Alternative Pre-mRNA Splicing in Mammals and Teleost Fish: A Effective Strategy for the Regulation of Immune Responses Against Pathogen Infection
Abstract
1. Introduction
2. Alternative Splicing and Immune Function of Peptidoglycan Recognition Proteins
3. Alternative Splicing and Immune Function of Nucleotide Binding and Oligomerization Domain-Like Receptors
3.1. NLRA Subfamily
3.2. NLRC Subfamily
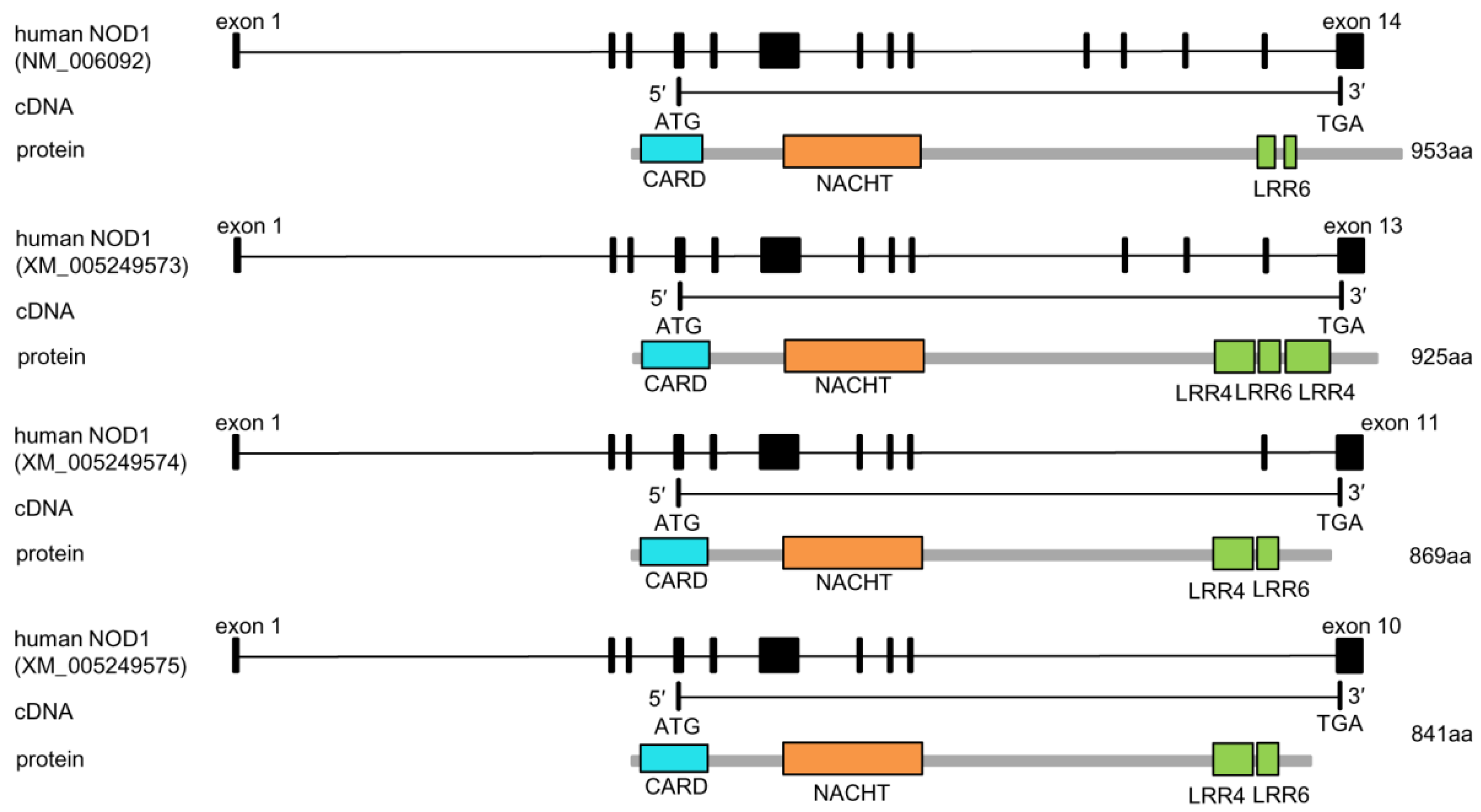
3.2.1. Nucleotide-Binding Oligomerization Domain-Containing Protein 1 and 2
3.2.2. NLR Family CARD Domain Containing 3
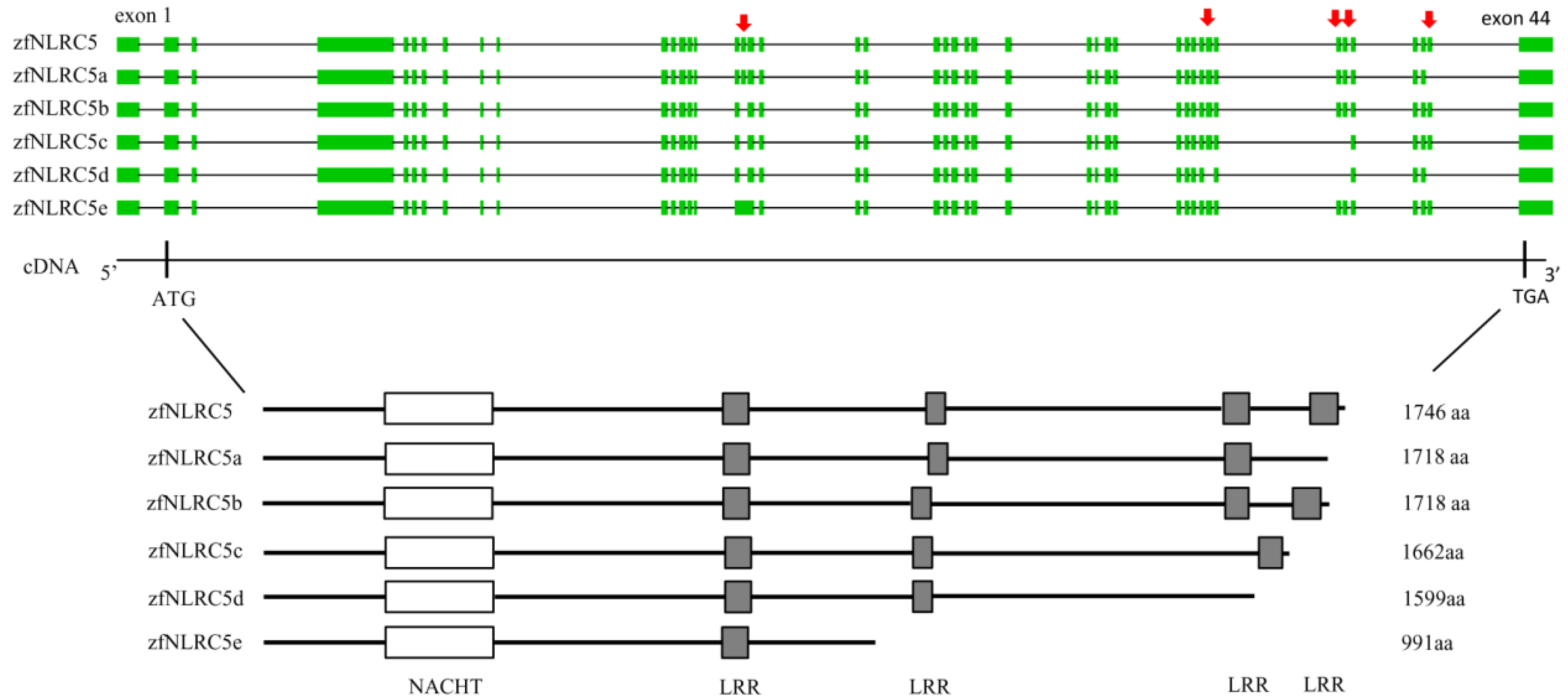
3.2.3. NLR Family CARD Domain Containing 5
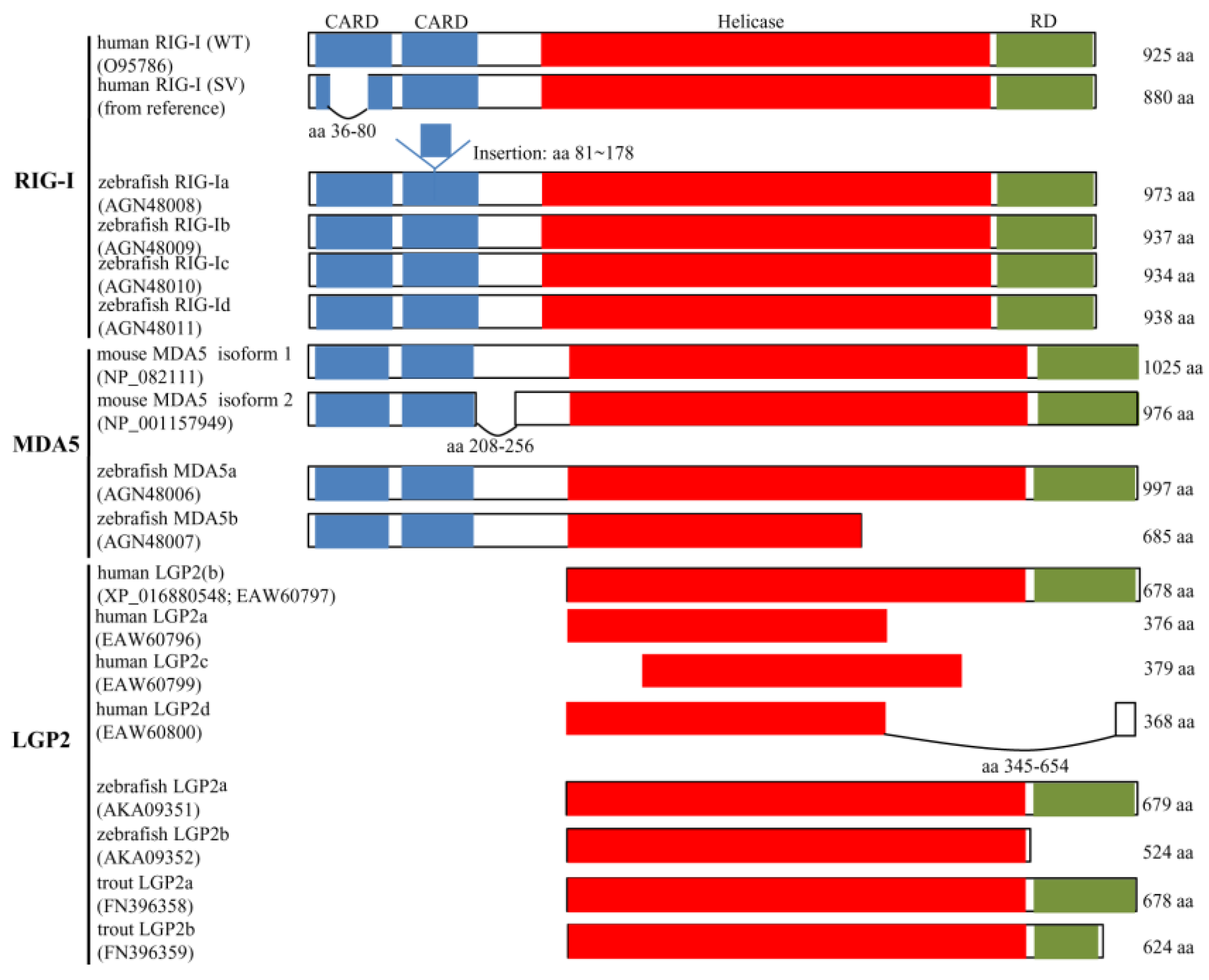
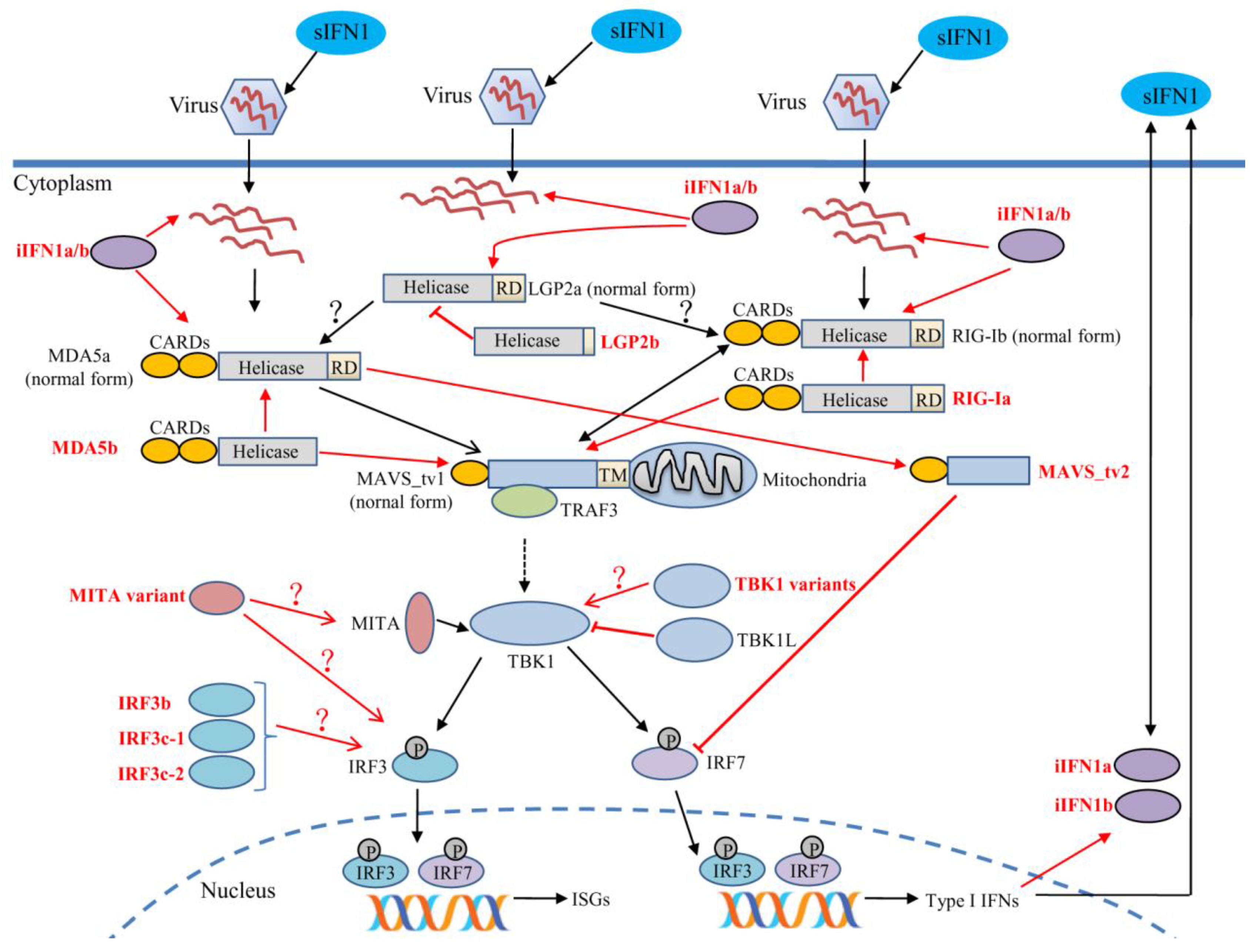
4. Alternative Splicing and Immune Function of Retinoic Acid-Inducible Gene-I-Like Receptors
5. Alternative Splicing and Immune Function of Downstream Signaling Molecules
5.1. Mitochondrial Antiviral Signaling Protein
5.2. Stimulator of Interferon Genes
5.3. TRAF Family Member-Associated NF-kappaB Activator (TANK) Binding Kinase 1
5.4. Interferon Regulatory Factor 3
5.5. Interferons and Their Receptors
6. Conclusions and Prospects
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Palm, N.W.; Medzhitov, R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009, 227, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Brennan, K.; Bowie, A.G. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 2010, 13, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016, 16, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, P. Alternative splicing takes shape during neuronal development. Curr. Opin. Genet. Dev. 2011, 21, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.K.; Black, D.L.; Zheng, S. The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 2016, 17, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Martinez, N.M.; Lynch, K.W. Control of alternative splicing in immune responses: Many regulators, many predictions, much still to learn. Immunol. Rev. 2013, 253, 216–236. [Google Scholar] [CrossRef] [PubMed]
- Faustino, N.A.; Cooper, T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003, 17, 419–437. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.F.; Yu, J.C.; Ho, L.I.; Chiu, S.C.; Harn, H.J. Molecular studies into the role of CD44 variants in metastasis in gastric cancer. Mol. Pathol. 1999, 52, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Olsson, E.; Honeth, G.; Bendahl, P.O.; Saal, L.H.; Gruvberger-Saal, S.; Ringnér, M.; Vallon-Christersson, J.; Jönsson, G.; Holm, K.; Lövgren, K.; et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer. 2011, 11, 418. [Google Scholar] [CrossRef] [PubMed]
- Prochazka, L.; Tesarik, R.; Turanek, J. Regulation of alternative splicing of CD44 in cancer. Cell Signal. 2014, 26, 2234–2239. [Google Scholar] [CrossRef] [PubMed]
- Moors, M.; Vudattu, N.K.; Abel, J.; Krämer, U.; Rane, L.; Ulfig, N.; Ceccatelli, S.; Seyfert-Margolies, V.; Fritsche, E.; Maeurer, M.J. Interleukin-7 (IL-7) and IL-7 splice variants affect differentiation of human neural progenitor cells. Genes Immun. 2010, 11, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, A.; Im, S.H. Interleukin and interleukin receptor diversity: Role of alternative splicing. Int. Rev. Immunol. 2010, 29, 77–109. [Google Scholar] [CrossRef] [PubMed]
- Shakola, F.; Suri, P.; Ruggiu, M. Splicing Regulation of pro-inflammatory cytokines and chemokines: At the interface of the neuroendocrine and immune systems. Biomolecules 2015, 5, 2073–2100. [Google Scholar] [CrossRef] [PubMed]
- Kurata, S. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 2014, 42, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Liu, G.; Kang, D.; Ekengren, S.; Steiner, H.; Hultmark, D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13772–13777. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Borge-Renberg, K.; Mellroth, P.; Steiner, H.; Hultmark, D. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J. Biol. Chem. 2003, 278, 26319–26322. [Google Scholar] [CrossRef] [PubMed]
- Neyen, C.; Poidevin, M.; Roussel, A.; Lemaitre, B. Tissue- and ligand-specific sensing of gram-negative infection in drosophila by PGRP-LC isoforms and PGRP-LE. J. Immunol. 2012, 189, 1886–1897. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Li, X.; Cocklin, R.R.; Wang, M.; Wang, M.; Fukase, K.; Inamura, S.; Kusumoto, S.; Gupta, D.; Dziarski, R. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-l-alanine amidase. J. Biol. Chem. 2003, 278, 49044–49052. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Gupta, D. Mammalian PGRPs: Novel antibacterial proteins. Cell Microbiol. 2006, 8, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006, 7, 232. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, M.; Qi, J.; Wang, H.; Li, X.; Gupta, D.; Dziarski, R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 2006, 281, 5895–5907. [Google Scholar] [CrossRef] [PubMed]
- Kibardin, A.V.; Mirkina, I.I.; Baranova, E.V.; Zakeyeva, I.R.; Georgiev, G.P.; Kiselev, S.L. The differentially spliced mouse tagL gene, homolog of tag7/PGRP gene family in mammals and Drosophila, can recognize gram-positive and gram-negative bacterial cell wall independently of T phage lysozyme homology domain. J. Mol. Biol. 2003, 326, 467–474. [Google Scholar] [CrossRef]
- Li, X.; Wang, S.; Qi, J.; Echtenkamp, S.F.; Chatterjee, R.; Wang, M.; Boons, G.J.; Dziarski, R.; Gupta, D. Zebrafish peptidoglycan recognition proteins are bactericidal amidases essential for defense against bacterial infections. Immunity 2007, 27, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Nie, P. RNAi suppression of zebrafish peptidoglycan recognition protein 6 (zfPGRP6) mediated differentially expressed genes involved in Toll-like receptor signaling pathway and caused increased susceptibility to Flavobacterium columnare. Vet. Immunol. Immunopathol. 2008, 124, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Wang, Y.P.; Nie, P. Zebrafish peptidoglycan recognition protein SC (zfPGRP-SC) mediates multiple intracellular signaling pathways. Fish Shellfish Immunol. 2009, 26, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Nie, P.; Wei, L.L. Short and long peptidoglycan recognition proteins (PGRPs) in zebrafish, with findings of multiple PGRP homologs in teleost fish. Mol. Immunol. 2007, 44, 3005–3023. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.L.; Li, J.H.; Xue, N.N.; Nie, P.; Chang, M.X. Expression and functional characterization of PGRP6 splice variants in grass carp Ctenopharyngodon idella. Dev. Comp. Immunol. 2014, 47, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Yu, Z.L.; Xue, N.N.; Zou, P.F.; Hu, J.Y.; Nie, P.; Chang, M.X. Molecular cloning and functional characterization of peptidoglycan recognition protein 6 in grass carp Ctenopharyngodon idella. Dev. Comp. Immunol. 2014, 42, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, Y.; Toma, C.; Higa, N.; Nohara, T.; Nakasone, N.; Suzuki, T. Inflammasome activation via intracellular NLRs triggered by bacterial infection. Cell Microbiol. 2012, 14, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.R.; Damania, B. NLRs, inflammasomes, and viral infection. J. Leukoc. Biol. 2012, 92, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Miao, E.A.; Ting, J.P. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity 2013, 39, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Lupfer, C.; Kanneganti, T.D. The expanding role of NLRs in antiviral immunity. Immunol. Rev. 2013, 255, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Parlato, M.; Yeretssian, G. NOD-like receptors in intestinal homeostasis and epithelial tissue repair. Int. J. Mol. Sci. 2014, 15, 9594–9627. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, L.A.; Travassos, L.H. The interplay between NLRs and autophagy in immunity and inflammation. Front. Immunol. 2013, 4, 361. [Google Scholar] [CrossRef] [PubMed]
- Van Gorp, H.; Kuchmiy, A.; Van Hauwermeiren, F.; Lamkanfi, M. NOD-like receptors interfacing the immune and reproductive systems. FEBS. J. 2014, 281, 4568–4582. [Google Scholar] [CrossRef] [PubMed]
- Ting, J.P.; Lovering, R.C.; Alnemri, E.S.; Bertin, J.; Boss, J.M.; Davis, B.K.; Flavell, R.A.; Girardin, S.E.; Godzik, A.; Harton, J.A.; et al. The NLR gene family: A standard nomenclature. Immunity 2008, 28, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.; Caccamo, M.; Laird, G.; Leptin, M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007, 8, R251. [Google Scholar] [CrossRef] [PubMed]
- Steimle, V.; Otten, L.A.; Zufferey, M.; Mach, B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 1993, 75, 135–146. [Google Scholar] [CrossRef]
- Muhlethaler-Mottet, A.; Otten, L.A.; Steimle, V.; Mach, B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO. J. 1997, 16, 2851–2860. [Google Scholar] [CrossRef] [PubMed]
- Riley, J.L.; Westerheide, S.D.; Price, J.A.; Brown, J.A.; Boss, J.M. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA). Immunity 1995, 2, 533–543. [Google Scholar] [CrossRef]
- Peijnenburg, A.; Van den Berg, R.; Van Eggermond, M.J.; Sanal, O.; Vossen, J.M.; Lennon, A.M.; Alcaïde-Loridan, C.; Van den Elsen, P.J. Defective MHC class II expression in an MHC class II deficiency patient is caused by a novel deletion of a splice donor site in the MHC class II transactivator gene. Immunogenetics 2000, 51, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Chiu, B.L.; Li, C.H.; Chang, C.C. Selective modulation of MHC class II chaperons by a novel IFN-γ-inducible class II transactivator variant in lung adenocarcinoma A549 cells. Biochem. Biophys. Res. Commun. 2013, 440, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Day, N.E.; Ugai, H.; Yokoyama, K.K.; Ichiki, A.T. K-562 cells lack MHC class II expression due to an alternatively spliced CIITA transcript with a truncated coding region. Leuk. Res. 2003, 27, 1027–1038. [Google Scholar] [CrossRef]
- Rajendran, K.V.; Zhang, J.; Liu, S.; Kucuktas, H.; Wang, X.; Liu, H.; Sha, Z.; Terhune, J.; Peatman, E.; Liu, Z. Pathogen recognition receptors in channel catfish: I Identification, phylogeny and expression of NOD-like receptors. Dev. Comp. Immunol. 2012, 37, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Meng, Y.; Wang, Q.; Sha, Z. Class II, major histocompatibility complex, transactivator (CIITA) in channel catfish: Identification and expression patterns responding to different pathogens. Mol. Biol. Rep. 2012, 39, 11041–11050. [Google Scholar] [CrossRef] [PubMed]
- LeibundGut-Landmann, S.; Waldburger, J.M.; Krawczyk, M.; Otten, L.A.; Suter, T.; Fontana, A.; Acha-Orbea, H.; Reith, W. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur. J. Immunol. 2004, 34, 1513–1525. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.W.; Wu, X.M.; Ren, S.S.; Cao, L.; Nie, P.; Chang, M.X. NOD1 deficiency impairs CD44a/LCK as well as PI3K/Akt pathway. Sci. Rep. 2017, 7, 2979. [Google Scholar] [CrossRef] [PubMed]
- Tattoli, I.; Travassos, L.H.; Carneiro, L.A.; Magalhaes, J.G.; Girardin, S.E. The Nodosome: NOD1 and NOD2 control bacterial infections and inflammation. Semin. Immunopathol. 2007, 29, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Clay, G.M.; Sutterwala, F.S.; Wilson, M.E. NLR proteins and parasitic disease. Immunol. Res. 2014, 59, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Coutermarsh-Ott, S.; Eden, K.; Allen, I.C. Beyond the inflammasome: Regulatory NOD-like receptor modulation of the host immune response following virus exposure. J. Gen. Virol. 2016, 97, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Inohara, N.; Benito, A.; Chen, F.F.; Yamaoka, S.; Nunez, G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 2001, 276, 4812–4818. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.; Hong, J.; Fraser, A.; Krissansen, G.W. Splicing of NOD2 (CARD15) RNA transcripts. Mol. Immunol. 2007, 44, 284–294. [Google Scholar] [CrossRef] [PubMed]
- King, K.; Bagnall, R.; Fisher, S.A.; Sheikh, F.; Cuthbert, A.; Tan, S.; Mundy, N.I.; Rosenstiel, P.; Schreiber, S.; Mathew, C.G.; et al. Identification, evolution, and association study of a novel promoter and first exon of the human NOD2 (CARD15) gene. Genomics 2007, 90, 493–501. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rosenstiel, P.; Huse, K.; Franke, A.; Hampe, J.; Reichwald, K.; Platzer, C.; Roberts, R.G.; Mathew, C.G.; Platzer, M.; Schreiber, S. Functional characterization of two novel 5′ untranslated exons reveals a complex regulation of NOD2 protein expression. BMC Genomics 2007, 8, 472. [Google Scholar] [CrossRef] [PubMed]
- Rosenstiel, P.; Huse, K.; Till, A.; Hampe, J.; Hellmig, S.; Sina, C.; Billmann, S.; von Kampen, O.; Waetzig, G.H.; Platzer, M.; et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc. Natl. Acad. Sci. USA 2006, 103, 3280–3285. [Google Scholar] [CrossRef] [PubMed]
- Oehlers, S.H.; Flores, M.V.; Hall, C.J.; Swift, S.; Crosier, K.E.; Crosier, P.S. The inflammatory bowel disease (IBD) susceptibility genes NOD1 and NOD2 have conserved anti-bacterial roles in zebrafish. Dis. Model. Mech. 2011, 4, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Wang, T.; Nie, P.; Zou, J.; Secombes, C.J. Cloning of two rainbow trout nucleotide-binding oligomerization domain containing 2 (NOD2) splice variants and functional characterization of the NOD2 effector domains. Fish Shellfish Immunol. 2011, 30, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Zimmermann, A.G.; Roberts, R.A.; Zhang, L.; Swanson, K.V.; Wen, H.; Davis, B.K.; Allen, I.C.; Holl, E.K.; Ye, Z.; et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat. Immunol. 2012, 13, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mo, J.; Swanson, K.V.; Wen, H.; Petrucelli, A.; Gregory, S.M.; Zhang, Z.; Schneider, M.; Jiang, Y.; Fitzgerald, K.A.; et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor sting. Immunity 2014, 40, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Conti, B.J.; Davis, B.K.; Zhang, J.; O′connor, W.; Williams, K.L.; Ting, J.P. CATERPILLER 16.2 (CLR16.2), a novel NBD/LRR family member that negatively regulates T cell function. J. Biol. Chem. 2005, 280, 18375–18385. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Man, S.M.; Malireddi, R.K.; Kesavardhana, S.; Zhu, Q.; Burton, A.R.; Sharma, B.R.; Qi, X.; Pelletier, S.; Vogel, P.; et al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature 2016. [Google Scholar] [CrossRef] [PubMed]
- Leavy, O. Tumour immunology: NLRC3 inhibits mTOR in colorectal cancer. Nat. Rev. Immunol. 2017, 17, 79. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Ye, Z.; Zhang, D.; Gao, C.; Su, B.; Song, L.; Tan, F.; Song, H.; Wang, Y.; Li, C. Characterization and expression profiling of NOD-like receptor C3 (NLRC3) in mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish Shellfish Immunol. 2017, 66, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, C.A.; Ramírez-Cepeda, F.; Santana, P.; Torres, E.; Cortés, J.; Guzmán, F.; Schmitt, P.; Mercado, L. Insights into the diversity of NOD-like receptors: Identification and expression analysis of NLRC3, NLRC5 and NLRX1 in rainbow trout. Mol. Immunol. 2017, 87, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Paria, A.; Deepika, A.; Sreedharan, K.; Makesh, M.; Chaudhari, A.; Purushothaman, C.S.; Thirunavukkarasu, A.R.; Rajendran, K.V. Identification of Nod like receptor C3 (NLRC3) in Asian seabass, Lates calcarifer: Characterisation, ontogeny and expression analysis after experimental infection and ligand stimulation. Fish Shellfish Immunol. 2016, 55, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, X.; Hao, G.; Geng, X.; Zhan, W.; Sun, J. Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus) head kidney macrophages. Fish Shellfish Immunol. 2016, 50, 79–90. [Google Scholar] [PubMed]
- Li, J.; Kong, L.; Gao, Y.; Wu, C.; Xu, T. Characterization of NLR-A subfamily members in miiuy croaker and comparative genomics revealed NLRX1 underwent duplication and lose in actinopterygii. Fish Shellfish Immunol. 2015, 47, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Abernathy, J.W.; Wang, S.; Li, P.; Kucuktas, H.; Liu, H.; Peatman, E.; Liu, Z. NOD-like subfamily of the nucleotide-binding domain and leucine-rich repeat containing family receptors and their expression in channel catfish. Dev. Comp. Immunol. 2009, 33, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, Q.L.; Lu, Y.; Chen, S.L.; Li, Q.; Sha, Z.X. Expression profiles of NODs in channel catfish (Ictalurus punctatus) after infection with Edwardsiella tarda, Aeromonas hydrophila, Streptococcus iniae and channel catfish hemorrhage reovirus. Fish Shellfish Immunol. 2012, 33, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Shiau, C.E.; Monk, K.R.; Joo, W.; Talbot, W.S. An anti-inflammatory NOD-like receptor is required for microglia development. Cell Rep. 2013, 5, 1342–1352. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhu, L.; Xia, X.; Wang, H.Y.; Legras, X.; Hong, J.; Ji, J.; Shen, P.; Zheng, S.; Chen, Z.J.; et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 2010, 141, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Pandey, S.; Zou, J.; Kumagai, Y.; Takahashi, K.; Akira, S.; Kawai, T. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J. Immunol. 2011, 186, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Kuenzel, S.; Till, A.; Winkler, M.; Häsler, R.; Lipinski, S.; Jung, S.; Grötzinger, J.; Fickenscher, H.; Schreiber, S.; Rosenstiel, P. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J. Immunol. 2010, 184, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- Neerincx, A.; Lautz, K.; Menning, M.; Kremmer, E.; Zigrino, P.; Hösel, M.; Büning, H.; Schwarzenbacher, R.; Kufer, T.A. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J. Biol. Chem. 2010, 285, 26223–26232. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, P.; Singh, N.; Kumar, A.; Neerincx, A.; Kremmer, E.; Cao, W.; Davis, W.G.; Katz, J.M.; Gangappa, S.; Lin, R.; et al. NLRC5 interacts with RIG-I to induce a robust antiviral response against influenza virus infection. Eur. J. Immunol. 2015, 45, 758–772. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.K.; Roberts, R.A.; Huang, M.T.; Willingham, S.B.; Conti, B.J.; Brickey, W.J.; Barker, B.R.; Kwan, M.; Taxman, D.J.; Accavitti-Loper, M.A.; et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J. Immunol. 2011, 186, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Triantafilou, K.; Kar, S.; van Kuppeveld, F.J.; Triantafilou, M. Rhinovirus-induced calcium flux triggers NLRP3 and NLRC5 activation in bronchial cells. Am. J. Respir. Cell Mol. Biol. 2013, 49, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Meissner, T.B.; Li, A.; Kobayashi, K.S. NLRC5: A newly discovered MHC class I transactivator (CITA). Microbes Infect. 2012, 14, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.S.; van den Elsen, P.J. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 2012, 12, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Ludigs, K.; Seguín-Estévez, Q.; Lemeille, S.; Ferrero, I.; Rota, G.; Chelbi, S.; Mattmann, C.; MacDonald, H.R.; Reith, W.; Guarda, G. NLRC5 exclusively transactivates MHC class I and related genes through a distinctive SXY module. PLoS Genet. 2015, 11, e1005088. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Hu, Y.W.; Xue, N.N.; Ren, S.S.; Chen, S.N.; Nie, P.; Chang, M.X. Role of zebrafish NLRC5 in antiviral response and transcriptional regulation of MHC related genes. Dev. Comp. Immunol. 2017, 68, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.M.; Horvath, C.M. Activation of RIG-I-like receptor signal transduction. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Ramos, H.J.; Gale, M., Jr. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 2011, 1, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Kato, H.; Kumagai, Y.; Yoneyama, M.; Sato, S.; Matsushita, K.; Tsujimura, T.; Fujita, T.; Akira, S.; Takeuchi, O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. USA 2010, 107, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Rothenfusser, S.; Goutagny, N.; DiPerna, G.; Gong, M.; Monks, B.G.; Schoenemeyer, A.; Yamamoto, M.; Akira, S.; Fitzgerald, K.A. The RNA helicase LGP2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005, 175, 5260–5268. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Hirai, R.; Loo, Y.M.; Owen, D.; Johnson, C.L.; Sinha, S.C.; Akira, S.; Fujita, T.; Gale, M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 2007, 104, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Desalle, R.; Fisher, P.B. Evolution of MDA-5/RIG-I-dependent innate immunity: Independent evolution by domain grafting. Proc. Natl. Acad. Sci. USA 2008, 105, 17040–17045. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, Y.B.; Liu, T.K.; Shi, J.; Wang, B.; Gui, J.F. Fish MITA serves as a mediator for distinct fish IFN gene activation dependent on IRF3 or IRF7. J. Immunol. 2011, 187, 2531–2539. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Liu, H.; Kong, R.; Wang, L.; Wang, Y.; Hu, W.; Guo, Q. Expression profiles of carp IRF-3/-7 correlate with the up-regulation of RIG-I/MAVS/TRAF3/TBK1, four pivotal molecules in RIG-I signaling pathway. Fish Shellfish Immunol. 2011, 30, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Yan, J.; Chen, H.; Li, J.; Tian, Y.; Feng, H. LGP2 of black carp plays an important role in the innate immune response against SVCV and GCRV. Fish Shellfish Immunol. 2016, 57, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Xiao, J.; Chen, H.; Lu, L.; Wang, X.; Tian, Y.; Feng, H. The antiviral signaling mediated by black carp MDA5 is positively regulated by LGP2. Fish Shellfish Immunol. 2017, 66, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Huang, T.; Dong, J.; Heng, J.; Zhang, R.; Peng, L. Molecular cloning and immune responsive expression of MDA5 gene, a pivotal member of the RLR gene family from grass carp Ctenopharyngodon idella. Fish Shellfish Immunol. 2010, 28, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Su, J.; Heng, J.; Dong, J.; Zhang, R.; Zhu, H. Identification and expression profiling analysis of grass carp Ctenopharyngodon idella LGP2 cDNA. Fish Shellfish Immunol. 2010, 29, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Su, J.; Huang, T.; Zhang, R.; Peng, L. Identification of a retinoic acid-inducible gene I from grass carp (Ctenopharyngodon idella) and expression analysis in vivo and in vitro. Fish Shellfish Immunol. 2011, 30, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.F.; Chang, M.X.; Xue, N.N.; Liu, X.Q.; Li, J.H.; Fu, J.P.; Chen, S.N.; Nie, P. Melanoma differentiation-associated gene 5 in zebrafish provoking higher interferon-promoter activity through signalling enhancing of its shorter splicing variant. Immunology 2014, 141, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.F.; Chang, M.X.; Li, Y.; Zhang, S.H.; Fu, J.P.; Chen, S.N.; Nie, P. Higher antiviral response of RIG-I through enhancing RIG-I/MAVS-mediated signaling by its long insertion variant in zebrafish. Fish Shellfish Immunol. 2015, 43, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Zhang, Y.S.; Dong, W.R.; Xiang, L.X.; Shao, J.Z. Involvement of zebrafish RIG-I in NF-κB and IFN signaling pathways: Insights into functional conservation of RIG-I in antiviral innate immunity. Dev. Comp. Immunol. 2015, 48, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Asim, M.; Yi, L.; Hegazy, A.M.; Hu, X.; Zhou, Y.; Ai, T.; Lin, L. Abortive infection of snakehead fish vesiculovirus in ZF4 cells was associated with the RLRs pathway activation by viral replicative intermediates. Int. J. Mol. Sci. 2015, 16, 6235–6250. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, K.V.; Zhang, J.; Liu, S.; Peatman, E.; Kucuktas, H.; Wang, X.; Liu, H.; Wood, T.; Terhune, J.; Liu, Z. Pathogen recognition receptors in channel catfish: II Identification, phylogeny and expression of retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs). Dev. Comp. Immunol. 2012, 37, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Huang, Y.; Yang, Y.; Wang, S.; Yang, M.; Huang, X.; Qin, Q. Negative regulation of the antiviral response by grouper LGP2 against fish viruses. Fish Shellfish Immunol. 2016, 56, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yu, Y.; Yang, Y.; Yang, M.; Zhou, L.; Huang, X.; Qin, Q. Antiviral function of grouper MDA5 against iridovirus and nodavirus. Fish Shellfish Immunol. 2016, 54, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Nerbøvik, I.G.; Solheim, M.A.; Eggestøl, H.Ø.; Rønneseth, A.; Jakobsen, R.A.; Wergeland, H.I.; Haugland, G.T. Molecular cloning of MDA5, phylogenetic analysis of RIG-I-like receptors (RLRs) and differential gene expression of RLRs, interferons and proinflammatory cytokines after in vitro challenge with IPNV, ISAV and SAV in the salmonid cell line TO. J. Fish. Dis. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Collet, B.; Nie, P.; Lester, K.; Campbell, S.; Secombes, C.J.; Zou, J. Expression and functional characterization of the RIG-I-like receptors MDA5 and LGP2 in Rainbow trout (Oncorhynchus mykiss). J. Virol. 2011, 85, 8403–8412. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Hu, Y.; Zhang, S.; Zheng, J.; Zeng, L.; Zhang, J.; Zhu, A.; Wu, C. Molecular characterization and expression analyses of three RIG-I-like receptor signaling pathway genes (MDA5, LGP2 and MAVS) in Larimichthys crocea. Fish Shellfish Immunol. 2016, 55, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.; Paria, A.; Deepika, A.; Sreedharan, K.; Makesh, M.; Bedekar, M.K.; Purushothaman, C.S.; Rajendran, K.V. Molecular cloning, characterisation and expression analysis of melanoma differentiation associated gene 5 (MDA5) of green chromide, Etroplus suratensis. Gene 2015, 557, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Zhang, J.; Jin, Y.; Zeng, L.; Jia, K.; Yi, M. Characterization and expression analysis of laboratory of genetics and physiology 2 gene in sea perch, Lateolabrax japonicus. Fish Shellfish Immunol. 2015, 47, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Jia, K.; Chen, L.; Le, Y.; Jin, Y.; Zhang, J.; Zhu, L.; Zhang, L.; Yi, M. Identification and characterization of the melanoma differentiation-associated gene 5 in sea perch, Lateolabrax japonicus. Dev. Comp. Immunol. 2016, 61, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Hikima, J.; Kondo, H.; Hirono, I.; Jung, T.S.; Aoki, T. Evolutional conservation of molecular structure and antiviral function of a viral RNA receptor, LGP2, in Japanese flounder, Paralichthys olivaceus. J. Immunol. 2010, 185, 7507–7517. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Kirchhofer, A.; Shin, Y.C.; Inn, K.S.; Liang, C.; Cui, S.; Myong, S.; Ha, T.; Hopfner, K.P.; Jung, J.U. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. USA 2008, 105, 16743–16748. [Google Scholar] [CrossRef] [PubMed]
- Biacchesi, S.; LeBerre, M.; Lamoureux, A.; Louise, Y.; Lauret, E.; Boudinot, P.; Brémont, M. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J. Virol. 2009, 83, 7815–7827. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, Y.; Chu, Q.; Xu, T. The evolution and functional characterization of Miiuy croaker cytosolic gene LGP2 involved in immune response. Fish Shellfish Immunol. 2016, 58, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Wan, Q.; Yang, C.; Su, J. Grass carp laboratory of genetics and physiology 2 serves as a negative regulator in retinoic acid-inducible gene I- and melanoma differentiation-associated gene 5-mediated antiviral signaling in resting state and early stage of grass carp reovirus infection. Front Immunol. 2017, 8, 352. [Google Scholar] [PubMed]
- Lad, S.P.; Yang, G.; Scott, D.A.; Chao, T.H.; Correia, J.d.S.; de la Torre, J.C.; Li, E. Identification of MAVS splicing variants that interfere with RIGI/MAVS pathway signaling. Mol. Immunol. 2008, 45, 2277–2287. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.W.; Gauthier, A.E.; Mills, E.W.; Ingolia, N.T.; Kagan, J.C. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 2014, 156, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Jin, Y.; Chen, L.; Zhang, J.; Jia, K.; Yi, M. Molecular characterization and expression analysis of mitochondrial antiviral signaling protein gene in sea perch, Lateolabrax japonicus. Dev. Comp. Immunol. 2016, 55, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhou, J.; Lv, Y.; Qu, Y.; Chi, M.; Li, J.; Feng, H. Identification and characterization of MAVS from black carp Mylopharyngodon piceus. Fish Shellfish Immunol. 2015, 43, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Qi, L.; Chen, W.; Dong, C.; Liu, Z.; Liu, D.; Huang, M.; Li, W.; Yang, G.; Weng, S.; et al. Characterization of a TnMAVS protein from Tetraodon nigroviridis. Dev. Comp. Immunol. 2011, 35, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Simora, R.M.; Ohtani, M.; Hikima, J.; Kondo, H.; Hirono, I.; Jung, T.S.; Aoki, T. Molecular cloning and antiviral activity of IFN-β promoter stimulator-1 (IPS-1) gene in Japanese flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 2010, 29, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Huang, T.; Yang, C.; Zhang, R. Molecular cloning, characterization and expression analysis of interferon-β promoter stimulator 1 (IPS-1) gene from grass carp Ctenopharyngodon idella. Fish Shellfish Immunol. 2011, 30, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Hu, Y.W.; Zou, P.F.; Ren, S.S.; Nie, P.; Chang, M.X. MAVS splicing variants contribute to the induction of interferon and interferon-stimulated genes mediated by RIG-I-like receptors. Dev Comp Immunol. 2015, 49, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.F.; Li, S.; Lu, X.B.; Zhang, Y.A. Functions of the two zebrafish MAVS variants are opposite in the induction of IFN1 by targeting IRF7. Fish Shellfish Immunol. 2015, 45, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, Y.; Chen, L.; Chen, H.; You, F.; Zhou, X.; Zhou, Y.; Zhai, Z.; Chen, D.; Jiang, Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA 2009, 106, 8653–8658. [Google Scholar] [CrossRef] [PubMed]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Burdette, D.L.; Monroe, K.M.; Sotelo-Troha, K.; Iwig, J.S.; Eckert, B.; Hyodo, M.; Hayakawa, Y.; Vance, R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011, 478, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Yi, G.; Watts, T.; Kao, C.C.; Li, P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 2012, 19, 722–724. [Google Scholar] [CrossRef] [PubMed]
- Shang, G.; Zhu, D.; Li, N.; Zhang, J.; Zhu, C.; Lu, D.; Liu, C.; Yu, Q.; Zhao, Y.; Xu, S.; et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 2012, 19, 725–727. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Song, X.; Wang, Y.; Ru, H.; Shaw, N.; Jiang, Y.; Niu, F.; Zhu, Y.; Qiu, W.; Parvatiyar, K.; et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 2012, 36, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Tian, Y.; Kabaleeswaran, V.; Jiang, X.; Tu, D.; Eck, M.J.; Chen, Z.J.; Wu, H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 2012, 46, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Härtlova, A.; Erttmann, S.F.; Raffi, F.A.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kröger, A.; Nilsson, J.A.; et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 2015, 42, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, L.; Keating, S.E.; Baran, M.; Horan, K.A.; Jensen, S.B.; Sharma, S.; Sirois, C.M.; Jin, T.; Latz, E.; Xiao, T.S.; et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010, 11, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, S.; Maestre, A.M.; Pagni, S.; Patel, J.R.; Savage, T.; Gutman, D.; Maringer, K.; Bernal-Rubio, D.; Shabman, R.S.; Simon, V.; et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012, 8, e1002934. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Chang, T.H.; Liang, J.J.; Chiang, R.L.; Lee, Y.L.; Liao, C.L.; Lin, Y.L. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012, 8, e1002780. [Google Scholar] [CrossRef] [PubMed]
- Nitta, S.; Sakamoto, N.; Nakagawa, M.; Kakinuma, S.; Mishima, K.; Kusano-Kitazume, A.; Kiyohashi, K.; Murakawa, M.; Nishimura-Sakurai, Y.; Azuma, S.; et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 2013, 57, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Pei, R.; Zhu, W.; Zeng, R.; Wang, Y.; Wang, Y.; Lu, M.; Chen, X. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J. Immunol. 2014, 192, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhao, K.; Su, X.; Lu, L.; Zhao, H.; Zhang, X.; Wang, Y.; Wu, C.; Chen, J.; Zhou, Y.; et al. MITA/STING and its alternative splicing isoform MRP restrict hepatitis B virus replication. PLoS ONE 2017, 12, e0169701. [Google Scholar] [CrossRef] [PubMed]
- Biacchesi, S.; Mérour, E.; Lamoureux, A.; Bernard, J.; Brémont, M. Both STING and MAVS fish orthologs contribute to the induction of interferon mediated by RIG-I. PLoS ONE 2012, 7, e47737. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ouyang, Z.; Wang, W.; Yu, Y.; Li, P.; Zhou, S.; Wei, S.; Wei, J.; Huang, X.; Qin, Q. Antiviral role of grouper STING against iridovirus infection. Fish Shellfish Immunol. 2015, 47, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.F.; Li, S.; Wang, Z.X.; Du, S.Q.; Chen, D.D.; Nie, P.; Zhang, Y.A. Grass carp reovirus VP41 targets fish MITA to abrogate the IFN response. J. Virol. 2017, JVI-00390. [Google Scholar]
- Perry, A.K.; Chow, E.K.; Goodnough, J.B.; Yeh, W.C.; Cheng, G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to Toll-like receptor activation and viral infection. J. Exp. Med. 2004, 199, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.C.; Giani, J.F.; Mayer, M.A.; Toblli, J.E.; Turyn, D.; Dominici, F.P. TANK-binding kinase 1 mediates phosphorylation of insulin receptor at serine residue 994: A potential link between inflammation and insulin resistance. J. Endocrinol. 2009, 201, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Miyahira, A.K.; Shahangian, A.; Hwang, S.; Sun, R.; Cheng, G. TANK-binding kinase-1 plays an important role during in vitro and in vivo type I IFN responses to DNA virus infections. J. Immunol. 2009, 182, 2248–2257. [Google Scholar] [CrossRef] [PubMed]
- Weidberg, H.; Elazar, Z. TBK1 mediates crosstalk between the innate immune response and autophagy. Sci. Signal. 2011, 4, pe39. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Yi, Y.S.; Yang, Y.; Oh, J.; Jeong, D.; Cho, J.Y. The pivotal role of TBK1 in inflammatory responses mediated by macrophages. Mediators Inflamm. 2012, 2012, 979105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 2013, 587, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Shi, M.; Han, M.; Zhong, J.; Li, Z.; Li, W.; Hu, Y.; Yan, L.; Wang, J.; He, Y.; et al. Negative regulation of virus-triggered IFN-β signaling pathway by alternative splicing of TBK1. J. Biol. Chem. 2008, 283, 35590–35597. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Su, J.; Yang, C.; Yan, N.; Rao, Y.; Chen, X. Molecular characterizations of grass carp (Ctenopharyngodon idella) TBK1 gene and its roles in regulating IFN-I pathway. Dev. Comp. Immunol. 2014, 45, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lu, L.F.; Wang, Z.X.; Lu, X.B.; Chen, D.D.; Nie, P.; Zhang, Y.A. The P protein of spring viremia of carp virus negatively regulates the fish interferon response by inhibiting the kinase activity of TANK-binding kinase 1. J. Virol. 2016, 90, 10728–10737. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, W.Q.; Hu, Y.W.; Wu, X.M.; Nie, P.; Chang, M.X. TBK1-like transcript negatively regulates the production of IFN and IFN-stimulated genes through RLRs-MAVS-TBK1 pathway. Fish Shellfish Immunol. 2016, 54, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Yu, D.H.; Chen, J.; Fan, S.; Wang, Z.Y. Expression profiles and interaction suggest TBK1 can be regulated by Nrdp1 in response to immune stimulation in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. 2015, 46, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.M.; Maniatis, T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, J. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 2007, 282, 15325–15329. [Google Scholar] [CrossRef] [PubMed]
- Karpova, A.Y.; Howley, P.M.; Ronco, L.V. Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes Dev. 2000, 14, 2813–2818. [Google Scholar] [CrossRef] [PubMed]
- Karpova, A.Y.; Ronco, L.V.; Howley, P.M. Functional characterization of interferon regulatory factor 3a (IRF-3a), an alternative splice isoform of IRF-3. Mol. Cell Biol. 2001, 21, 4169–4176. [Google Scholar] [CrossRef] [PubMed]
- Marozin, S.; Altomonte, J.; Stadler, F.; Thasler, W.E.; Schmid, R.M.; Ebert, O. Inhibition of the IFN-β response in hepatocellular carcinoma by alternative spliced isoform of IFN regulatory factor-3. Mol. Ther. 2008, 16, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ma, L.; Chen, X. Interferon regulatory factor 3-CL, an isoform of IRF3, antagonizes activity of IRF3. Cell. Mol. Immunol. 2011, 8, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Xu, H.G.; Lu, C.; Jin, R.; Zou, L.; Wang, Y.; Zhou, G.P. The characterization of two novel IRF-3 transcripts starting from intron 2 of the wild type of IRF-3. Mol. Biol. Rep. 2011, 38, 4415–4421. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, X.; Song, Y.; Lu, Z.; Ning, T.; Cai, H.; Ke, Y. Identification of novel alternative splicing variants of interferon regulatory factor 3. Biochim. Biophys. Acta 2011, 1809, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Chu, Q.; Bi, D.; Wang, Y.; Xu, T. Identification and functional characterization of miiuy croaker IRF3 as an inducible protein involved regulation of IFN response. Fish Shellfish Immunol. 2016, 54, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lai, Q.; Gu, M.; Liu, D.; Hou, Q.; Liu, X.; Mi, Y.; Sun, Z.; Wang, H.; Lin, G.; et al. Fish IRF3 up-regulates the transcriptional level of IRF1, IRF2, IRF3 and IRF7 in CIK cells. Fish Shellfish Immunol. 2015, 47, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.F.; Wei, Q.; Tang, S.J.; Chen, X.W.; Zhao, J.L. Molecular characterization and functional analysis of IRF3 in tilapia (Oreochromis niloticus). Dev. Comp. Immunol. 2016, 55, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.X.; Hu, Y.H. Molecular characterization and expression analysis of eleven interferon regulatory factors in half-smooth tongue sole, Cynoglossus semilaevis. Fish Shellfish Immunol. 2015, 44, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Huang, W.S.; Nie, P. Cloning and expression analyses of interferon regulatory factor (IRF) 3 and 7 genes in European eel, Anguilla anguilla with the identification of genes involved in IFN production. Fish Shellfish Immunol. 2014, 37, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.L.; Huang, X.N.; Fan, Z.; Kong, P.; Wang, Z.Y. Cloning and expression analysis of interferon regulatory factor (IRF) 3 and 7 in large yellow croaker, Larimichthys crocea. Fish Shellfish Immunol. 2012, 32, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Hikima, J.; Hwang, S.D.; Morita, T.; Suzuki, Y.; Kato, G.; Kondo, H.; Hirono, I.; Jung, T.S.; Aoki, T. Transcriptional regulation of type I interferon gene expression by interferon regulatory factor-3 in Japanese flounder, Paralichthys olivaceus. Dev. Comp. Immunol. 2012, 36, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004, 202, 8–32. [Google Scholar] [CrossRef] [PubMed]
- Bonjardim, C.A.; Ferreira, P.C.; Kroon, E.G. Interferons: Signaling, antiviral and viral evasion. Immunol. Lett. 2009, 122, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Novick, D.; Cohen, B.; Rubinstein, M. The human interferon α/β receptor: Characterization and molecular cloning. Cell 1994, 77, 391–400. [Google Scholar] [CrossRef]
- Farrar, M.A.; Schreiber, R.D. The molecular cell biology of interferon-γ and its receptor. Annu. Rev. Immunol. 1993, 11, 571–611. [Google Scholar] [CrossRef] [PubMed]
- Witte, K.; Witte, E.; Sabat, R.; Wolk, K. IL-28A, IL-28B, and IL-29: Promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 2010, 21, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Lutfalla, G.; Holland, S.J.; Cinato, E.; Monneron, D.; Reboul, J.; Rogers, N.C.; Smith, J.M.; Stark, G.R.; Gardiner, K.; Mogensen, K.E.; et al. Mutant U5A cells are complemented by an interferon-α β receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J. 1995, 14, 5100–5108. [Google Scholar] [PubMed]
- Domanski, P.; Colamonici, O.R. The type-I interferon receptor. The long and short of it. Cytokine Growth Factor Rev. 1996, 7, 143–151. [Google Scholar] [CrossRef]
- Gazziola, C.; Cordani, N.; Carta, S.; De Lorenzo, E.; Colombatti, A.; Perris, R. The relative endogenous expression levels of the IFNAR2 isoforms influence the cytostatic and pro-apoptotic effect of IFNα on pleomorphic sarcoma cells. IntJ. Oncol. 2005, 26, 129–140. [Google Scholar] [CrossRef]
- McKenna, S.D.; Vergilis, K.; Arulanandam, A.R.; Weiser, W.Y.; Nabioullin, R.; Tepper, M.A. Formation of human IFN-β complex with the soluble type I interferon receptor IFNAR-2 leads to enhanced IFN stability, pharmacokinetics, and antitumor activity in xenografted SCID mice. J. Interferon Cytokine Res. 2004, 24, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.R.; Cleary, C.M.; Mariano, T.M.; Izotova, L.; Pestka, S. Differential responsiveness of a splice variant of the human type I interferon receptor to interferons. J. Biol. Chem. 1996, 271, 13448–13453. [Google Scholar] [CrossRef] [PubMed]
- Gilli, F. Role of differential expression of interferon receptor isoforms on the response of multiple sclerosis patients to therapy with interferon α. J Interferon Cytokine Res. 2010, 30, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J. Teleost fish interferons and their role in immunity. Dev. Comp. Immunol. 2011, 35, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Robertsen, B. The interferon system of teleost fish. Fish Shellfish Immunol. 2006, 20, 172–191. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Robertsen, B.; Wang, Z.; Liu, B. Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev. Comp. Immunol. 2009, 33, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Laing, K.J.; Woodson, J.C.; Thorgaard, G.H.; Hansen, J.D. Characterization of the interferon genes in homozygous rainbow trout reveals two novel genes, alternate splicing and differential regulation of duplicated genes. Fish Shellfish Immunol. 2009, 26, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Zou, J.; Nie, P.; Huang, B.; Yu, Z.; Collet, B.; Secombes, C.J. Intracellular interferons in fish: A unique means to combat viral infection. PLoS Pathog. 2013, 9, e1003736. [Google Scholar] [CrossRef] [PubMed]
- Milev-Milovanovic, I.; Long, S.; Wilson, M.; Bengten, E.; Miller, N.W.; Chinchar, V.G. Identification and expression analysis of interferon γ genes in channel catfish. Immunogenetics 2006, 58, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Chakraborty, T.; Miyagawa, S.; Zhou, L.; Ohta, K.; Iguchi, T.; Nagahama, Y. Steroid responsive regulation of IFNγ2 alternative splicing and its possible role in germ cell proliferation in medaka. Mol. Cell. Endocrinol. 2015, 400, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Belosevic, M. Molecular characterization of novel interferon γ receptor 1 isoforms in zebrafish (Danio rerio) and goldfish (Carassius auratus L.). Mol. Immunol. 2009, 46, 3050–3059. [Google Scholar] [CrossRef] [PubMed]
- Yabu, T.; Toda, H.; Shibasaki, Y.; Araki, K.; Yamashita, M.; Anzai, H.; Mano, N.; Masuhiro, Y.; Hanazawa, S.; Shiba, H.; et al. Antiviral protection mechanisms mediated by ginbuna crucian carp interferon γ isoforms 1 and 2 through two distinct interferon γ-receptors. J. Biochem. 2011, 150, 635–648. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, M.X.; Zhang, J. Alternative Pre-mRNA Splicing in Mammals and Teleost Fish: A Effective Strategy for the Regulation of Immune Responses Against Pathogen Infection. Int. J. Mol. Sci. 2017, 18, 1530. https://doi.org/10.3390/ijms18071530
Chang MX, Zhang J. Alternative Pre-mRNA Splicing in Mammals and Teleost Fish: A Effective Strategy for the Regulation of Immune Responses Against Pathogen Infection. International Journal of Molecular Sciences. 2017; 18(7):1530. https://doi.org/10.3390/ijms18071530
Chicago/Turabian StyleChang, Ming Xian, and Jie Zhang. 2017. "Alternative Pre-mRNA Splicing in Mammals and Teleost Fish: A Effective Strategy for the Regulation of Immune Responses Against Pathogen Infection" International Journal of Molecular Sciences 18, no. 7: 1530. https://doi.org/10.3390/ijms18071530
APA StyleChang, M. X., & Zhang, J. (2017). Alternative Pre-mRNA Splicing in Mammals and Teleost Fish: A Effective Strategy for the Regulation of Immune Responses Against Pathogen Infection. International Journal of Molecular Sciences, 18(7), 1530. https://doi.org/10.3390/ijms18071530






