T-DNA Tagging-Based Gain-of-Function of OsHKT1;4 Reinforces Na Exclusion from Leaves and Stems but Triggers Na Toxicity in Roots of Rice Under Salt Stress
Abstract
:1. Introduction
2. Results
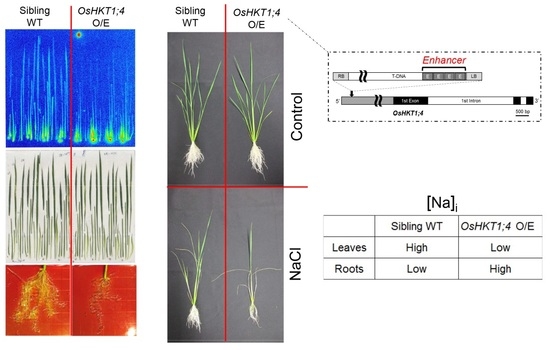
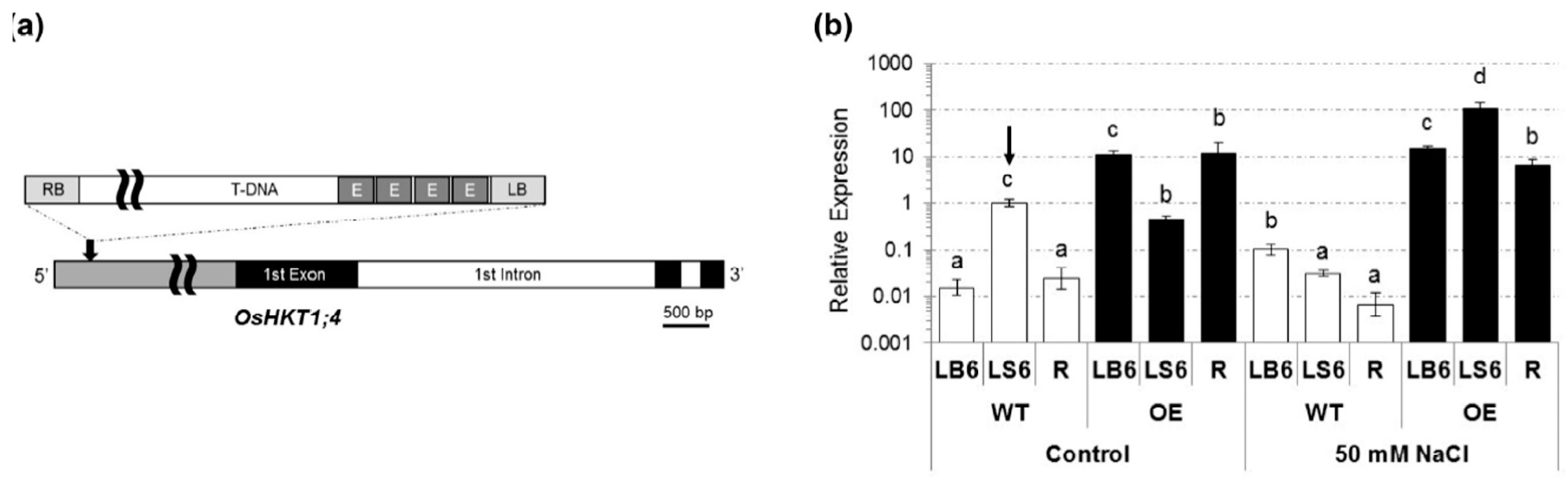
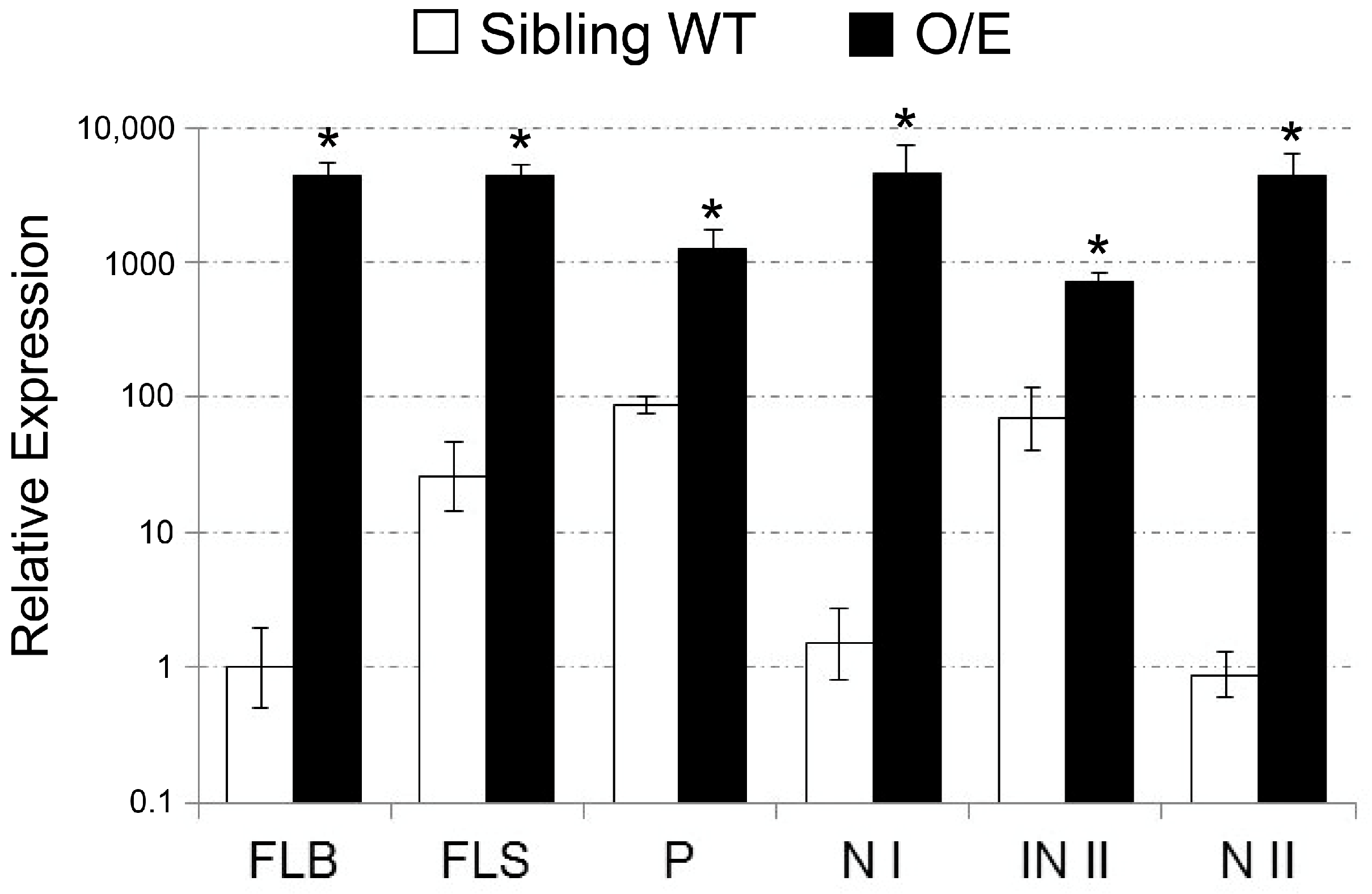
2.1. Isolation of a T-DNA Insertion Mutant of Rice Overexpressing OsHKT1;4
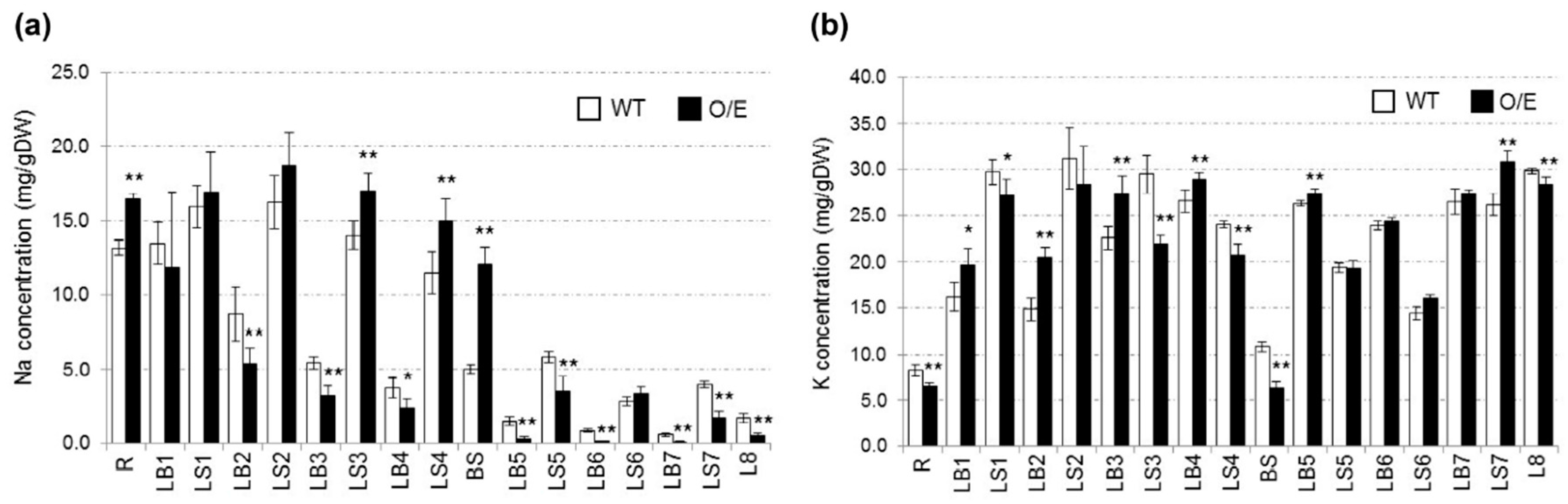
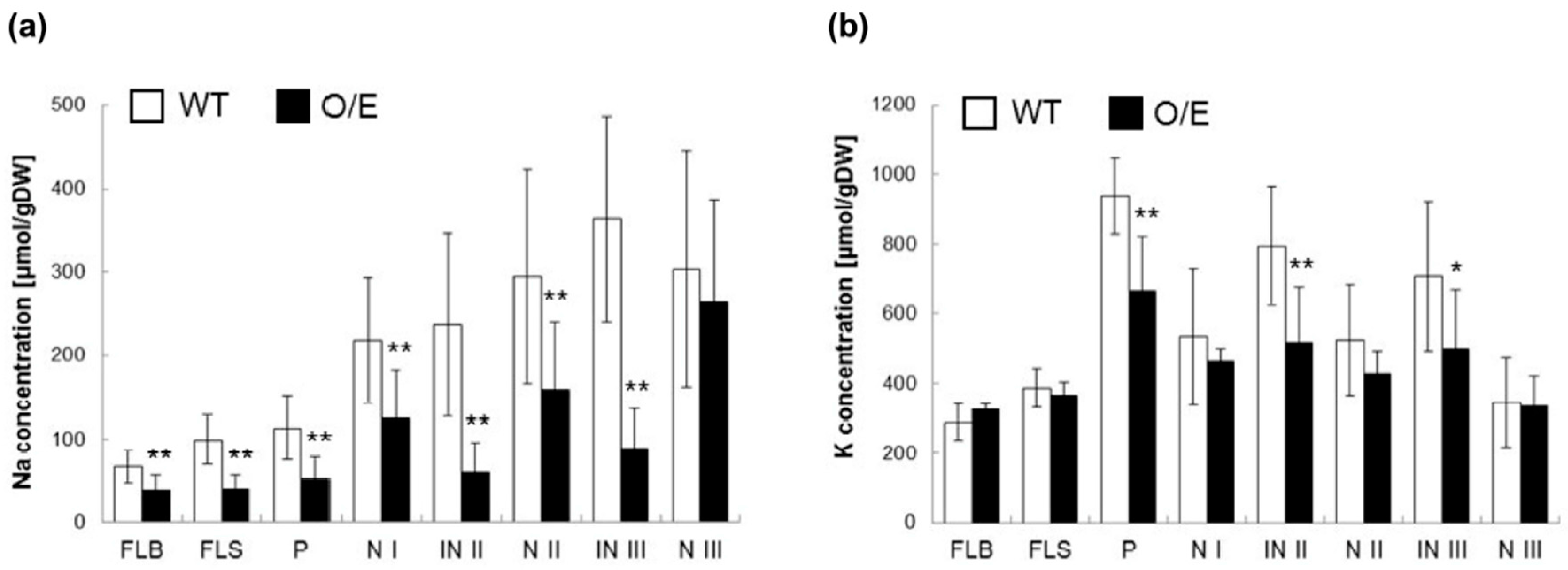
2.2. OsHKT1;4 Overexpression Changes the Patterns of Na and K Accumulations in Tissues of Rice
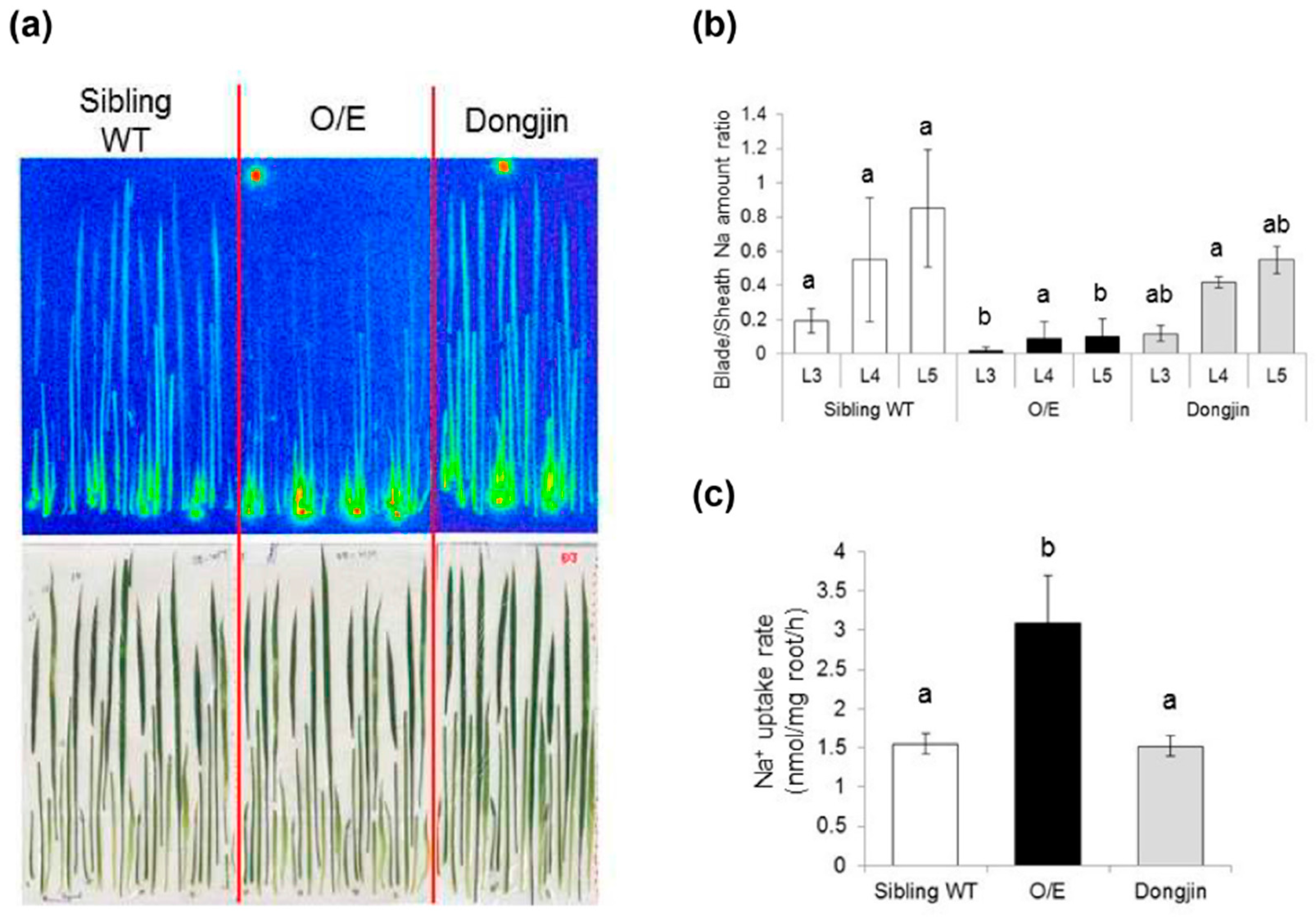
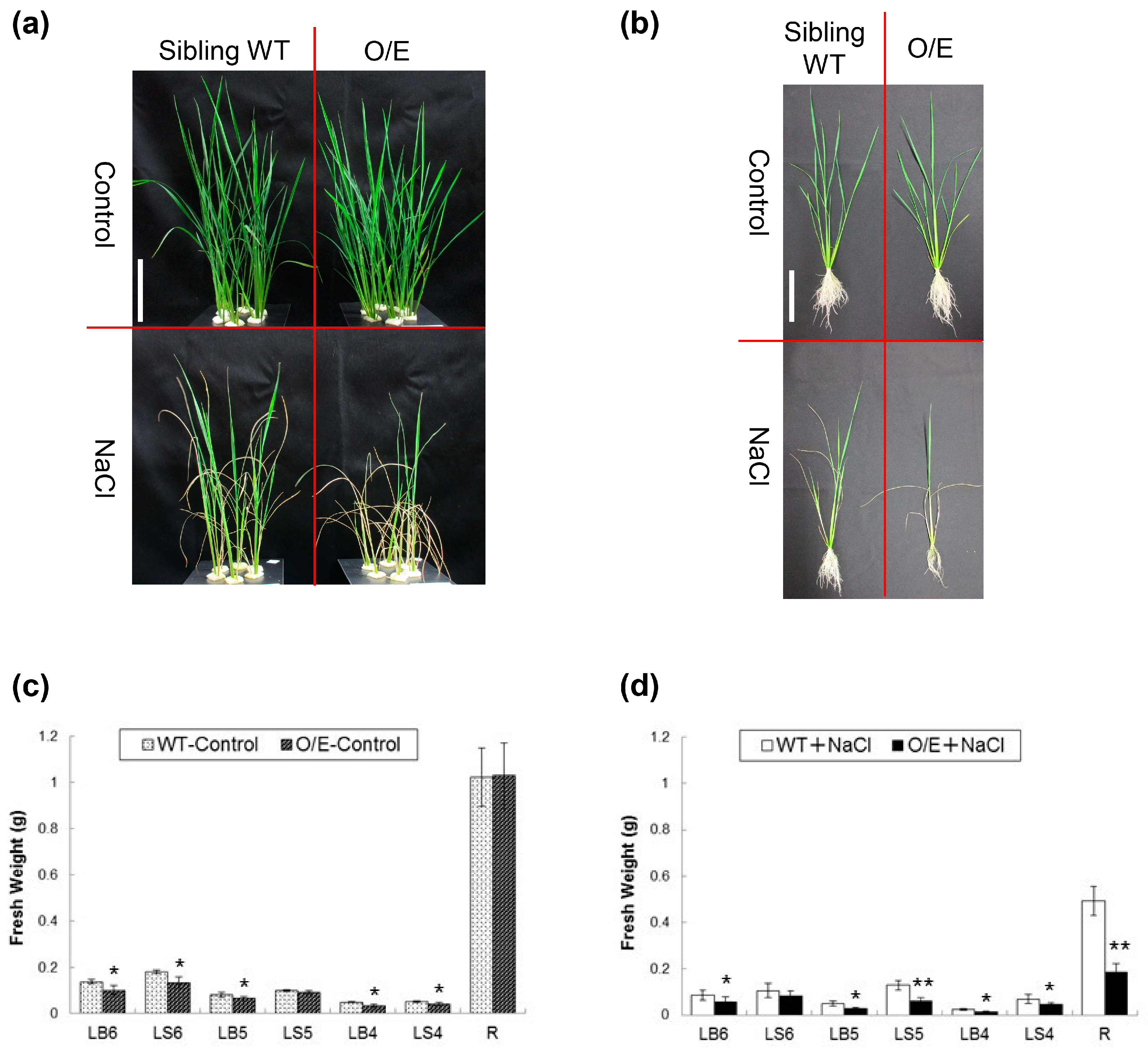
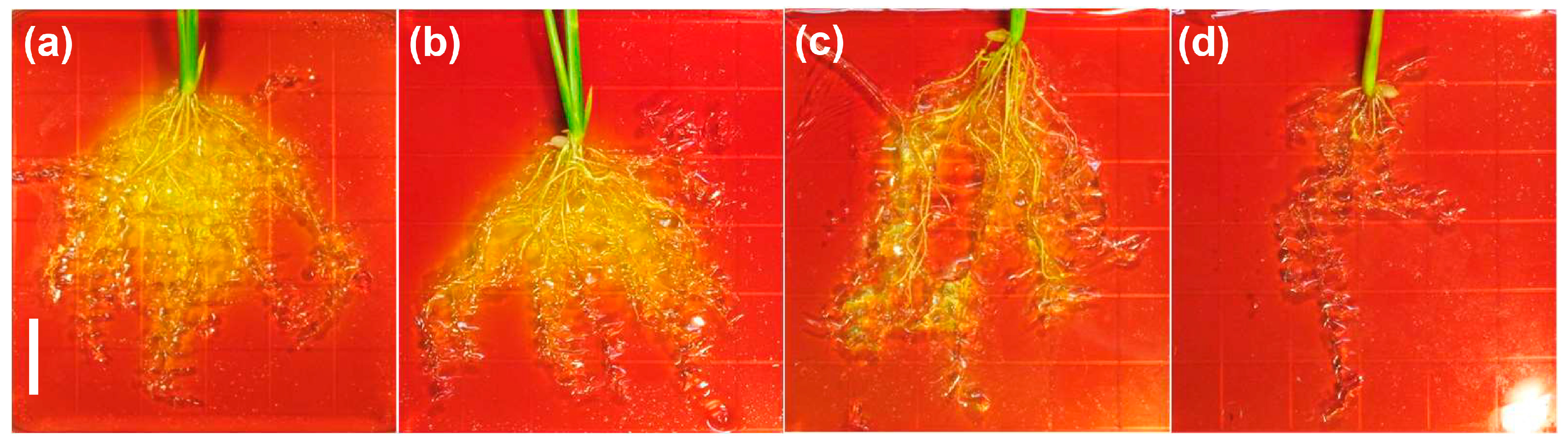
2.3. OsHKT1;4 O/E Rice Shows Increased Sensitivity to Salt Stress
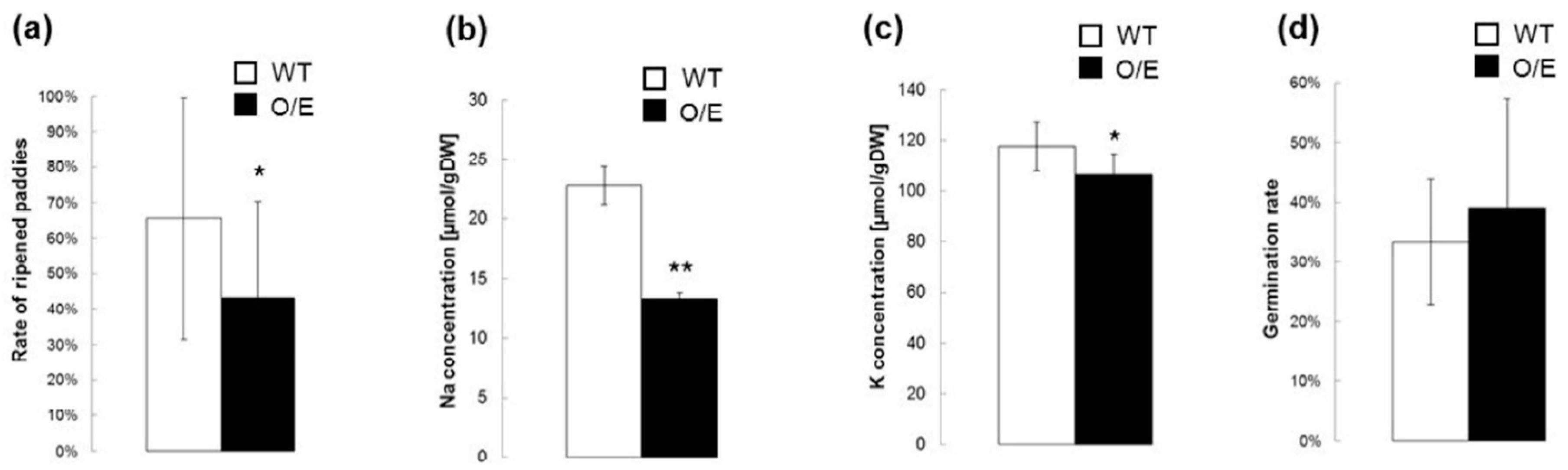
2.4. Influences of OsHKT1;4 Overexpression on Phenotypes of Rice Lines That Are in the Reproductive Growth Stage Under Salt Stress
3. Discussion
3.1. Influences of OsHKT1;4 Overexpression on the Salt Sensitivity and the Distribution of Na+ and K+ of the Rice Mutant Under Salt Stress
3.2. Implications toward the Breeding of Salt Tolerant Crops by the Reinforcement of the HKT1-Mediated Na+ Exclusion System
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Total RNA Extraction and Real-Time PCR analysis
4.3. Measurements of Na and K Concentrations in Tissues and Seeds of Rice
4.4. 22Na Tracer Experiment
4.5. Detection of Rhizosphere Acidification Using a pH Indicator
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HKT | high affinity K+ transporter |
| ICP-MS | inductively coupled plasma-mass spectrometry |
| ICP-OES | inductively coupled plasma-optical emission spectrometry |
| PCR | polymerase chain reaction |
| QTL | quantitative trait locus |
| RNAi | RNA interference |
| T-DNA | transfer DNA |
References
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1123. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice 2012, 5. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Ann. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Ann. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Berthomieu, P.; Conejero, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F.; et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003, 22, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Mäser, P.; Eckelman, B.; Vaidyanathan, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, K.; Yamaguchi, K.; Nishimura, M.; Uozumi, N.; et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsubota, A.; Bakker, E.P.; Nakamura, T.; Schroeder, J.I. The Arabidopsis HKT1 gene homologue mediates inward Na+ currents in Xenopus oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000, 121, 1249–1259. [Google Scholar] [CrossRef]
- Davenport, R.J.; Munoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.J.; Coates, J.C.; Haseloff, J.; Tester, M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar] [CrossRef] [PubMed]
- Sunarpi; Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.Y.; Leung, H.Y.; Hattori, K.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 44, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef] [PubMed]
- James, R.A.; Davenport, R.J.; Munns, R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 2006, 142, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.; Munns, R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007, 143, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Spielmeyer, W.; Lagudah, E.S.; James, R.A.; Platten, J.D.; Dennis, E.S.; Munns, R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006, 142, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Jabnoune, M.; Espeout, S.; Mieulet, D.; Fizames, C.; Verdeil, J.L.; Conejero, G.; Rodriguez-Navarro, A.; Sentenac, H.; Guiderdoni, E.; Abdelly, C.; et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009, 150, 1955–1971. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.T.; Bandillo, N.; Al Shiblawi, F.R.A.; Sharma, S.; Liu, K.; Du, Q.; Schmitz, A.J.; Zhang, C.; Very, A.A.; Lorenz, A.J.; et al. Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLoS Genet. 2017, 13, e1006823. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- Cotsaftis, O.; Plett, D.; Shirley, N.; Tester, M.; Hrmova, M. A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 2012, 7, e39865. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Yamaji, N.; Costa, A.; Okuma, E.; Kobayashi, N.I.; Kashiwagi, T.; Katsuhara, M.; Wang, C.; Tanoi, K.; Murata, Y.; et al. OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 2016, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.H.; An, S.; Kang, H.G.; Moon, S.; Han, J.J.; Park, S.; Lee, H.S.; An, K.; An, G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002, 130, 1636–1644. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K/Na ratio in leaves during salinity stress. Plant Cell Environ. 2010, 33, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Cuin, T.A. Potassium transport and plant salt tolerance. Physiol. Plant 2008, 133, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Shabala, L.; Zhou, M.; Shabala, S. Durum and bread wheat differ in their ability to retain potassium in leaf mesophyll: Implications for salinity stress tolerance. Plant Cell Physiol. 2014, 55, 1749–1762. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhu, M.; Shabala, L.; Zhou, M.; Shabala, S. K+ retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: A case study for barley. J. Integr. Plant Biol. 2015, 57, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Ma, J.F. The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci. 2014, 19, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Janda, T.; Darko, E.; Shehata, S.; Kovacs, V.; Pal, M.; Szalai, G. Salt acclimation processes in wheat. Plant Physiol. Biochem. 2016, 101, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Waters, S.; Byrt, C.S.; Plett, D.; Tyerman, S.D.; Tester, M.; Munns, R.; Hrmova, M.; Gilliham, M. Structural variations in wheat HKT1;5 underpin differences in Na+ transport capacity. Cell. Mol. Life Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Yoshida, K.; Nakayama, H.; Yamada, K.; Oiki, S.; Shinmyo, A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001, 27, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Sasaki, A.; Xia, J.X.; Yokosho, K.; Ma, J.F. A node-based switch for preferential distribution of manganese in rice. Nat. Commun. 2013, 4, 2442. [Google Scholar] [CrossRef] [PubMed]
- Rao, T.P.; Yano, K.; Iijima, M.; Yamauchi, A.; Tatsumi, J. Regulation of rhizosphere acidification by photosynthetic activity in cowpea (Vigna unguiculata L. walp.) seedlings. Ann. Bot. 2002, 89, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhu, Y.; Muller, C.; Zorb, C.; Schubert, S. Adaptation of H+-pumping and plasma membrane H+ ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiol. 2002, 129, 50–63. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oda, Y.; Kobayashi, N.I.; Tanoi, K.; Ma, J.F.; Itou, Y.; Katsuhara, M.; Itou, T.; Horie, T. T-DNA Tagging-Based Gain-of-Function of OsHKT1;4 Reinforces Na Exclusion from Leaves and Stems but Triggers Na Toxicity in Roots of Rice Under Salt Stress. Int. J. Mol. Sci. 2018, 19, 235. https://doi.org/10.3390/ijms19010235
Oda Y, Kobayashi NI, Tanoi K, Ma JF, Itou Y, Katsuhara M, Itou T, Horie T. T-DNA Tagging-Based Gain-of-Function of OsHKT1;4 Reinforces Na Exclusion from Leaves and Stems but Triggers Na Toxicity in Roots of Rice Under Salt Stress. International Journal of Molecular Sciences. 2018; 19(1):235. https://doi.org/10.3390/ijms19010235
Chicago/Turabian StyleOda, Yuuka, Natsuko I. Kobayashi, Keitaro Tanoi, Jian Feng Ma, Yukiko Itou, Maki Katsuhara, Takashi Itou, and Tomoaki Horie. 2018. "T-DNA Tagging-Based Gain-of-Function of OsHKT1;4 Reinforces Na Exclusion from Leaves and Stems but Triggers Na Toxicity in Roots of Rice Under Salt Stress" International Journal of Molecular Sciences 19, no. 1: 235. https://doi.org/10.3390/ijms19010235
APA StyleOda, Y., Kobayashi, N. I., Tanoi, K., Ma, J. F., Itou, Y., Katsuhara, M., Itou, T., & Horie, T. (2018). T-DNA Tagging-Based Gain-of-Function of OsHKT1;4 Reinforces Na Exclusion from Leaves and Stems but Triggers Na Toxicity in Roots of Rice Under Salt Stress. International Journal of Molecular Sciences, 19(1), 235. https://doi.org/10.3390/ijms19010235