Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function
Abstract
1. Introduction
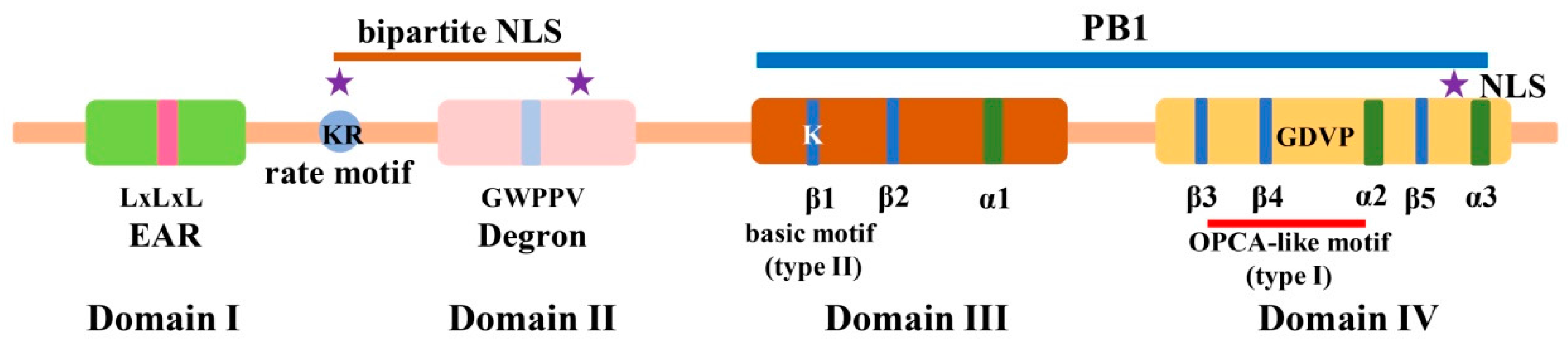
2. Identification, Gene Duplication, and Molecular Structure of Aux/IAA Proteins in Plants
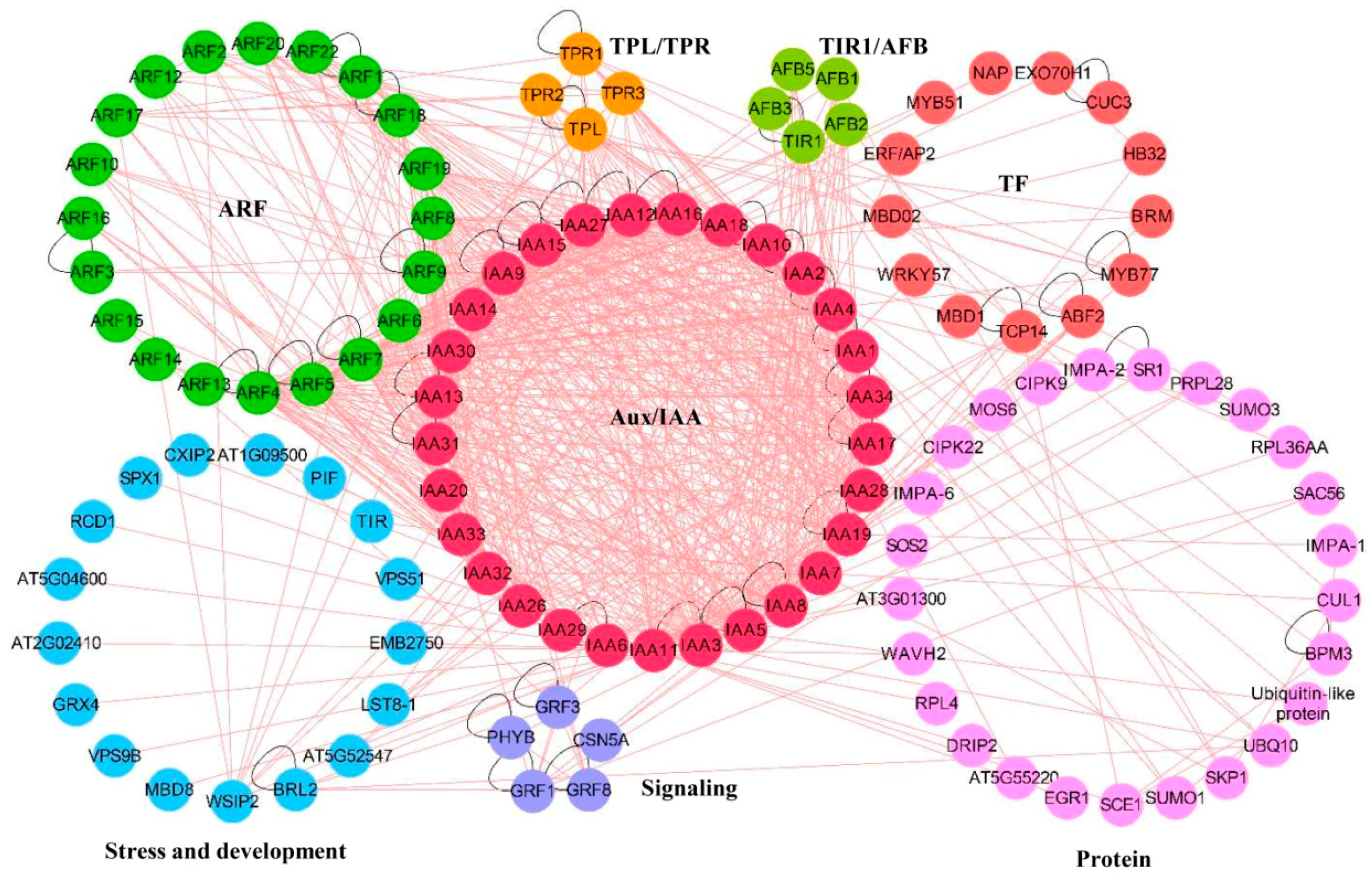
3. Interaction of Aux/IAA Proteins in Plants
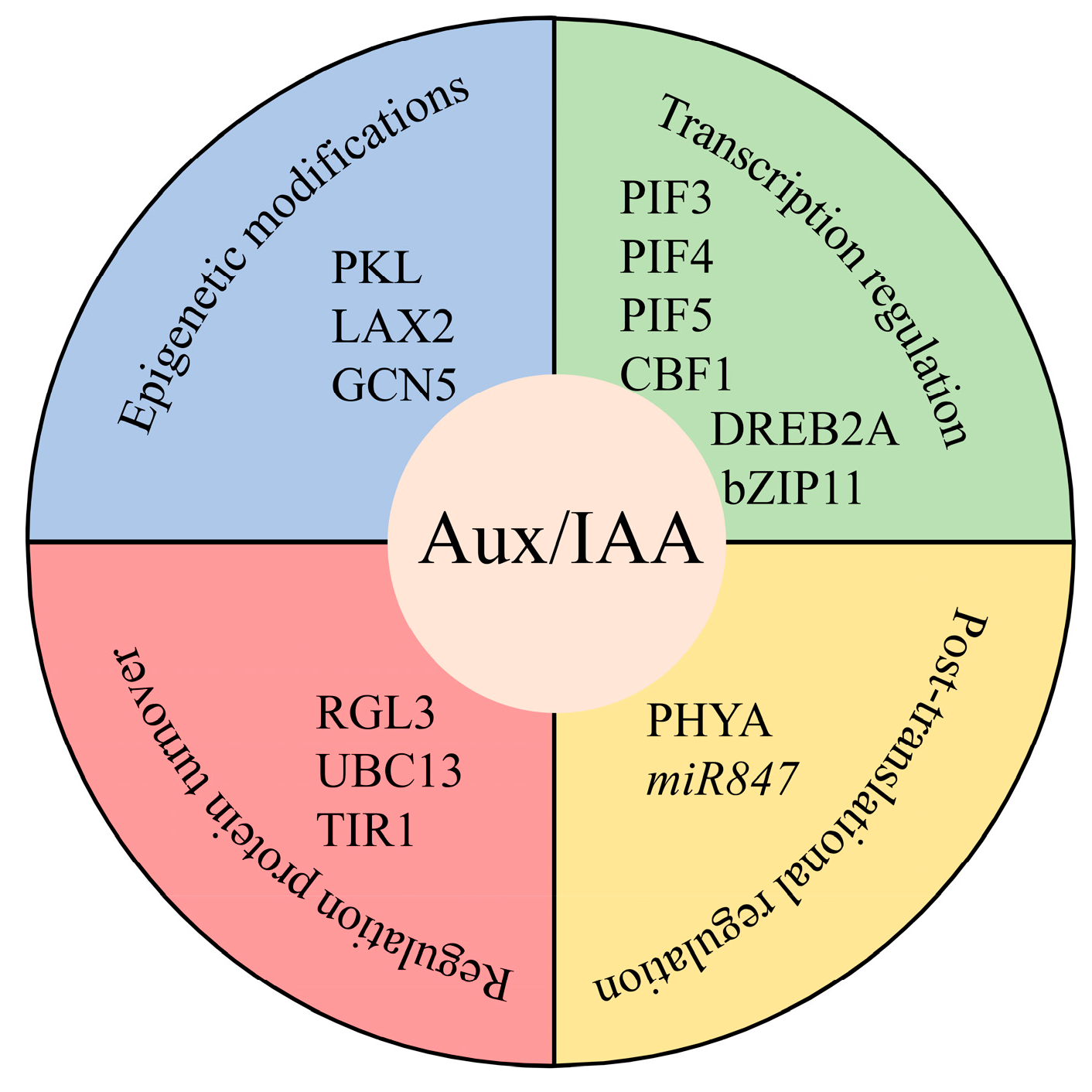
4. Aux/IAA Proteins Might be Regulated at Different Levels
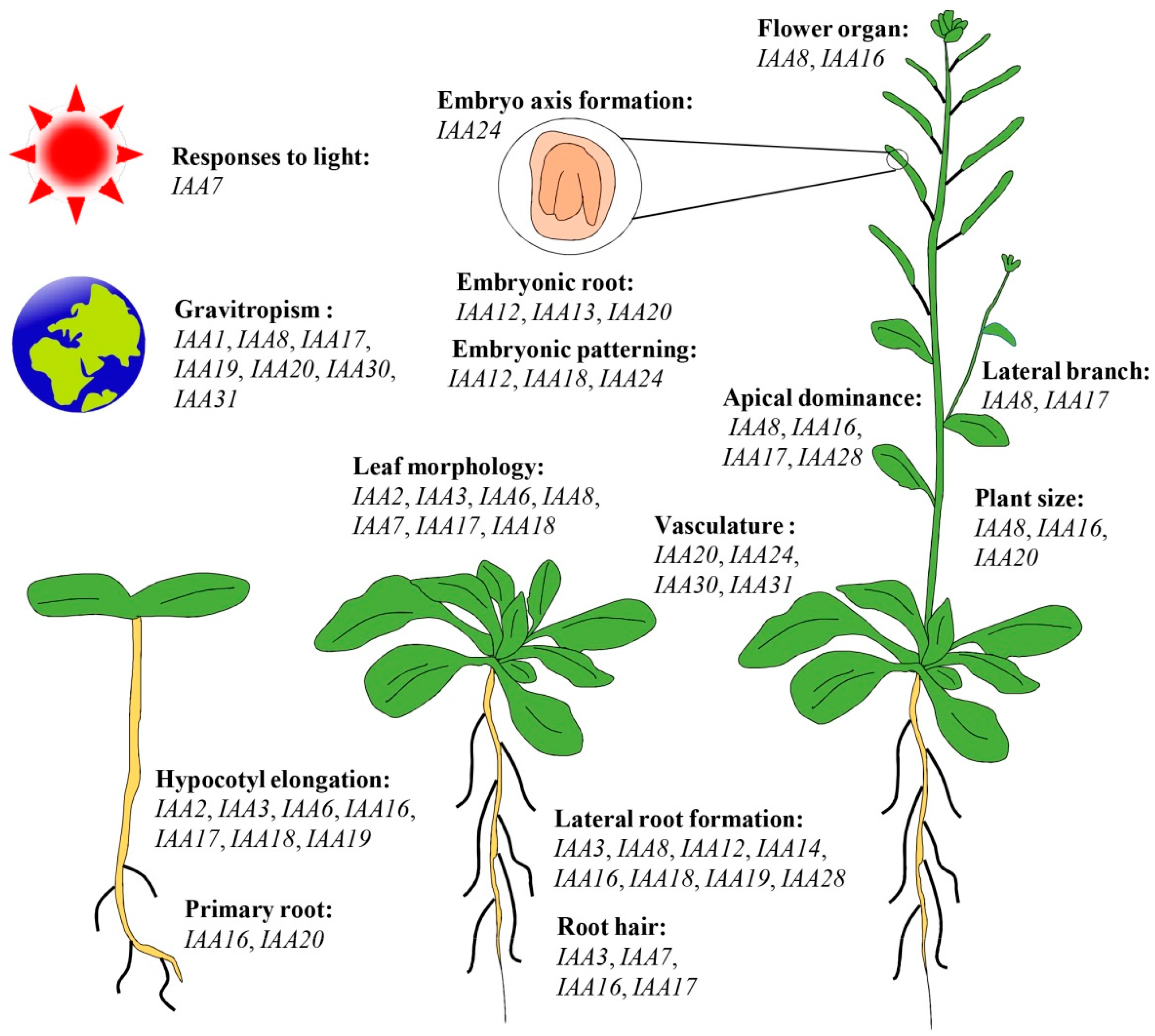
5. Functional Roles of Aux/IAA Genes during Plant Growth and Development Processes
6. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goldfarb, B.; Lanz-Garcia, C.; Lian, Z.; Whetten, R. Aux/IAA gene family is conserved in the gymnosperm, loblolly pine (Pinus taeda). Tree Physiol. 2003, 23, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, E.; Østergaard, L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb. Perspect. Biol. 2009, 1, a001628. [Google Scholar] [CrossRef] [PubMed]
- Goldental-Cohen, S.; Israeli, A.; Ori, N.; Yasuor, H. Auxin response dynamics during wild-type and entire flower development in tomato. Plant Cell Physiol. 2017, 58, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Perrotrechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Nguyen, M.D.; Theologis, A. The PS-IAA4/5-like family of early Auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 1995, 251, 533–549. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Oeller, P.W.; Theologis, A. Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Dreher, K.A.; Brown, J.; Saw, R.E.; Callis, J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 2006, 18, 699–714. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Benkova, E.; Jäger, K.E.; Schlereth, A.; Hamann, T.; Kientz, M.; Wilmoth, J.C.; Reed, J.W.; Jürgens, G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005, 24, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Parry, G.; Calderon-Villalobos, L.I.; Prigge, M.; Peret, B.; Dharmasiri, S.; Itoh, H.; Lechner, E.; Gray, W.M.; Bennett, M.; Estelle, M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 2009, 106, 22540–22545. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Calderon Villalobos, L.I.; Lee, S.; De Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Trenner, J.; Poeschl, Y.; Grau, J.; Gogol-Döring, A.; Quint, M.; Delker, C. Auxin-induced expression divergence between Arabidopsis species may originate within the TIR1/AFB-AUX/IAA-ARF module. J. Exp. Bot. 2017, 68, 539. [Google Scholar] [PubMed]
- Tian, Q.; Reed, J.W. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 1999, 126, 711–721. [Google Scholar] [PubMed]
- Rinaldi, M.A.; Liu, J.; Enders, T.A.; Bartel, B.; Strader, L.C. A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol. Biol. 2012, 79, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, D.-W.; Yuan, T.-T.; Gao, X.; Lu, Y.-T. A gain-of-function mutation in IAA8 alters Arabidopsis floral organ development by change of jasmonic acid level. Plant Mol. Biol. 2013, 82, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Overvoorde, P.J.; Okushima, Y.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Liu, A.; Onodera, C.; Quach, H.; et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 2005, 17, 3282–3300. [Google Scholar] [CrossRef] [PubMed]
- Liscum, E.; Reed, J. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.-I.; Jones, A.M.; Ogino, K.; Yamazoe, A.; Oono, Y.; Inoguchi, M.; Kondo, H.; Nozaki, H. Yokonolide B, A novel inhibitor of auxin action, blocks degradation of AUX/IAA factors. J. Biol. Chem. 2003, 278, 23797–23806. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Nagpal, P.; Reed, J.W. Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J. 2003, 36, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Soler, M.; Clemente, H.; Mila, I.; Paiva, J.A.P.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J.; Cassan-Wang, H. Comprehensive genome-wide analysis of the Aux/IAA gene family in eucalyptus: Evidence for the role of EgrIAA4 in wood formation. Plant Cell Physiol. 2015, 56, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Gan, D.; Zhuang, D.; Ding, F.; Yu, Z.; Zhao, Y. Identification and expression analysis of primary auxin-responsive Aux/IAA gene family in cucumber (Cucumis sativus). J. Genet. 2013, 92, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, U.C.; DiFazio, S.P.; Brunner, A.M.; Tuskan, G.A. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, Y.; Zhang, Y.; Hochholdinger, F. The maize (Zea mays L.) AUXIN/INDOLE-3-ACETIC ACID gene family: Phylogeny, synteny, and unique root-type and tissue-specific expression patterns during development. PLoS ONE 2013, 8, e78859. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liu, Y.; Wang, Y.; Li, H.; Liu, J.; Tan, J.; He, J.; Bai, J.; Ma, H. Evolution analysis of the Aux/IAA gene family in plants shows dual origins and variable nuclear localization signals. Int. J. Mol. Sci. 2017, 18, 2107. [Google Scholar] [CrossRef] [PubMed]
- Oeller, P.W.; Keller, J.A.; Parks, J.E.; Silbert, J.E.; Theologis, A. Structural Characterization of the early indoleacetic acid-inducible genes, PS-IAA4/5 and PS-IAA6, of Pea (Pisum sativum L.). J. Mol. Biol. 1993, 233, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Theologis, A.; Huynh, T.V.; Davis, R.W. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J. Mol. Biol. 1985, 183, 53–68. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Pang, S.; Ma, Y.; Deng, L.; He, S.; Yi, S.; Lv, Q.; Zheng, Y. The ARF, AUX/IAA and GH3 gene families in citrus: Genome-wide identification and expression analysis during fruitlet drop from abscission zone A. Mol. Genet. Genom. 2015, 290, 2089–2105. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peng, Z.; Liu, S.; He, Y.; Cheng, L.; Kong, F.; Wang, J.; Lu, G. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genom. 2012, 287, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Çakir, B.; Kiliçkaya, O.; Olcay, A.C. Genome-wide analysis of Aux/IAA genes in Vitis vinifera: Cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses. Acta Physiol. Plant. 2013, 35, 365–377. [Google Scholar] [CrossRef]
- Wang, B.; King, G.J.; Li, H.; Wang, J.; Liu, K.; Zhang, Q. Genome-wide analysis of the auxin/indoleacetic acid (Aux/IAA) gene family in allotetraploid rapeseed (Brassica napus L.). BMC Plant Biol. 2017, 17, 204. [Google Scholar]
- Singh, V.K.; Jain, M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front. Plant Sci. 2015, 6, 918. [Google Scholar] [CrossRef] [PubMed]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Agarwal, P.; Pareek, A.; Tyagi, A.K.; Sharma, A.K. Genomic Survey, Gene Expression, and Interaction Analysis Suggest Diverse Roles of ARF and Aux/IAA Proteins in Solanaceae. Plant Mol. Biol. Rep. 2015, 33, 1552–1572. [Google Scholar] [CrossRef]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F. Insights into land plant evolution garnered from the Marchantia polymorpha Genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Ishizaki, K.; Kouno, M.; Shirakawa, M.; Bowman, J.L.; Nishihama, R.; Kohchi, T. Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. PLoS Genet. 2015, 11, e1005084. [Google Scholar]
- Singh, V.K.; Rajkumar, M.; Garg, R.; Jain, M. Genome-wide identification and co-expression network analysis provide insights into the roles of auxin response factor gene family in chickpea. Sci. Rep. 2017, 7, 10895. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Wagner, D. Transcriptional responses to the auxin hormone. Ann. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, K. Evolution of land plants: Insights from molecular studies on basal lineages. Biosci. Biotechnol. Biochem. 2017, 81, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Dhandapani, V.; Rameneni, J.J.; Li, X.; Sivanandhan, G.; Choi, S.R.; Pang, W.; Im, S.; Lim, Y.P. Genome-wide analysis and characterization of Aux/IAA family genes in Brassica rapa. PLoS ONE 2016, 11, e0151522. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yuan, C.; Feng, S.; Zhong, S.; Li, H.; Zhong, J.; Shen, C.; Liu, J. Genome-wide analysis and characterization of Aux/IAA family genes related to fruit ripening in papaya (Carica papaya L.). BMC Genom. 2017, 18, 351. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Yue, R.; Yang, Y.; Zhang, L.; Sun, T.; Xu, L.; Tie, S.; Wang, H. Genome-wide identification and expression profiling analysis of the Aux/IAA gene family in Medicago truncatula during the early phase of Sinorhizobium meliloti infection. PLoS ONE 2014, 9, e107495. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zhang, X.; Han, X.; Zhang, L.; Li, X.; Zhan, H.; Ma, J.; Luo, P.; Zhang, W.; Cui, L.; et al. A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front. Plant Sci. 2015, 6, 770. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.A.; Zenser, N.; Leyser, O.; Callis, J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 2001, 13, 2349–2360. [Google Scholar] [CrossRef] [PubMed]
- Moss, B.L.; Mao, H.; Guseman, J.M.; Hinds, T.R.; Hellmuth, A.; Kovenock, M.; Noorassa, A.; Lanctot, A.; Villalobos, L.I.A.C.; Zheng, N. Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol. 2015, 169, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Mironova, V.; Teale, W.; Shahriari, M.; Dawson, J.; Palme, K. The systems biology of auxin in developing embryos. Trends Plant Sci. 2017, 22, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Winkler, M.; Niemeyer, M.; Hellmuth, A.; Janitza, P.; Christ, G.; Samodelov, S.L.; Wilde, V.; Majovsky, P.; Trujillo, M.; Zurbriggen, M.D.; et al. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 2017, 8, 15706. [Google Scholar] [CrossRef] [PubMed]
- Morgan, K.E.; Zarembinski, T.I.; Theologis, A.; Abel, S. Biochemical characterization of recombinant polypeptides corresponding to the predicted βαα fold in Aux/IAA proteins. FEBS Lett. 1999, 454, 283–287. [Google Scholar] [PubMed]
- Kim, J.; Harter, K.; Theologis, A. Protein–protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 11786–11791. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Getting a grasp on domain III/IV responsible for auxin response factor–IAA protein interactions. Plant Sci. 2012, 190, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Chatterjee, S.; Tonelli, M.; Dashti, H.; Lee, S.G.; Westfall, C.S.; Fulton, D.B.; Andreotti, A.H.; Amarasinghe, G.K.; Strader, L.C.; et al. Defining a two-pronged structural model for PB1 (Phox/Bem1p) domain interaction in plant auxin responses. J. Biol. Chem. 2015, 290, 12868–12878. [Google Scholar] [CrossRef] [PubMed]
- Noda, Y.; Kohjima, M.; Izaki, T.; Ota, K.; Yoshinaga, S.; Inagaki, F.; Ito, T.; Sumimoto, H. Molecular recognition in dimerization between PB1 domains. J. Biol. Chem. 2003, 278, 43516–43524. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-Based auxin perception: Mechanism and role in plant growth and development. Plant Cell 2015, 27, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin Signaling. Plant Physiol. 2017, 176. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J. The PB1 domain in auxin response factor and Aux/IAA proteins: A versatile protein interaction module in the Auxin response. Plant Cell 2015, 27, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Parcy, F.; Vernoux, T.; Dumas, R. A Glimpse beyond structures in auxin-Dependent transcription. Trends Plant Sci. 2016, 21, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Fukaki, H.; Onoda, M.; Li, L.; Li, C.; Tasaka, M.; Furutani, M. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. USA 2016, 113, 6562–6567. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, D.; Villalobos, L.A.; Abel, S. Structural biology of nuclear auxin action. Trends Plant Sci. 2016, 21, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Brunoud, G.; Wells, D.M.; Oliva, M.; Larrieu, A.; Mirabet, V.; Burrow, A.H.; Beeckman, T.; Kepinski, S.; Traas, J.; Bennett, M.J.; et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 2012, 482, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhang, L.; Ding, Z. 26S Proteasome: Hunter and prey in auxin signaling. Trends Plant Sci. 2016, 21, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.-J.; Han, X.-X.; Yin, L.-L.; Xing, M.-Q.; Xu, Z.-H.; Xue, H.-W. Arabidopsis proteasome regulator1 is required for auxin-mediated suppression of proteasome activity and regulates auxin signalling. Nat. Commun. 2016, 7, 11388. [Google Scholar] [CrossRef] [PubMed]
- Gilkerson, J.; Kelley, D.R.; Tam, R.; Estelle, M.; Callis, J. Lysine residues are not required for proteasome-mediated proteolysis of the auxin/indole acidic acid protein IAA1. Plant Physiol. 2015, 168, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Park, Y.; Kim, I.; Kim, E.-H.; Yu, T.-K.; Rhee, S.; Suh, J.-Y. Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc. Natl. Acad. Sci. USA 2014, 111, 18613–18618. [Google Scholar] [CrossRef] [PubMed]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Dezfulian, M.H.; Jalili, E.; Roberto, D.A.; Moss, B.L.; Khoo, K.; Nemhauser, J.L.; Crosby, W.L. Oligomerization of SCFTIR1 is essential for aux/iaa degradation and auxin signaling in Arabidopsis. PLoS Genet. 2016, 12, e1006301. [Google Scholar] [CrossRef] [PubMed]
- Chatr-aryamontri, A.; Oughtred, R.; Boucher, L.; Rust, J.; Chang, C.; Kolas, N.K.; O’Donnell, L.; Oster, S.; Theesfeld, C.; Sellam, A. The BioGRID interaction database: 2017 update. Nucl. Acids Res. 2017, 45, D369–D379. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Estelle, M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, W.; Wei, Y.; Ye, T. Integration of auxin/indole-3-acetic acid 17 and RGA-LIKE3 confers salt stress resistance through stabilization by nitric oxide in Arabidopsis. J. Exp. Bot. 2017, 68, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Terrile, M.C.; París, R.; Calderón-Villalobos, L.I.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongué, C.A. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Wang, S.; Xiang, D.; Venglat, P.; Shi, X.; Zang, Y.; Datla, R.; Xiao, W.; Wang, H. UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J. 2014, 80, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Fukaki, H.; Taniguchi, N.; Tasaka, M. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 2006, 48, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Zhu, J.K.; Yang, Z. Epigenetic modifications and plant hormone action. Mol. Plant 2016, 9, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Benhamed, M.; Bertrand, C.; Servet, C.; Zhou, D.-X. Arabidopsis GCN5, HD1, and TAF1/HAF2 Interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 2006, 18, 2893–2903. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Qi, L.; Li, Y.; Zhai, Q.; Li, C. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 2013, 25, 2102–2114. [Google Scholar] [CrossRef] [PubMed]
- Sellaro, R.; Yanovsky, M.J.; Casal, J.J. Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J. 2011, 68, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Sellaro, R.; Zurbriggen, M.D.; Casal, J.J. Multiple links between shade avoidance and auxin networks. J. Exp. Bot. 2017, 69, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid—And auxin-mediated signaling in jasmonic acid—Induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Shani, E.; Salehin, M.; Zhang, Y.; Sanchez, S.E.; Doherty, C.; Wang, R.; Mangado, C.; Song, L.; Tal, I.; Pisanty, O.; et al. Plant stress tolerance requires auxin-sensitive Aux/IAA transcriptional repressors. Curr. Biol. 2017, 27, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Weiste, C.; Pedrotti, L.; Selvanayagam, J.; Muralidhara, P.; Fröschel, C.; Novák, O.; Ljung, K.; Hanson, J.; Dröge-Laser, W. The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 2017, 13, e1006607. [Google Scholar] [CrossRef] [PubMed]
- Herud, O.; Weijers, D.; Lau, S.; Jürgens, G. Auxin responsiveness of the MONOPTEROS-BODENLOS module in primary root initiation critically depends on the nuclear import kinetics of the Aux/IAA inhibitor BODENLOS. Plant J. 2016, 85, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Q.; Sang, S.; Wei, Z.; Wang, P. Rice inositol polyphosphate kinase (OsIPK2) directly interacts with OsIAA11 to regulate lateral root formation. Plant Cell Physiol. 2017, 58, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Zhang, Z.; Wang, L.; Zheng, L.; Mao, W.; Li, M.; Wu, Y.; Wu, P.; Mo, X. OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 2013, 74, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Liu, H.; Ruan, S.; Ali, B.; Gill, R.; Ma, H.; Zheng, Z.; Zhou, W. A zinc finger protein, interacted with cyclophilin, affects root development via IAA pathway in rice. J. Integr. Plant Biol. 2017, 59, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Qin, Q.; Wang, Y.; Pu, Y.; Liu, L.; Wen, X.; Ji, S.; Wu, J.; Wei, C.; Ding, B.; et al. Rice Dwarf Virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 2016, 12, e1005847. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Hwang, I. Integration of multiple signaling pathways shapes the auxin response. J. Exp. Bot. 2017, 69, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Colón-Carmona, A.; Chen, D.L.; Yeh, K.-C.; Abel, S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000, 124, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Yang, X.; Zhang, J.; Liu, X.; Zheng, H.; Dong, G.; Nian, J.; Feng, J.; Xia, B.; Qian, Q. Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 2015, 6, 7395. [Google Scholar] [CrossRef] [PubMed]
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; de-la-Peña, C. Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-J.; Guo, H.-S. Cleavage of indole-3-acetic acid inducible28 mRNA by microRNA847 upregulates auxin signaling to modulate cell proliferation and lateral organ growth in Arabidopsis. Plant Cell 2015, 27, 574–590. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lee, S.; So, J.H.; Dharmasiri, S.; Dharmasiri, N.; Ge, L.; Jensen, C.; Hangarter, R.; Hobbie, L.; Estelle, M. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J. 2004, 40, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.C.; Soh, M.S.; Kang, B.J.; Furuya, M.; Nam, H.G. Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 1996, 9, 441–456. [Google Scholar] [CrossRef] [PubMed]
- Guseman, J.M.; Hellmuth, A.; Lanctot, A.; Feldman, T.P.; Moss, B.L.; Klavins, E.; Calderón Villalobos, L.I.A.; Nemhauser, J.L. Auxin-induced degradation dynamics set the pace for lateral root development. Development 2015, 142, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Walker, L.M.; Young, J.C.; Sonawala, A.; Timpte, C.; Estelle, M.; Reed, J.W. AXR2 Encodes a Member of the Aux/IAA Protein Family. Plant Physiol. 2000, 123, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Arase, F.; Nishitani, H.; Egusa, M.; Nishimoto, N.; Sakurai, S.; Sakamoto, N.; Kaminaka, H. IAA8 involved in lateral root formation interacts with the TIR1 auxin receptor and ARF transcription factors in Arabidopsis. PLoS ONE 2012, 7, e43414. [Google Scholar] [CrossRef] [PubMed]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.-E.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2009, 51, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T.; Benkova, E.; Bäurle, I.; Kientz, M.; Jürgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef] [PubMed]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Rouse, D.; Mackay, P.; Stirnberg, P.; Estelle, M.; Leyser, O. Changes in auxin response from mutations in an AUX/IAA gene. Science 1998, 279, 1371–1373. [Google Scholar] [CrossRef] [PubMed]
- Leyser, H.; Pickett, F.B.; Dharmasiri, S.; Estelle, M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 1996, 10, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Konishi, N.; Kanno, K.; Yamaya, T.; Kojima, S. Transcriptional repressor IAA17 is involved in nitrogen use by modulating cytosolic glutamine synthetase GLN1; 2 in Arabidopsis roots. Soil Sci. Plant Nutr. 2017, 63, 163–170. [Google Scholar] [CrossRef]
- Ploense, S.E.; Wu, M.-F.; Nagpal, P.; Reed, J.W. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 2009, 136, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Notaguchi, M.; Wolf, S.; Lucas, W.J. Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 2012, 54, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Tatematsu, K.; Kumagai, S.; Muto, H.; Sato, A.; Watahiki, M.K.; Harper, R.M.; Liscum, E.; Yamamoto, K.T. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 2004, 16, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Rogg, L.E.; Lasswell, J.; Bartel, B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 2001, 13, 465–480. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; Vassileva, V.; Parizot, B.; Demeulenaere, M.; Grunewald, W.; Audenaert, D.; Van Campenhout, J.; Overvoorde, P.; Jansen, L.; Vanneste, S. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010, 20, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Yamamoto, K.T. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 2008, 133, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Chaabouni, S.; Jones, B.; Delalande, C.; Wang, H.; Li, Z.; Mila, I.; Frasse, P.; Latche, A.; Pech, J.-C.; Bouzayen, M. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J. Exp. Bot. 2009, 60, 1349–1362. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, B.; Visser, R.; Bachem, C. Isolation and characterization of a novel potato Auxin/Indole-3-Acetic Acid family member (StIAA2) that is involved in petiole hyponasty and shoot morphogenesis. Plant Physiol. Biochem. 2006, 44, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Karlberg, A.; Antti, H.; Lopez-Vernaza, M.; Mellerowicz, E.; Perrot-Rechenmann, C.; Sandberg, G.; Bhalerao, R.P. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell 2008, 20, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, T.; Wang, R.; Zhao, Y. Recent advances in auxin research in rice and their implications for crop improvement. J. Exp. Bot. 2017, 69, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Mazzucato, A.; Cellini, F.; Bouzayen, M.; Zouine, M.; Mila, I.; Minoia, S.; Petrozza, A.; Picarella, M.E.; Ruiu, F.; Carriero, F. A TILLING allele of the tomato Aux/IAA9 gene offers new insights into fruit set mechanisms and perspectives for breeding seedless tomatoes. Mol. Breed. 2015, 35, 22. [Google Scholar] [CrossRef]
- Wang, H.; Jones, B.; Li, Z.; Frasse, P.; Delalande, C.; Regad, F.; Chaabouni, S.; Latché, A.; Pech, J.-C.; Bouzayen, M. The tomato Aux/IAA transcription factor IAA9 Is involved in fruit development and leaf morphogenesis. Plant Cell 2005, 17, 2676–2692. [Google Scholar] [CrossRef] [PubMed]
- Koenig, D.; Bayer, E.; Kang, J.; Kuhlemeier, C.; Sinha, N. Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 2009, 136, 2997–3006. [Google Scholar] [CrossRef] [PubMed]
- Ueta, R.; Abe, C.; Watanabe, T.; Sugano, S.S.; Ishihara, R.; Ezura, H.; Osakabe, Y.; Osakabe, K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 2017, 7, 507. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Yang, Y.; Ren, Z.; Audran-Delalande, C.; Mila, I.; Wang, X.; Song, H.; Hu, Y.; Bouzayen, M.; Li, Z. The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol. 2012, 194, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Bassa, C.; Audran, C.; Mila, I.; Cheniclet, C.; Chevalier, C.; Bouzayen, M.; Roustan, J.-P.; Chervin, C. The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant Cell Physiol. 2014, 55, 1969–1976. [Google Scholar] [CrossRef] [PubMed]
- Bassa, C.; Mila, I.; Bouzayen, M.; Audran-Delalande, C. Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol. 2012, 53, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Guillotin, B.; Etemadi, M.; Audran, C.; Bouzayen, M.; Becard, G.; Combier, J.-P. Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom). New Phytol. 2017, 213, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. OsIAA1, an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res. 2001, 8, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xu, Z.-F. Ectopic Overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) Gene OsIAA4 in Rice Induces Morphological Changes and Reduces Responsiveness to Auxin. Int. J. Mol. Sci. 2013, 14, 13645–13656. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Lee, D.-K.; Choi, Y.; Kim, J.-K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.X.; Liu, Y.; Liu, S.J.; Mao, C.Z.; Wu, Y.R.; Wu, P. A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol. Plant 2012, 5, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Kitomi, Y.; Inahashi, H.; Takehisa, H.; Sato, Y.; Inukai, Y. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 2012, 190, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Jun, N.; Gaohang, W.; Zhenxing, Z.; Huanhuan, Z.; Yunrong, W.; Ping, W. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 2011, 68, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Paschold, A.; Marcon, C.; Liu, S.; Tai, H.; Nestler, J.; Yeh, C.-T.; Opitz, N.; Lanz, C.; Schnable, P.S.; et al. The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J. Exp. Bot. 2014, 65, 4919–4930. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hu, Q.; Luo, S.; Li, Q.; Yang, X.; Wang, X.; Wang, S. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 2015, 6, 388. [Google Scholar] [CrossRef] [PubMed]
| Species | Protein No. | Truncated Protein No. | Complete Protein No. | Reference |
|---|---|---|---|---|
| Amborella trichopoda | 16 | 8 (50%) | 8 (50%) | [29] |
| Arabidopsis thaliana | 29 | 11 (38%) | 18 (62%) | [19] |
| Brachypodium distachyon | 27 | 8 (30%) | 19 (70%) | [29] |
| Brassica rapa | 55 | 18 (33%) | 37 (67%) | [46] |
| Brassica napus | 119 | 75 (63%) | 44 (37%) | [36] |
| Carica papaya | 18 | 8 (44%) | 10 (56%) | [47] |
| Cicer arietinum | 22 | 10 (45%) | 12 (55%) | [37] |
| Citrus | 26 | 3 (12%) | 23 (88%) | [33] |
| Cucumis sativus | 27 | 9 (33%) | 18 (67%) | [25] |
| Eucalyptus grandis | 26 | 9 (35%) | 17 (65%) | [23] |
| Glycine max | 63 | 22 (35%) | 41 (65%) | [37] |
| Gossypium raimondii | 44 | 14 (32%) | 30 (68%) | [29] |
| Marchantia polymorpha | 1 | - | 1 (100%) | [41,42] |
| Medicago truncatula | 17 | 6 (35%) | 11 (65%) | [48] |
| Oryza sativa | 31 | 5 (16%) | 26 (84%) | [28] |
| Petunia hybrida | 16 | - | - | [40] |
| Phalaenopsis equestris | 16 | 8 (50%) | 8 (50%) | [29] |
| Physcomitrella patens | 3 | 1 (33%) | 2 (67%) | [29] |
| Picea abies | 31 | 23 (74%) | 8 (26%) | [29] |
| Populus trichocarpa | 35 | 11 (31%) | 24 (69%) | [26] |
| Prunus persica | 24 | 8 (33%) | 16 (67%) | [29] |
| Ricinus communis | 21 | 8 (38%) | 13 (62%) | [29] |
| Selaginella moellendorffii | 9 | 7 (77%) | 2 (22%) | [40] |
| Solanum lycopersicum | 26 | 3 (12%) | 23 (88%) | [24] |
| Solanum melongena | 16 | - | - | [40] |
| Solanum tuberosum | 26 | 5 (19%) | 21 (81%) | [34] |
| Sorghum bicolor | 26 | 2 (8%) | 24 (92%) | [49] |
| Triticum aestivum | 84 | 34 (40%) | 50 (60%) | [50] |
| Triticum urartu | 27 | - | - | [50] |
| Utricularia gibba | 25 | 7 (28%) | 18 (72%) | [29] |
| Vitis vinifera | 26 | 0 (0%) | 26 (100%) | [35] |
| Zea mays | 34 | 10 (29%) | 24 (71%) | [27] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. https://doi.org/10.3390/ijms19010259
Luo J, Zhou J-J, Zhang J-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. International Journal of Molecular Sciences. 2018; 19(1):259. https://doi.org/10.3390/ijms19010259
Chicago/Turabian StyleLuo, Jie, Jing-Jing Zhou, and Jin-Zhi Zhang. 2018. "Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function" International Journal of Molecular Sciences 19, no. 1: 259. https://doi.org/10.3390/ijms19010259
APA StyleLuo, J., Zhou, J.-J., & Zhang, J.-Z. (2018). Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. International Journal of Molecular Sciences, 19(1), 259. https://doi.org/10.3390/ijms19010259







