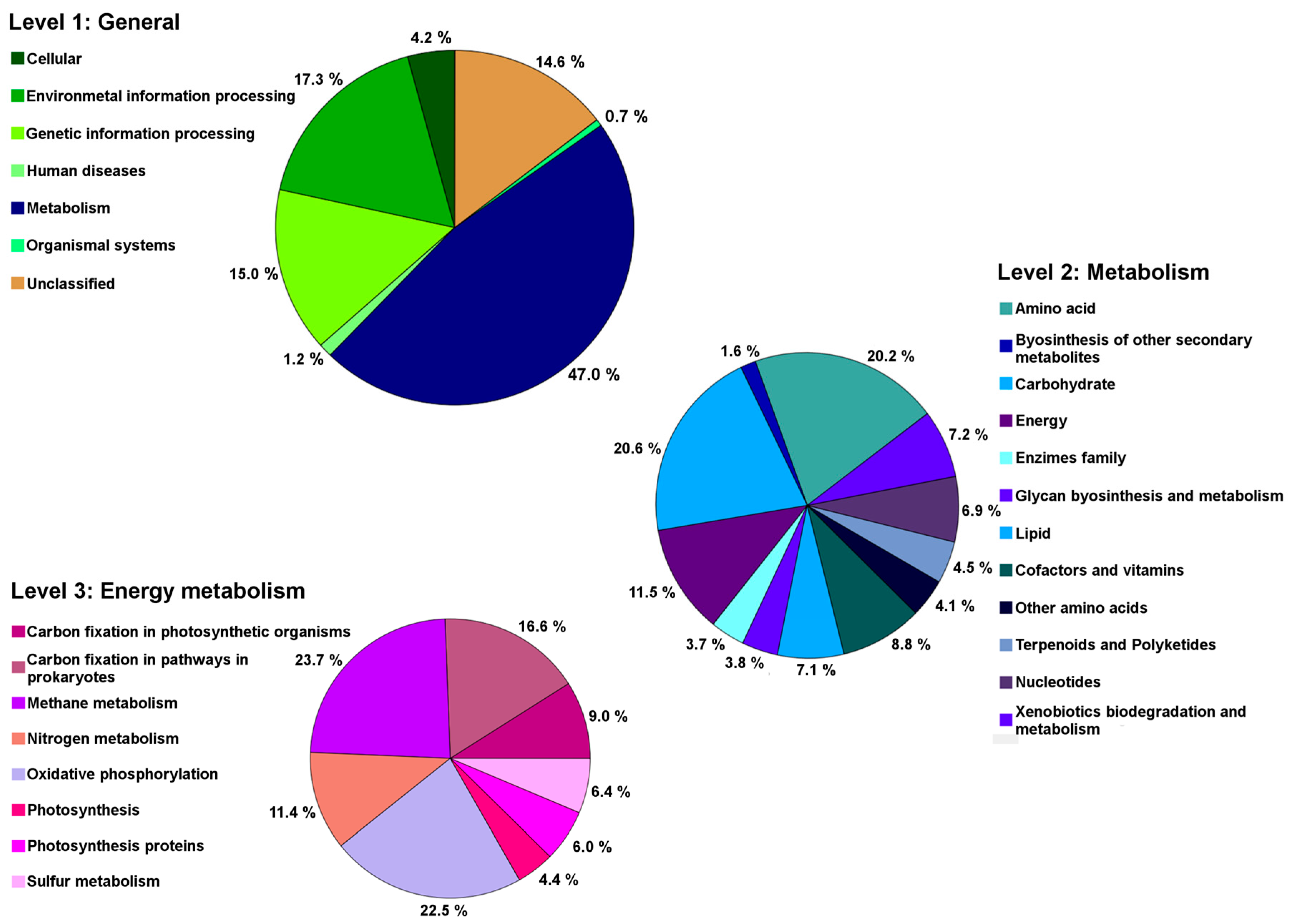
2.5. Genes Related to Cadmium Tolerance, Plant Colonisation and Plant-Growth Promotion
3. Discussion
4. Materials and Methods
4.1. Crotalaria pumila Seed Core Microbiome Analyses
4.2. Methylobacterium Cp3 Isolate Sanger Sequence
4.3. Plant Colonisation Experiment
4.4. Seed Sample Preparation and DNA-Extraction
4.5. Specific Automated Ribosomal Intergenic Spacer Analyses (ARISA)
4.6. Isolation of Methylobacteria and BOX-Fingerprint Analysis
4.7. Confocal Microscopy Analyses
4.8. Image Processing and Three-Dimensional (3D) Visualisation
4.9. In Vitro Plant Growth Promotion Traits Detection
4.10. Genome Sequencing, Assembly and Annotation
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tóth, G.; Hermann, T.; Da Silva, M.R.; Montanarella, L. Heavy metals in agricultural soils of the European Union with implications for food safety. Environ. Int. 2016, 88, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhong, T.; Liu, L.; Ouyang, X. Impact of soil heavy metal pollution on food safety in China. PLoS ONE 2015, 10, e0135182. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Puschenreiter, M. Endophytes and rhizosphere bacteria of plants growing in heavy metal-containing soils. In Microbiology of Extreme Soils Volume 13; Dion, P., Nautiyal, C.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Mench, M.; Lepp, N.; Bert, V.; Schwitzguébel, J.P.; Gawronski, S.W.; Schröder, P.; Vangronsveld, J. Successes and limitations of phytotechnologies at field scale: Outcomes, assessment and outlook from COST Action 859. J. Soils Sediments 2010, 10, 1039–1070. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Thijs, S.; Langill, T.; Vangronsveld, J. The bacterial and fungal microbiota of hyperaccumulator plants: Small organisms, large influence. Adv. Bot. Res. 2017, 83, 43–86. [Google Scholar]
- Visioli, G.; D’Egidio, S.; Sanangelantoni, A.M. The bacterial rhizobiome of hyperaccumulators: Future perspectives based on omics analysis and advanced microscopy. Front. Plant. Sci. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Kidd, P.S.; Álvarez-López, V.; Becerra-Castro, C.; Cabello-Conejo, M.; Prieto-Fernández, Á. Potential role of plant-associated bacteria in plant metal uptake and implications in phytotechnologies. Adv. Bot. Res. 2017, 83, 87–126. [Google Scholar]
- Kidd, P.; Barceló, J.; Bernal, M.P.; Navari-Izzo, F.; Poschenrieder, C.; Shilev, S.; Clemente, R.; Monterroso, C. Trace element behaviour at the root-soil interface: Implications in phytoremediation. Environ. Exp. Bot. 2009, 67, 243–259. [Google Scholar] [CrossRef]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Vangronsveld, J. Phytoremediation: Plant-endophyte partnerships take the challenge. Curr. Opin. Biotechnol. 2009, 20, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Montalbán, B.; Thijs, S.; Lobo, M.C.; Weyens, N.; Ameloot, M.; Vangronsveld, J.; Pérez-Sanz, A. Cultivar and metal-specific effects of endophytic bacteria in Helianthus tuberosus exposed to Cd and Zn. Int. J. Mol. Sci. 2017, 18, 2026. [Google Scholar] [CrossRef] [PubMed]
- Mastretta, C.; Taghavi, S.; van der Lelie, D.; Mengoni, A.; Galardi, F.; Gonnelli, C.; Barac, T.; Boulet, J.; Weyens, N.; Vangronsveld, J. Endophytic bacteria from seeds of Nicotiana tabacum can reduce cadmium phytotoxicity. Int. J. Phytoremediat. 2010, 11, 37–41. [Google Scholar]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Changes in the population of seed bacteria of transgenerationally Cd-exposed Arabidopsis thaliana. Plant Biol. 2013, 15, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Truyens, S.; Beckers, B.; Thijs, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Cadmium-induced and trans-generational changes in the cultivable and total seed endophytic community of Arabidopsis thaliana. Plant Biol. 2016, 18, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-López, A.S.; González-Chávez, M.D.C.A.; Carrillo-González, R.; Vangronsveld, J.; Díaz-Garduño, M. Wild flora of mine tailings: Perspectives for use in phytoremediation of potentially toxic elements in a semi-arid region in Mexico. Int. J. Phytoremediat. 2015, 17, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-López, A.S.; Thijs, S.; Beckers, B.; González-Chávez, M.C.; Weyens, N.; Carrillo-González, R.; Vangronsveld, J. Community structure and diversity of endophytic bacteria in seeds of three consecutive generations of Crotalaria pumila growing on metal mine residues. Plant Soil 2017. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial seed endophytes: Genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Dourado, M.N.; Camargo Neves, A.A.; Santos, D.S.; Araújo, W.L. Biotechnological and agronomic potential of endophytic pink-pigmented methylotrophic methylobacterium spp. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Idris, R.; Trifonova, R.; Puschenreiter, M.; Wenzel, W.W.; Sessitsch, A. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microbiol. 2004, 70, 2667–2677. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luo, S.; Chen, J.; Wan, Y.; Li, X.; Liu, C.; Liu, F. A comparative analysis of endophytic bacterial communities associated with hyperaccumulators growing in mine soils. Environ. Sci. Pollut. Res. 2014, 21, 7538–7547. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef] [PubMed]
- De Voogd, N.J.; Cleary, D.F.R.; Polónia, A.R.M.; Gomes, N.C.M. Bacterial community composition and predicted functional ecology of sponges, sediment and seawater from the thousand islands reef complex, West Java, Indonesia. FEMS Microbiol. Ecol. 2015, 91, fiv019. [Google Scholar] [CrossRef] [PubMed]
- Sy, A.; Timmers, A.C.J.; Knief, C.; Vorholt, J. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 2005, 71, 7245–7252. [Google Scholar] [CrossRef] [PubMed]
- Abanda-Nkpwatt, D.; Müsch, M.; Tschiersch, J.; Boettner, M.; Schwab, W. Molecular interaction between Methylobacterium extorquens and seedlings: Growth promotion, methanol consumption, and localization of the methanol emission site. J. Exp. Bot. 2006, 57, 4025–4032. [Google Scholar] [CrossRef] [PubMed]
- Marx, C.J.; Bringel, F.; Chistoserdova, L.; Moulin, L.; Farhan Ul Haque, M.; Fleischman, D.E.; Gruffaz, C.; Jourand, P.; Knief, C.; Lee, M.C.; et al. Complete genome sequences of six strains of the genus Methylobact. J. Bacteriol. 2012, 194, 4746–4748. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Martínez, J.; López-Díaz, S.; Rodríguez-Garay, B. Detection of the effects of Methylobacterium in Agave tequilana Weber var. azul by laser-induced fluorescence. Plant Sci. 2004, 166, 889–892. [Google Scholar] [CrossRef]
- Sundin, G.W.; Jacobs, J.L. Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.). Microb. Ecol. 1999, 38, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Umeno, D.; Tobias, A.V.; Frances, H.; Umeno, D.; Tobias, A.V.; Arnold, F.H. Diversifying carotenoid biosynthetic pathways by directed evolution diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 2005, 69, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Burbank, L.; Roper, M.C. Biological role of pigment production for the bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 2012, 78, 6859–6865. [Google Scholar] [CrossRef] [PubMed]
- Truyens, S.; Beckers, B.; Thijs, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. The effects of the growth substrate on cultivable and total endophytic assemblages of Arabidopsis thaliana. Plant Soil 2016, 405, 325–336. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Yergeau, E.; Sanschagrin, S.; Maynard, C.; St-Arnaud, M.; Greer, C.W. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 2014, 8, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Croes, S.; Weyens, N.; Janssen, J.; Vercampt, H.; Colpaert, J.V.; Carleer, R.; Vangronsveld, J. Bacterial communities associated with Brassica napus L. grown on trace element-contaminated and non-contaminated fields: A genotypic and phenotypic comparison. Microb. Biotechnol. 2013, 6, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Omer, Z.S.; Tombolini, R.; Broberg, A.; Gerhardson, B. Indole-3-acetic acid production by pink-pigmented facultative methylotrophic bacteria. Plant Growth Regul. 2004, 43, 93–96. [Google Scholar] [CrossRef]
- Poonguzhali, S.; Madhaiyan, M.; Yim, W.J.; Kim, K.A.; Sa, T.M. Colonization pattern of plant root and leaf surfaces visualized by use of green-fluorescent-marked strain of Methylobacterium suomiense and its persistence in rhizosphere. Appl. Microbiol. Biotechnol. 2008, 78, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Andreote, F.D.; Lacava, P.T.; Gai, C.S.; Araújo, W.L.; Maccheroni, W.J.; van Overbeek, L.S.; van Elsas, J.D.; Azevedo, J.L. Model plants for studying the interaction between Methylobacterium mesophilicum and Xylella fastidiosa. Can. J. Microbiol. 2006, 52, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.S.; Lacava, P.T.; Quecine, M.C.; Auriac, M.C.; Lopes, J.R.S.; Araújo, W.L.; Miller, T.A.; Azevedo, J.L. Transmission of Methylobacterium mesophilicum by Bucephalogonia xanthophis for paratransgenic control strategy of Citrus variegated chlorosis. J. Microbiol. 2009, 47, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Araújo, W.L.; Santos, D.S.; Dini-Andreote, F.; Salgueiro-Londoño, J.K.; Camargo-Neves, A.A.; Andreote, F.D.; Dourado, M.N. Genes related to antioxidant metabolism are involved in Methylobacterium mesophilicum-soybean interaction. Antonie Van Leeuwenhoek 2015, 108, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Rouws, L.F.M.; Meneses, C.H.S.G.; Guedes, H.V.; Vidal, M.S.; Baldani, J.I.; Schwab, S. Monitoring the colonization of sugarcane and rice plants by the endophytic diazotrophic bacterium Gluconacetobacter diazotrophicus marked with gfp and gusA reporter genes. Lett. Appl. Microbiol. 2010, 51, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, E.; Xiong, X.; Shen, D.; Feng, Y. Colonization of endophyte Pantoea agglomerans YS19 on host rice, with formation of multicellular symplasmata. World J. Microbiol. Biotechnol. 2010, 26, 1667–1673. [Google Scholar] [CrossRef]
- Prieto, P.; Schilirò, E.; Maldonado-González, M.M.; Valderrama, R.; Barroso-Albarracín, J.B.; Mercado-Blanco, J. Root hairs play a key role in the endophytic colonization of olive roots by Pseudomonas spp. with biocontrol activity. Microb. Ecol. 2011, 62, 435–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-López, A.S. Basis for the Remediation of Sites Polluted by Potentially Toxic Elements in Zimapan, Mexico: Interdisciplinary Approach. Ph.D. Thesis, Colegio de Postgraduados, Texcoco, Mexico, Hasselt University, Hasselt, Belgium, 2015. [Google Scholar]
- Khan, M.; Harun, N.; Rehman, A.H.N.A.; Elhussein, S.A. In vitro antioxidant evaluation of extracts of three wild Malaysian plants. Procedia Eng. 2013, 53, 29–36. [Google Scholar] [CrossRef]
- Villa-Ruano, N.; Zurita-Vásquez, G.G.; Pacheco-Hernández, Y.; Betancourt-Jiménez, M.G.; Cruz-Durán, R.; Duque-Bautista, H. Anti-Iipase and antioxidant properties of 30 medicinal plants used in Oaxaca, México. Biol. Res. 2013, 46, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Reiter, B.; Sessitsch, A.; Clément, C.; Barka, E.A.; Nowak, J. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 2005, 71, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Kaplan, H.; Sessitsch, A.; Nowak, J.; Ait Barka, E.; Clément, C. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: From the rhizosphere to inflorescence tissues. FEMS Microbiol. Ecol. 2008, 63, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Filho, A.S.F.; Quecine, M.C.; Bogas, A.C.; de Rossetto, P.B.; de Lima, A.O.S.; Lacava, P.T.; Azevedo, J.L.; Araújo, W.L. Endophytic Methylobacterium extorquens expresses a heterologous β-1,4-endoglucanase A (EglA) in Catharanthus roseus seedlings, a model host plant for Xylella fastidiosa. World J. Microbiol. Biotechnol. 2012, 28, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- De Procópio, R.E.L.; Araújo, W.L.; Andreote, F.D.; Azevedo, J.L. Characterization of a small cryptic plasmid from endophytic Pantoea agglomerans and its use in the construction of an expression vector. Genet. Mol. Biol. 2011, 34, 103–109. [Google Scholar]
- Anand, R.; Chanway, C.P. Detection of GFP-labeled Paenibacillus polymyxa in autofluorescing pine seedling tissues. Biol. Fertil. Soils 2013, 49, 111–118. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Singh, A.; Chowdhury, S.P.; Tripathi, A.K. Endophytic colonization ability of two deep-water rice endophytes, Pantoea sp. and Ochrobactrum sp. using green fluorescent protein reporter. Biotechnol. Lett. 2004, 26, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Quecine, M.C.; Lacava, P.T.; Oda, S.; Azevedo, J.L.; Araújo, W.L. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 2008, 287, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lin, L.; Zhu, Z.; Yang, X.; Wang, Y.; An, Q. Colonization and modulation of host growth and metal uptake by endophytic bacteria of Sedum alfredii. Int. J. Phytoremediat. 2013, 15, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Lundberg, D.S.; Lazarovits, G.; Reis, V.M.; Raizada, M.N. Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 2016, 405, 337–355. [Google Scholar] [CrossRef]
- Mitter, B.; Sessitsch, A.; Naveed, M. Method for Producing Plant Seed Containing Endophytic Micro-Organisms 2013. European Patent #2,676,536 A1, 26 November 2015. [Google Scholar]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R.; Hernandez, J.P. Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998–2013). Plant Soil 2014, 378, 1–33. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Taghavi, S.; Mergeay, M.; Vangronsveld, J.; Clijsters, H.; van der Lelie, D. The effect of recombinant heavy metal-resistant endophytic bacteria on heavy metal uptake by their host plant. Int. J. Phytoremediat. 2001, 3, 173–187. [Google Scholar] [CrossRef]
- Berg, G.; Zachow, C.; Müller, H.; Philipps, J.; Tilcher, R. Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 2013, 3, 648–656. [Google Scholar] [CrossRef]
- Keyser, C.A.; Thorup-Kristensen, K.; Meyling, N.V. Metarhizium seed treatment mediates fungal dispersal via roots and induces infections in insects. Fungal Ecol. 2014, 11, 122–131. [Google Scholar] [CrossRef]
- Gonzalez, F.; Tkaczuk, C.; Dinu, M.M.; Fiedler, Ż.; Vidal, S.; Zchori-Fein, E.; Messelink, G.J. New opportunities for the integration of microorganisms into biological pest control systems in greenhouse crops. J. Pest. Sci. 2016, 89, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Kuang, Z.; Wang, W.; Cao, L. Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol. Control 2016, 98, 27–33. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeets, K.; Ruytinx, J.; Van Belleghem, F.; Semane, B.; Lin, D.; Vangronsveld, J.; Cuypers, A. Critical evaluation and statistical validation of a hydroponic culture system for Arabidopsis thaliana. Plant. Physiol. Biochem. 2008, 46, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Knief, C.; Frances, L.; Cantet, F.; Vorholt, J.A. Cultivation-independent characterization of Methylobacterium populations in the plant phyllosphere by automated ribosomal intergenic spacer analysis. Appl. Environ. Microbiol. 2008, 74, 2218–2228. [Google Scholar] [CrossRef] [PubMed]
- Normand, P.; Ponsonnet, C.; Nesme, X.; Neyra, M.; Simonet, P. ITS analysis of prokaryotes. In Molecular Microbial Ecology Manual; Akkermans, D., van Elsas, J.D., de Bruijn, E.I., Eds.; Kluwer Academic Publishers: Amsterdam, The Netherlands, 1996. [Google Scholar]
- Michelland, R.J.; Dejean, S.; Combes, S.; Fortun-Lamothe, L.; Cauquil, L. StatFingerprints: A friendly graphical interface program for processing and analysis of microbial fingerprint profiles. Mol. Ecol. Resour. 2009, 9, 1359–1363. [Google Scholar] [CrossRef] [PubMed]
- Weyens, N.; Taghavi, S.; Barac, T.; van der Lelie, D.; Boulet, J.; Artois, T.; Carleer, R.; Vangronsveld, J. Bacteria associated with oak and ash on a TCE-contaminated site: Characterization of isolates with potential to avoid evapotranspiration of TCE. Environ. Sci. Pollut. Res. 2009, 16, 830–843. [Google Scholar] [CrossRef] [PubMed]
- Lagendijk, E.L.; Validov, S.; Lamers, G.E.M.; De Weert, S.; Bloemberg, G.V. Genetic tools for tagging Gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol. Lett. 2010, 305, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Linding-Cisneros, R.; Lara-Cabrera, S. Effect of scarification and growig media on seed germination of Crotalaria pumila (Ort.). Seed Sci. Technol. 2004, 32, 231–234. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Belimov, A.A.; Hontzeas, N.; Safronova, V.I.; Demchinskaya, S.V.; Piluzza, G.; Bullitta, S.; Glick, B.R. Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 2005, 37, 241–250. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of Inodoleacetic Acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Xie, G.H.; Cui, Z.; Yu, J.; Yan, J.; Hai, W.; Steinberger, Y. Identification of nif genes in N2-fixing bacterial strains isolated from rice fields along the Yangtze River Plain. J. Basic Microbiol. 2006, 46, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Romick, T.L.; Fleming, H.P. Acetoin production as an indicator of growth and metabolic inhibition of Listeria monocytogenes. J. Appl. Microbiol. 1998, 84, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-López, A.S.; Pintelon, I.; Stevens, V.; Imperato, V.; Timmermans, J.-P.; González-Chávez, C.; Carrillo-González, R.; Van Hamme, J.; Vangronsveld, J.; Thijs, S. Seed Endophyte Microbiome of Crotalaria pumila Unpeeled: Identification of Plant-Beneficial Methylobacteria. Int. J. Mol. Sci. 2018, 19, 291. https://doi.org/10.3390/ijms19010291
Sánchez-López AS, Pintelon I, Stevens V, Imperato V, Timmermans J-P, González-Chávez C, Carrillo-González R, Van Hamme J, Vangronsveld J, Thijs S. Seed Endophyte Microbiome of Crotalaria pumila Unpeeled: Identification of Plant-Beneficial Methylobacteria. International Journal of Molecular Sciences. 2018; 19(1):291. https://doi.org/10.3390/ijms19010291
Chicago/Turabian StyleSánchez-López, Ariadna S., Isabel Pintelon, Vincent Stevens, Valeria Imperato, Jean-Pierre Timmermans, Carmen González-Chávez, Rogelio Carrillo-González, Jonathan Van Hamme, Jaco Vangronsveld, and Sofie Thijs. 2018. "Seed Endophyte Microbiome of Crotalaria pumila Unpeeled: Identification of Plant-Beneficial Methylobacteria" International Journal of Molecular Sciences 19, no. 1: 291. https://doi.org/10.3390/ijms19010291
APA StyleSánchez-López, A. S., Pintelon, I., Stevens, V., Imperato, V., Timmermans, J.-P., González-Chávez, C., Carrillo-González, R., Van Hamme, J., Vangronsveld, J., & Thijs, S. (2018). Seed Endophyte Microbiome of Crotalaria pumila Unpeeled: Identification of Plant-Beneficial Methylobacteria. International Journal of Molecular Sciences, 19(1), 291. https://doi.org/10.3390/ijms19010291