Conditions Inducing Excessive O-GlcNAcylation Inhibit BMP2-Induced Osteogenic Differentiation of C2C12 Cells
Abstract
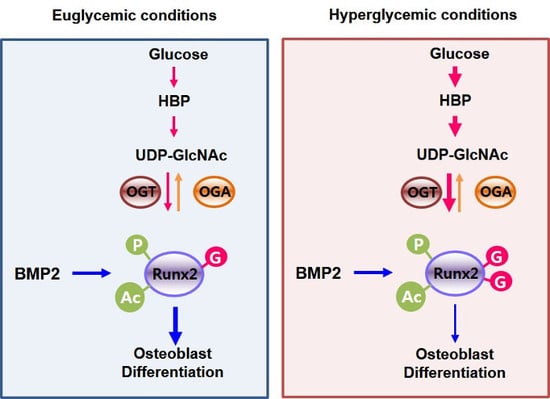
1. Introduction
2. Results
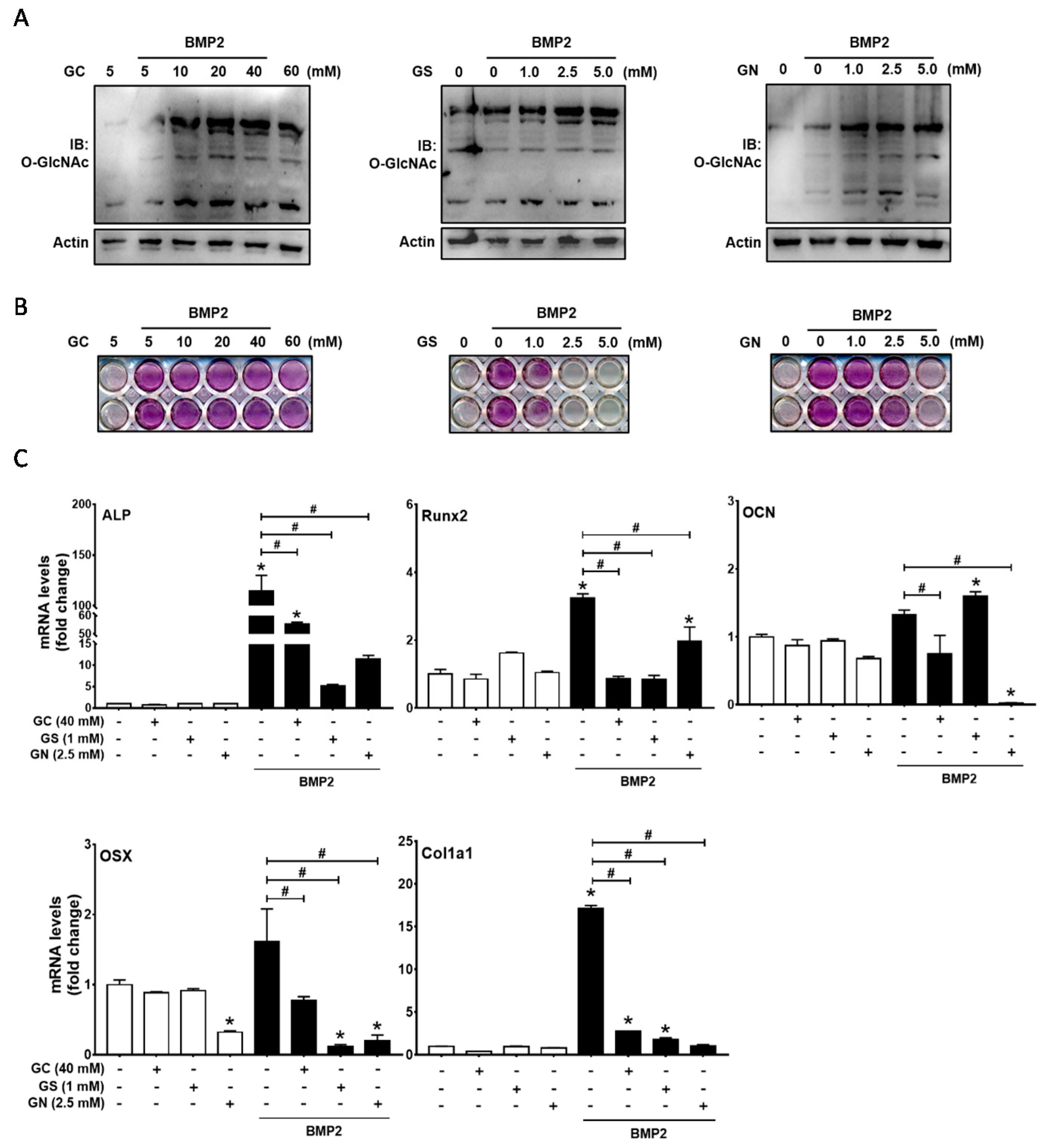
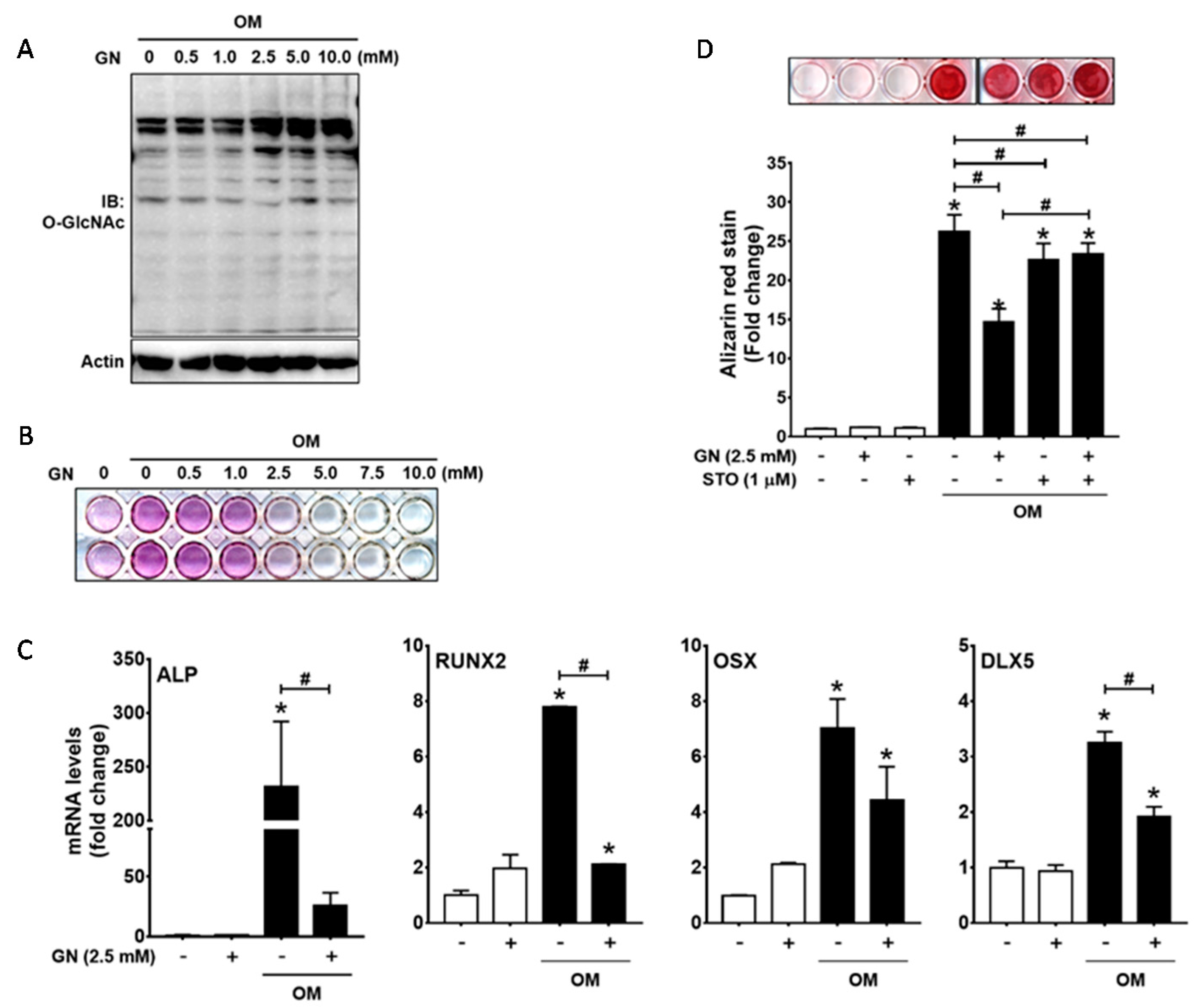
2.1. Excessive O-GlcNAcylation-Inducing Conditions Suppressed BMP2-Induced ALP Activity and Osteogenic Marker Gene Expression
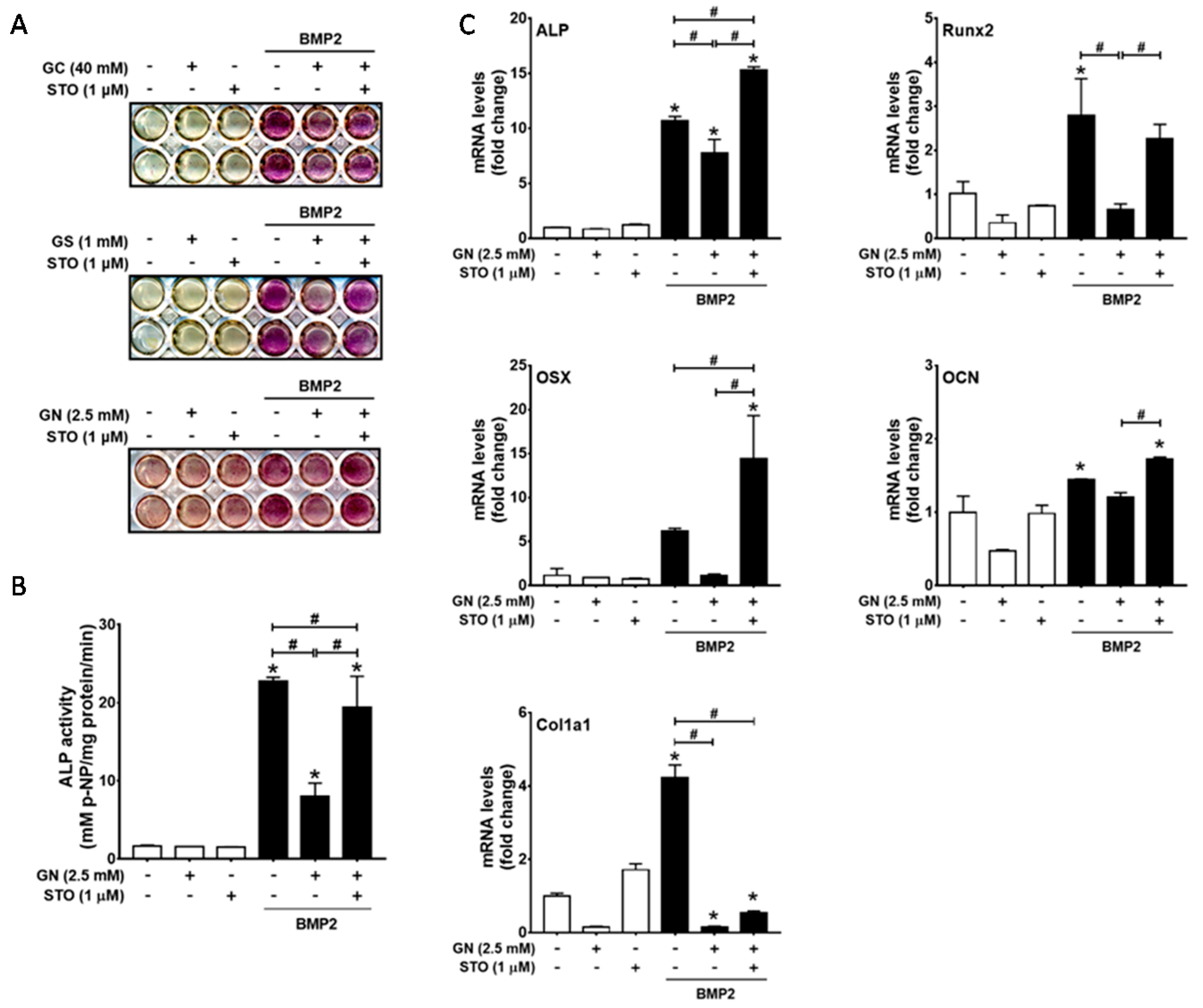
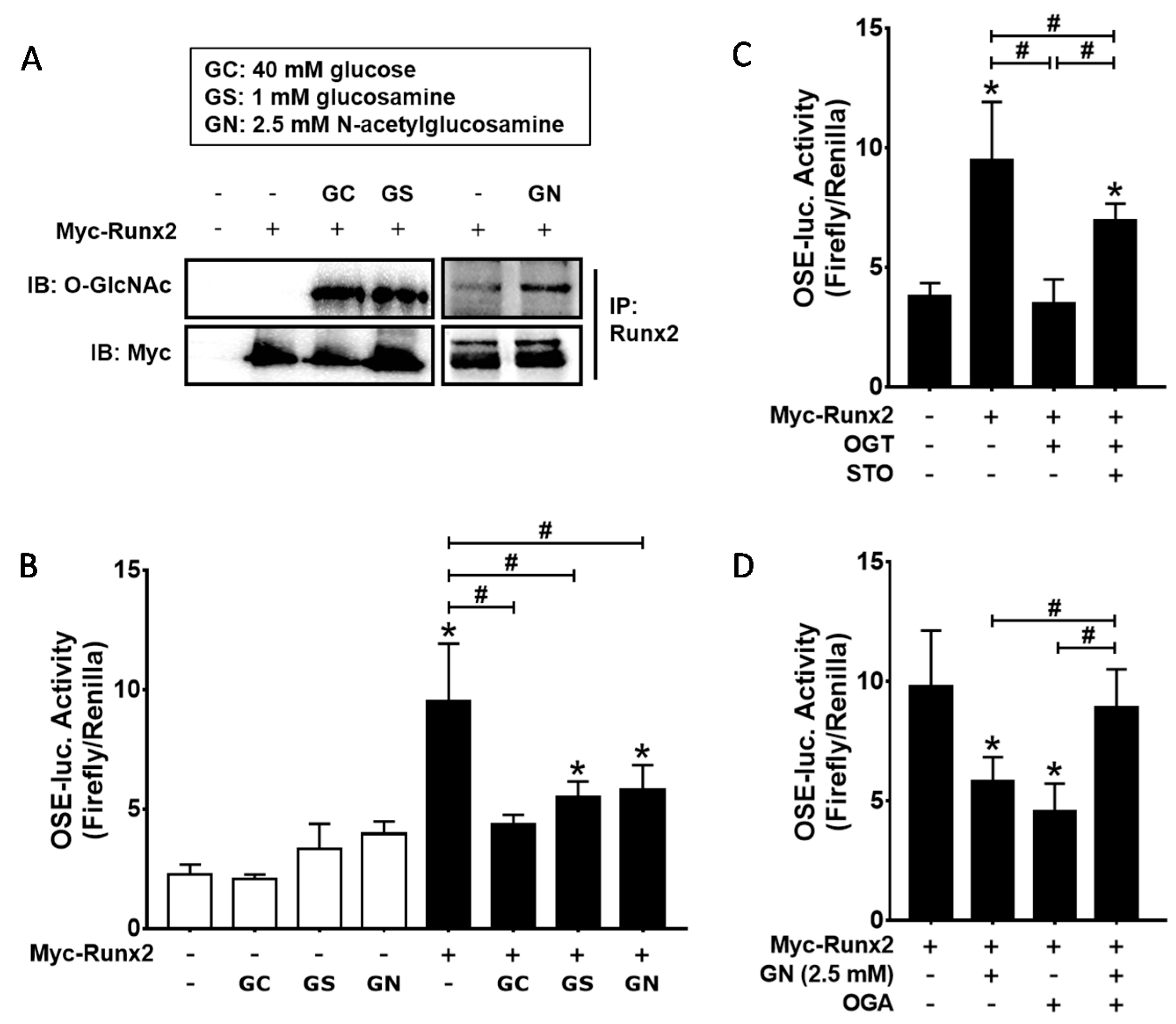
2.2. Inhibition of OGT Activity Rescued BMP2-Induced Osteogenic Differentiation that was Downregulated by O-GlcNAcylation Inducers
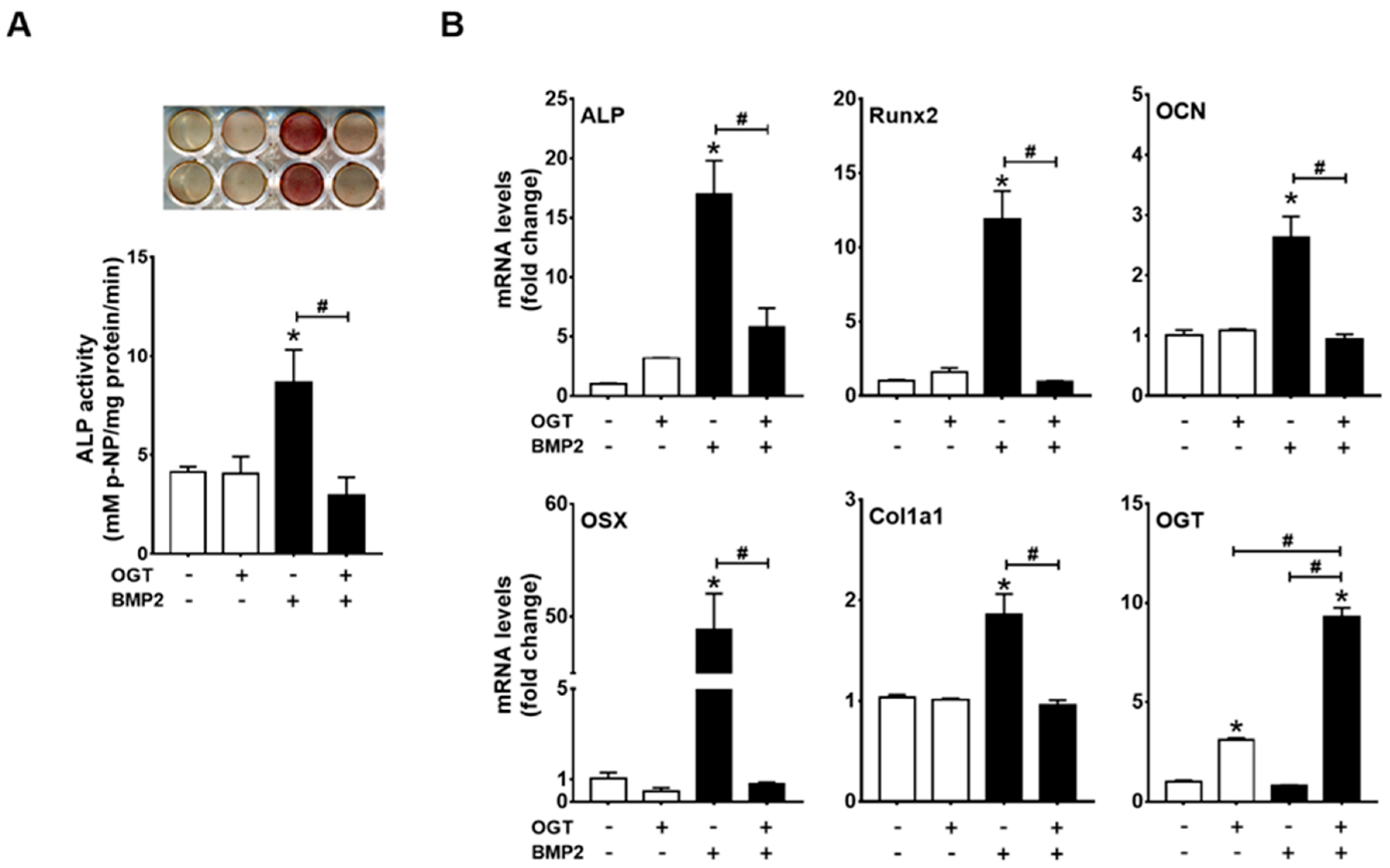
2.3. OGT Overexpression Inhibited BMP2-Induced Osteogenic Differentiation, whereas OGA Overexpression Rescued N-Acetylglucosamine-Inhibited Osteogenic Differentiation
2.4. Excessive O-GlcNAcylation Suppressed Runx2 Transcriptional Activity, Which Was Restored by OGT Inhibitor Addition or OGA Overexpression
2.5. Excessive O-GlcNAcylation Attenuated Osteogenic Differentiation and Matrix Mineralization of Human Periodontal Ligament Cells
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Reagents
4.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
4.3. Western Blot Analysis and Immunoprecipitation
4.4. ALP Staining, ALP Activity Assays, and Alizarin Red S Staining
4.5. Luciferase Reporter Assays
4.6. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Botolin, S.; McCabe, L.R. Chronic hyperglycemia modulates osteoblast gene expression through osmotic and non-osmotic pathways. J. Cell. Biochem. 2006, 99, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, V.; Romagnoli, E.; D’Erasmo, L.; D’Erasmo, E. Bone damage in type 2 diabetes mellitus. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Hart, G.W. Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu. Rev. Biochem. 1997, 66, 315–335. [Google Scholar] [CrossRef] [PubMed]
- Vosseller, K.; Sakabe, K.; Wells, L.; Hart, G.W. Diverse regulation of protein function by O-GlcNAc: A nuclear and cytoplasmic carbohydrate post-translational modification. Curr. Opin. Chem. Biol. 2002, 6, 851–857. [Google Scholar] [CrossRef]
- Ma, J.; Hart, G.W. O-GlcNAc profiling: From proteins to proteomes. Clin. Proteom. 2014, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Roquemore, E.P.; Chevrier, M.R.; Cotter, R.J.; Hart, G.W. Dynamic O-GlcNAcylation of the small heat shock protein alpha B-crystallin. Biochemistry 1996, 35, 3578–3586. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.S.; Lagerlof, O.; Hart, G.W. Roles of O-GlcNAc in chronic diseases of aging. Mol. Asp. Med. 2016, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Shafi, R.; Iyer, S.P.; Ellies, L.G.; O’Donnell, N.; Marek, K.W.; Chui, D.; Hart, G.W.; Marth, J.D. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 2000, 97, 5735–5739. [Google Scholar] [CrossRef] [PubMed]
- Keembiyehetty, C.; Love, D.C.; Harwood, K.R.; Gavrilova, O.; Comly, M.E.; Hanover, J.A. Conditional knock-out reveals a requirement for O-linked N-acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J. Biol. Chem. 2015, 290, 7097–7113. [Google Scholar] [CrossRef] [PubMed]
- Farook, V.S.; Bogardus, C.; Prochazka, M. Analysis of MGEA5 on 10q24.1-q24.3 encoding the beta-O-linked N-acetylglucosaminidase as a candidate gene for type 2 diabetes mellitus in Pima Indians. Mol. Genet. Metab. 2002, 77, 189–193. [Google Scholar] [PubMed]
- Lehman, D.M.; Fu, D.J.; Freeman, A.B.; Hunt, K.J.; Leach, R.J.; Johnson-Pais, T.; Hamlington, J.; Dyer, T.D.; Arya, R.; Abboud, H.; et al. A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-beta-d glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes 2005, 54, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Kudlow, J.E. The O-GlcNAcase theory of diabetes: Commentary on a candidate gene for diabetes. Mol. Genet. Metab. 2002, 77, 1–2. [Google Scholar] [CrossRef]
- Qin, C.X.; Sleaby, R.; Davidoff, A.J.; Bell, J.R.; De Blasio, M.J.; Delbridge, L.M.; Chatham, J.C.; Ritchie, R.H. Insights into the role of maladaptive hexosamine biosynthesis and O-GlcNAcylation in development of diabetic cardiac complications. Pharmacol. Res. 2017, 116, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, K.; Wells, L. Multiple tissue-specific roles for the O-GlcNAc post-translational modification in the induction of and complications arising from type ii diabetes. J. Biol. Chem. 2014, 289, 34466–34471. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shang, J.; Yao, Y.; Yin, X.; Liu, M.; Liu, H.; Zhou, Y. O-GlcNAcylation: A bridge between glucose and cell differentiation. J. Cell. Mol. Med. 2016, 20, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, Y.H.; Song, M.; An, S.H.; Byun, H.Y.; Heo, K.; Lim, S.; Oh, Y.S.; Ryu, S.H.; Suh, P.G. O-GlcNAc modification modulates the expression of osteocalcin via OSE2 and Runx2. Biochem. Biophys. Res. Commun. 2007, 362, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Nagel, A.K.; Ball, L.E. O-GlcNAc modification of the runt-related transcription factor 2 (Runx2) links osteogenesis and nutrient metabolism in bone marrow mesenchymal stem cells. Mol. Cell. Proteom. 2014, 13, 3381–3395. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Kamemura, K. Global increase in O-linked N-acetylglucosamine modification promotes osteoblast differentiation. Exp. Cell Res. 2015, 338, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Yamaguchi, A.; Komaki, M.; Abe, E.; Takahashi, N.; Ikeda, T.; Rosen, V.; Wozney, J.M.; Fujisawa-Sehara, A.; Suda, T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 1994, 127, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- King, G.L. The role of inflammatory cytokines in diabetes and its complications. J. Periodontol. 2008, 79, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Oei, L.; Rivadeneira, F.; Zillikens, M.C.; Oei, E.H. Diabetes, diabetic complications, and fracture risk. Curr. Osteoporos. Rep. 2015, 13, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Duarte, V.M.; Ramos, A.M.; Rezende, L.A.; Macedo, U.B.; Brandao-Neto, J.; Almeida, M.G.; Rezende, A.A. Osteopenia: A bone disorder associated with diabetes mellitus. J. Bone Miner. Metab. 2005, 23, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.V. Diabetes mellitus: Does it affect bone? Calcif. Tissue Int. 2003, 73, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhu, X.; Wang, Q.; Wang, L. Hyperglycemia induces endoplasmic reticulum stress-dependent CHOP expression in osteoblasts. Exp. Ther. Med. 2013, 5, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Boonanantanasarn, K.; Baek, K.; Woo, K.M.; Ryoo, H.M.; Baek, J.H.; Kim, G.S. Hyperglycemia increases the expression levels of sclerostin in a reactive oxygen species- and tumor necrosis factor-alpha-dependent manner. J. Periodontal Implant Sci. 2015, 45, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, B.; Li, Y.; Wang, D.; Lingling, E.; Bai, Y.; Liu, H. High glucose inhibits osteogenic differentiation through the BMP signaling pathway in bone mesenchymal stem cells in mice. EXCLI J. 2013, 12, 584–597. [Google Scholar] [PubMed]
- Komori, T. Runx2, a multifunctional transcription factor in skeletal development. J. Cell. Biochem. 2002, 87, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.R.; Kim, D.H.; Seo, Y.K.; Park, D.; Jang, H.J.; Choi, S.Y.; Lee, Y.H.; Lee, G.H.; Nakajima, K.; Taniguchi, N.; et al. Elevated O-GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF-κB signaling. Oncotarget 2015, 6, 12529–12542. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Yang, S.; Champattanachai, V.; Hu, S.; Chaudry, I.H.; Marchase, R.B.; Chatham, J.C. Glucosamine improves cardiac function following trauma-hemorrhage by increased protein O-GlcNAcylation and attenuation of NF-κB signaling. Am. J. Physiol. Heart Circ. Physiol. 2009, 296, H515–523. [Google Scholar] [CrossRef] [PubMed]
- Baudoin, L.; Issad, T. O-GlcNAcylation and inflammation: A vast territory to explore. Front. Endocrinol. 2014, 5, 235. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.D.; Hart, G.W. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J. Biol. Chem. 2008, 283, 13009–13020. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.D.; Sakabe, K.; Housley, M.P.; Dias, W.B.; Hart, G.W. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 2008, 283, 33935–33941. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Park, S.Y.; Nam, H.W.; Kim, D.H.; Kang, J.G.; Kang, E.S.; Kim, Y.S.; Lee, H.C.; Kim, K.S.; Cho, J.W. NF-kappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. USA 2008, 105, 17345–17350. [Google Scholar] [CrossRef] [PubMed]
- Boonanantanasarn, K.; Lee, H.L.; Baek, K.; Woo, K.M.; Ryoo, H.M.; Baek, J.H.; Kim, G.S. EGF inhibits Wnt/β-catenin-induced osteoblast differentiation by promoting β-catenin degradation. J. Cell. Biochem. 2015, 116, 2849–2857. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.H.; Yoon, W.J.; Seo, S.B.; Woo, K.M.; Kim, G.S.; Ryoo, H.M.; Baek, J.H. BMP2-activated Erk/MAP kinase stabilizes Runx2 by increasing p300 levels and histone acetyltransferase activity. J. Biol. Chem. 2010, 285, 36410–36419. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; Song, M.; Boonanantanasarn, K.; Baek, K.; Woo, K.M.; Ryoo, H.-M.; Baek, J.-H. Conditions Inducing Excessive O-GlcNAcylation Inhibit BMP2-Induced Osteogenic Differentiation of C2C12 Cells. Int. J. Mol. Sci. 2018, 19, 202. https://doi.org/10.3390/ijms19010202
Gu H, Song M, Boonanantanasarn K, Baek K, Woo KM, Ryoo H-M, Baek J-H. Conditions Inducing Excessive O-GlcNAcylation Inhibit BMP2-Induced Osteogenic Differentiation of C2C12 Cells. International Journal of Molecular Sciences. 2018; 19(1):202. https://doi.org/10.3390/ijms19010202
Chicago/Turabian StyleGu, Hanna, Mina Song, Kanitsak Boonanantanasarn, Kyunghwa Baek, Kyung Mi Woo, Hyun-Mo Ryoo, and Jeong-Hwa Baek. 2018. "Conditions Inducing Excessive O-GlcNAcylation Inhibit BMP2-Induced Osteogenic Differentiation of C2C12 Cells" International Journal of Molecular Sciences 19, no. 1: 202. https://doi.org/10.3390/ijms19010202
APA StyleGu, H., Song, M., Boonanantanasarn, K., Baek, K., Woo, K. M., Ryoo, H.-M., & Baek, J.-H. (2018). Conditions Inducing Excessive O-GlcNAcylation Inhibit BMP2-Induced Osteogenic Differentiation of C2C12 Cells. International Journal of Molecular Sciences, 19(1), 202. https://doi.org/10.3390/ijms19010202