Differential Effects of Histone Acetyltransferase GCN5 or PCAF Knockdown on Urothelial Carcinoma Cells
Abstract
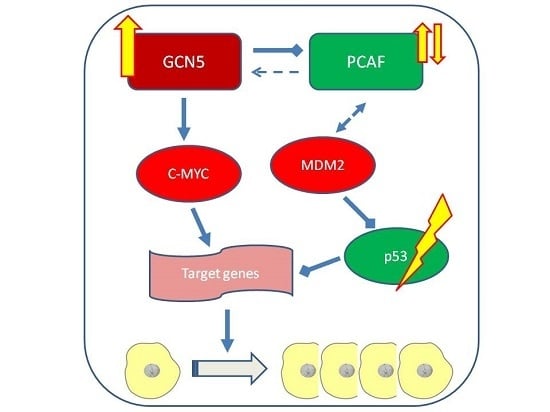
:1. Introduction
2. Results
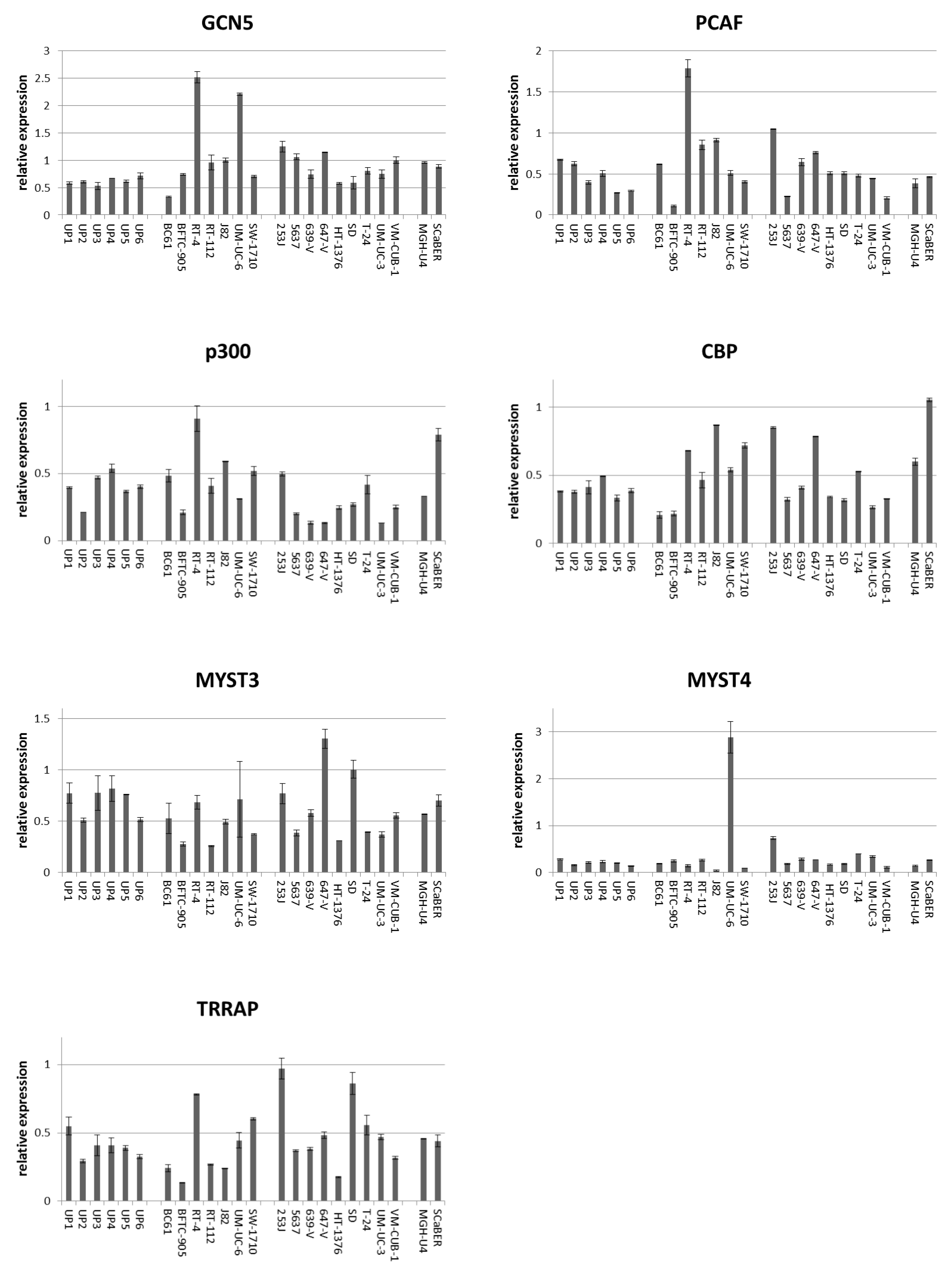
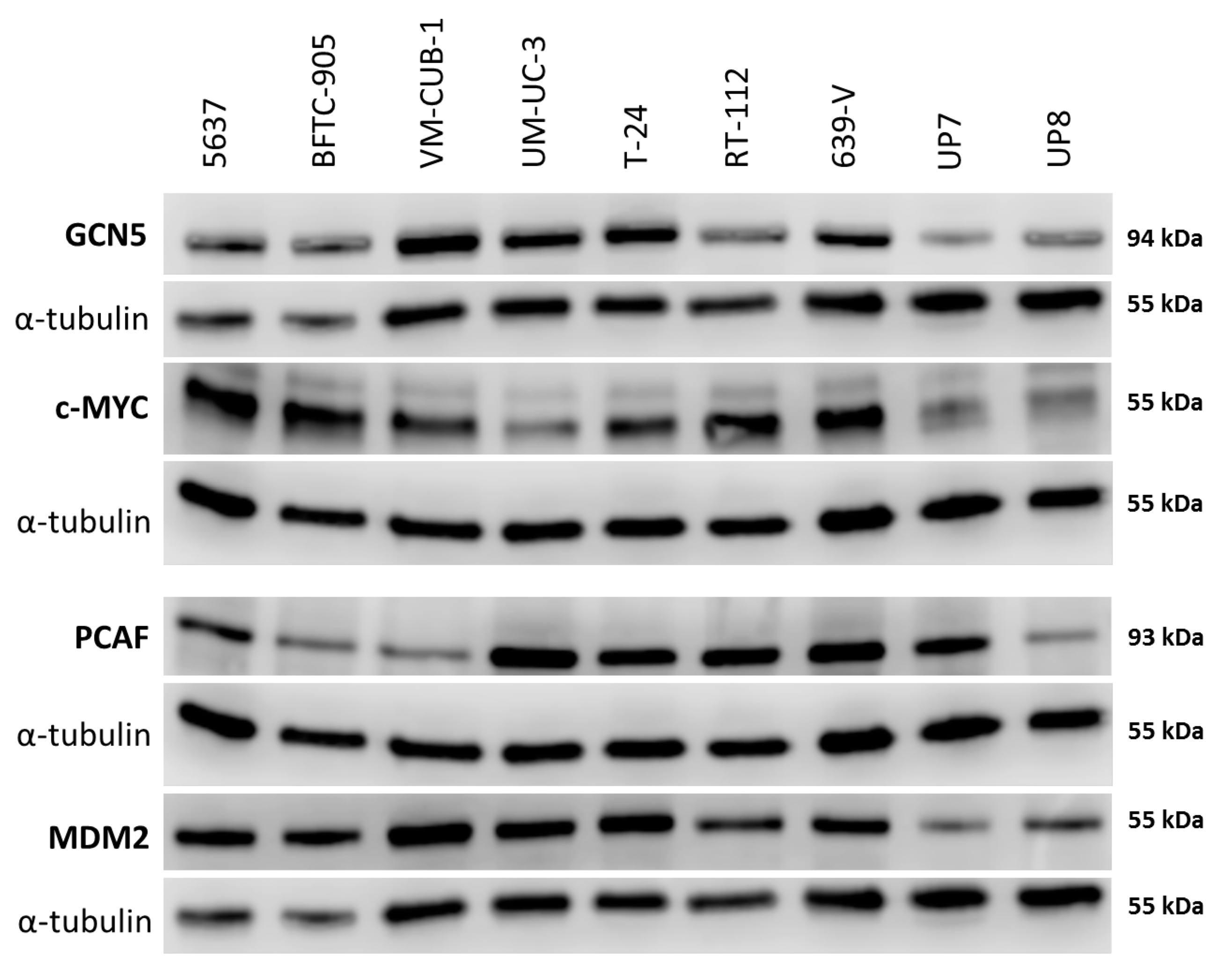
2.1. Expression of HATs in Urothelial Carcinoma Cell Lines and Non-Cancerous Cells
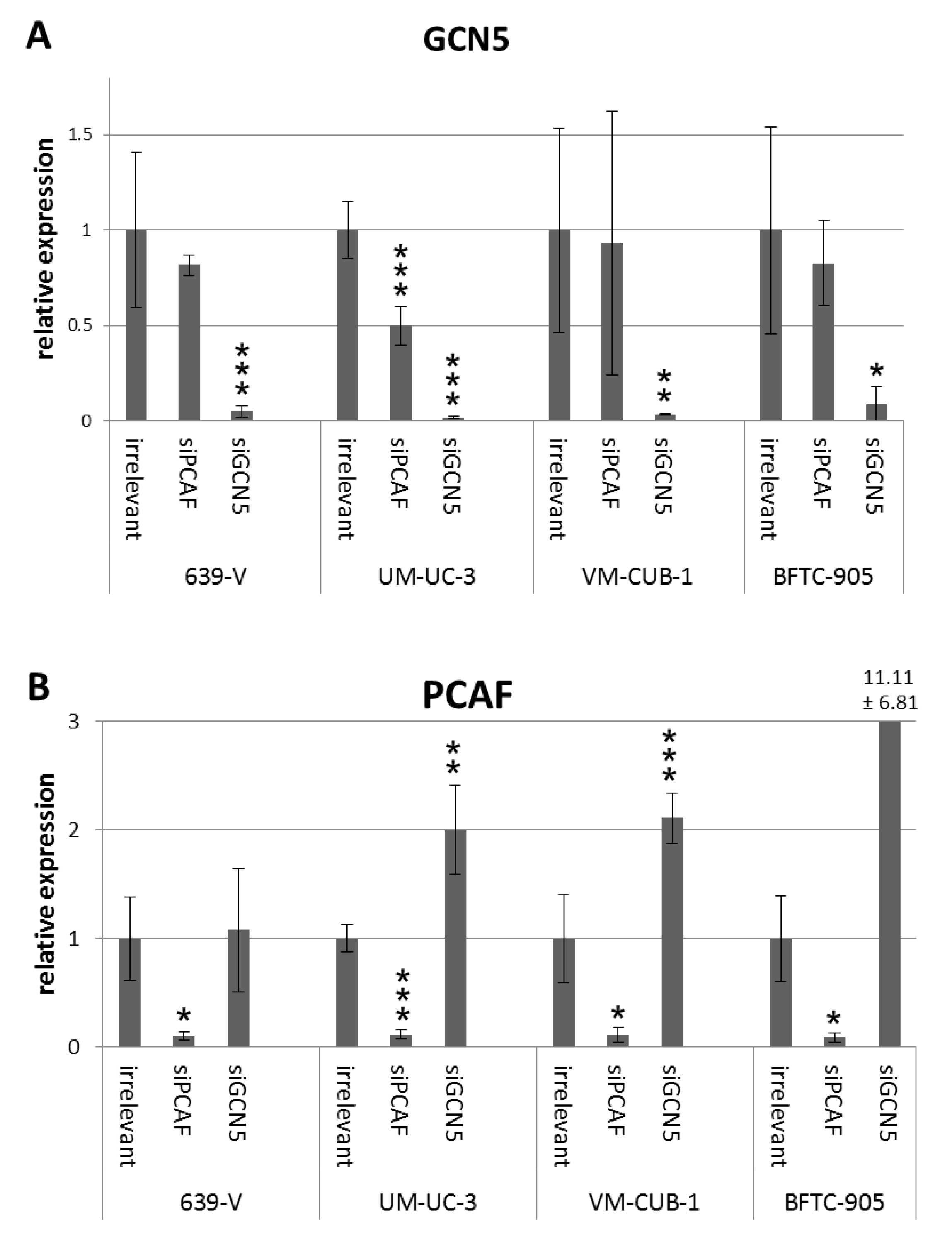
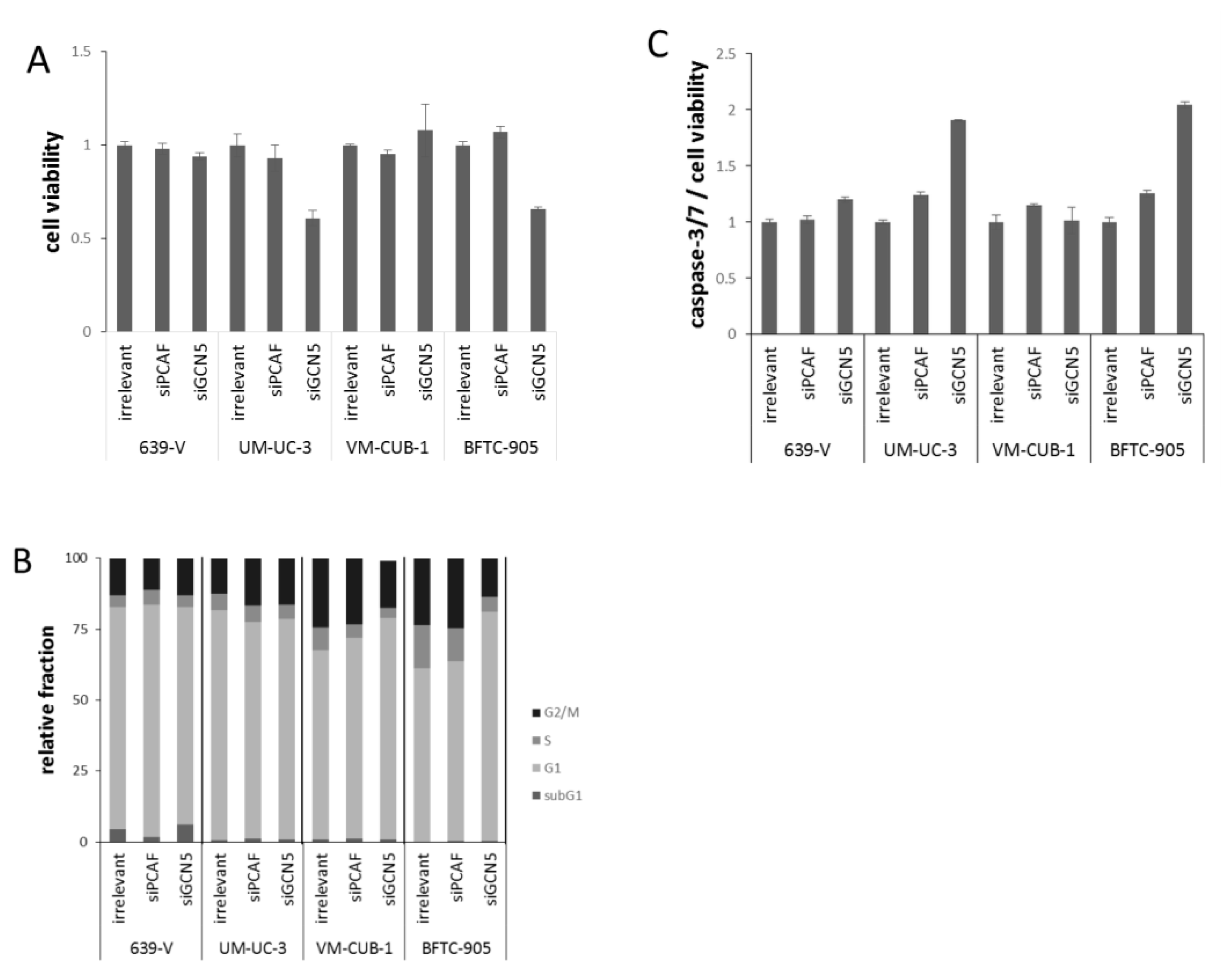
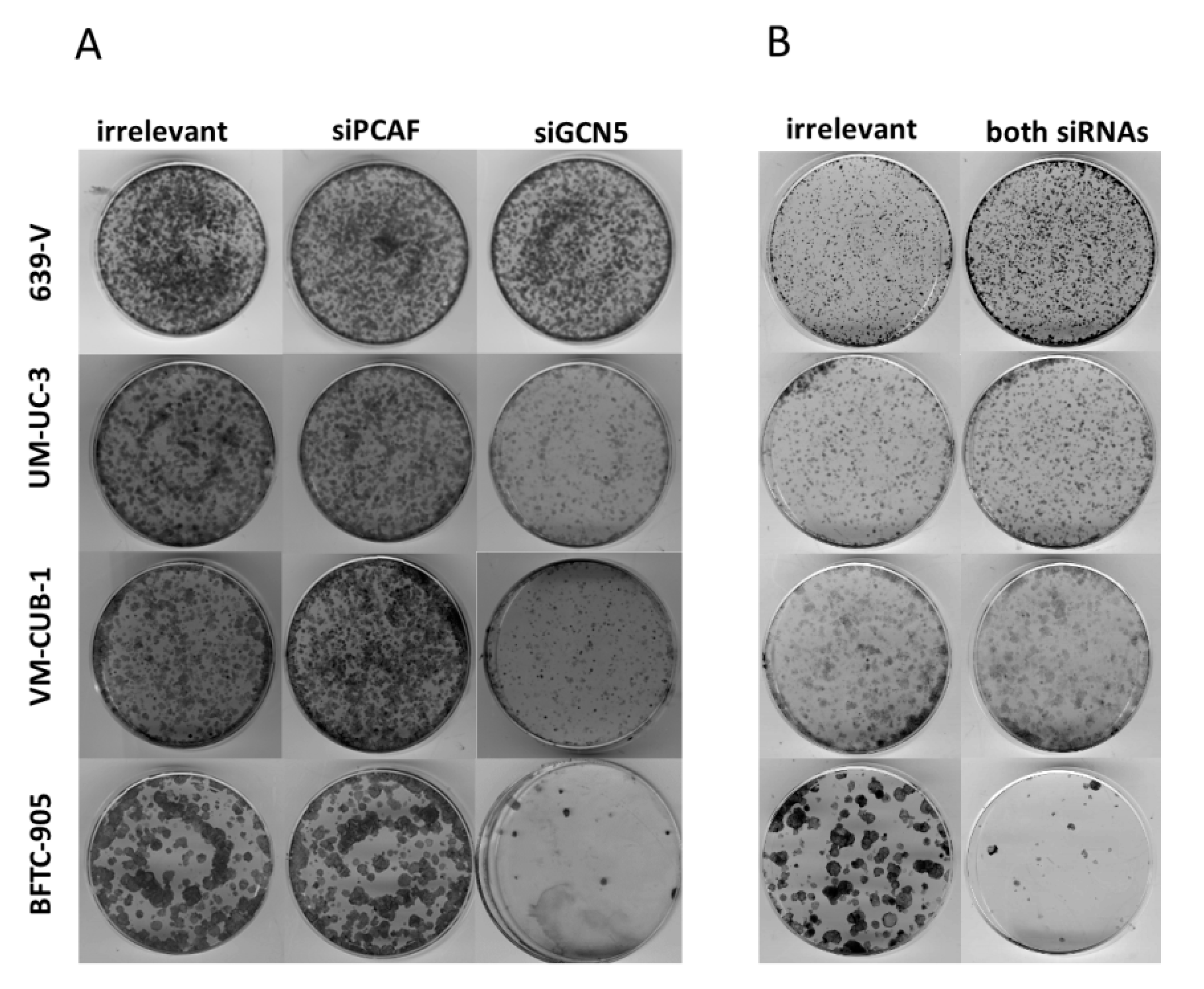
2.2. Effects of GCN5 and PCAF Single Knockdown
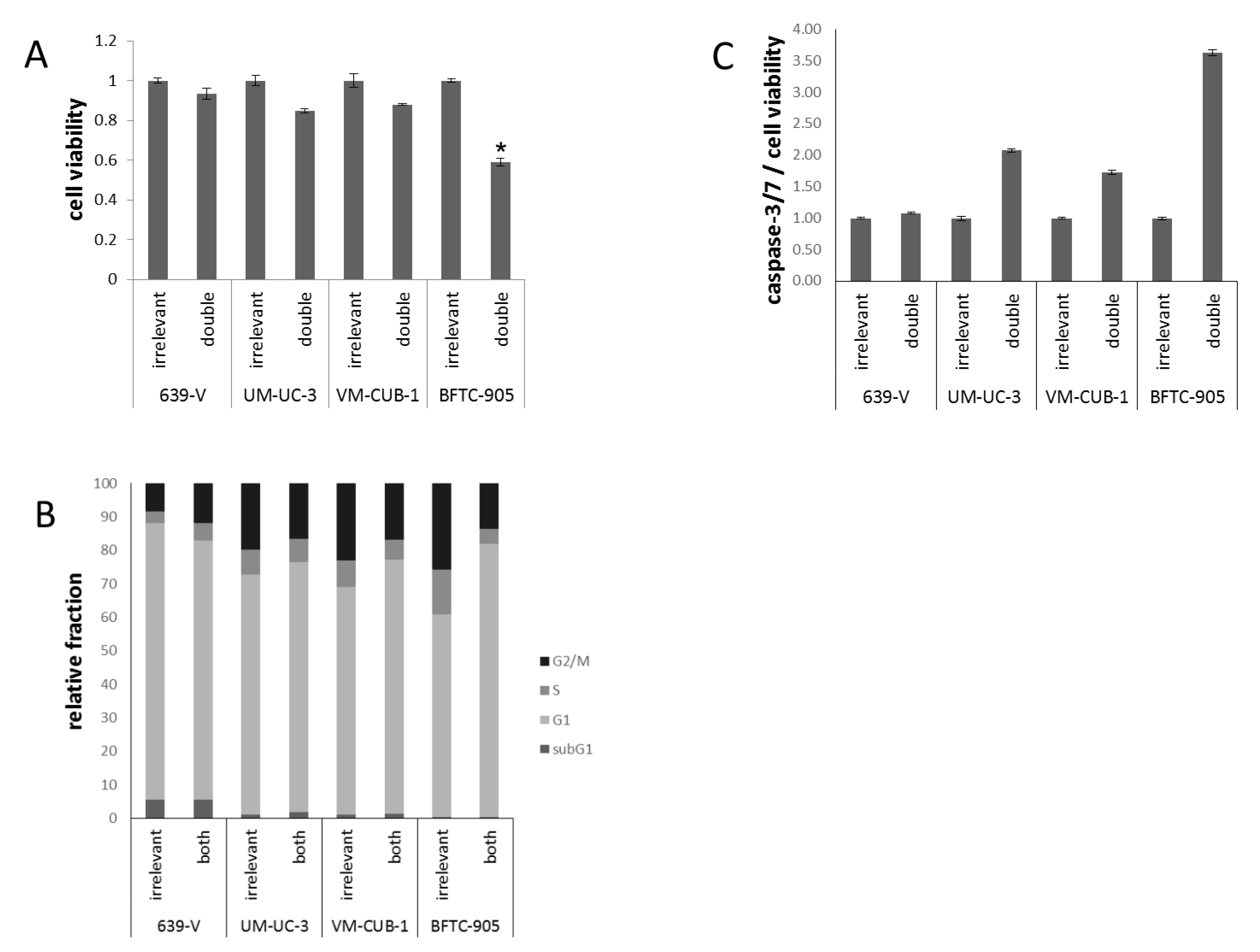
2.3. Effects of GCN5 and PCAF Double Knockdown
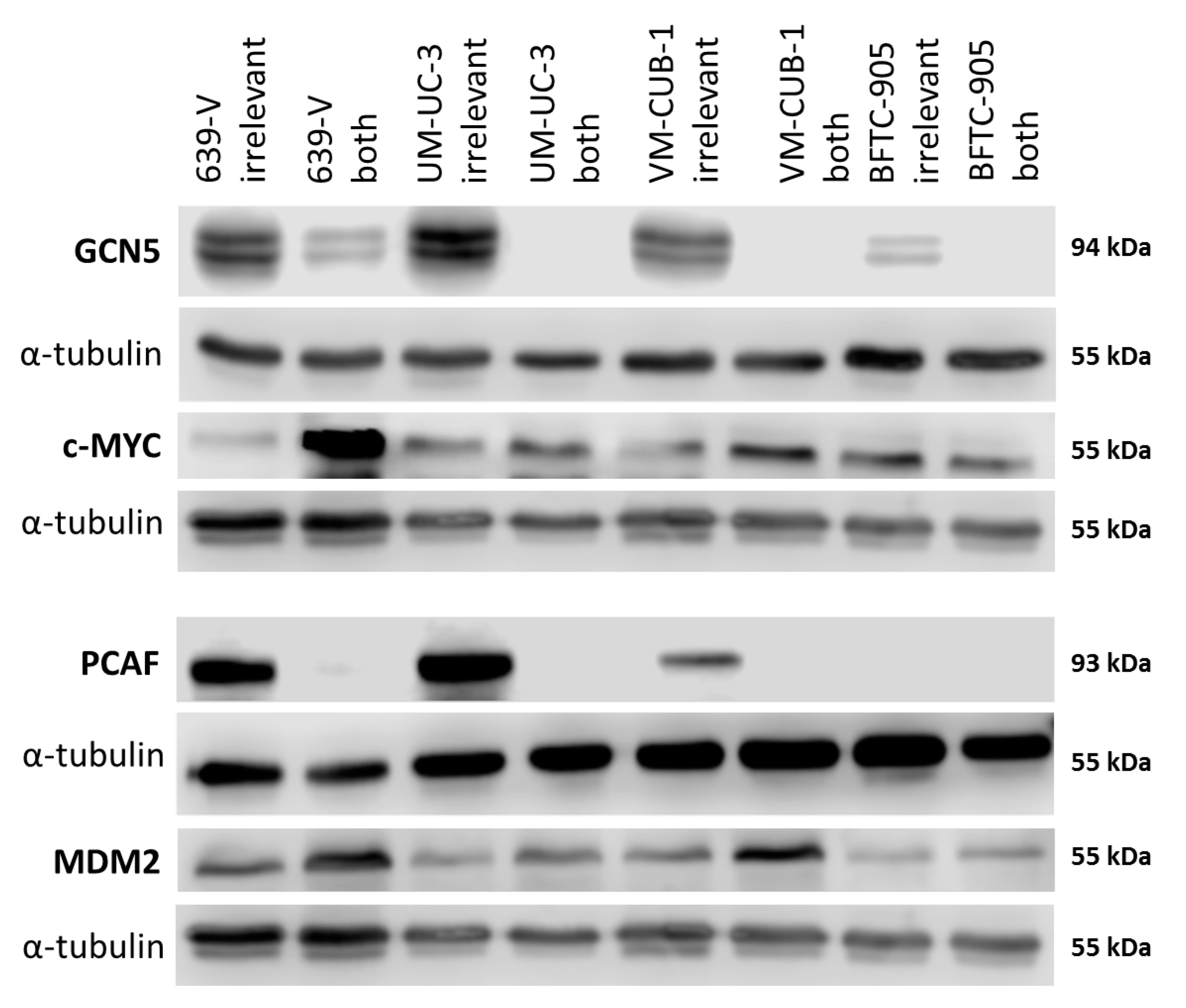
2.4. Effects of GCN5 and PCAF Double Knockdown on Expression of Potential Interactors
3. Discussion
4. Materials and Methods
4.1. Cell Culture and siRNA Transfection
4.2. Viability and Apoptosis Assays
4.3. Colony Forming Assay and Giemsa-Staining
4.4. Flow Cytometry
4.5. Senescence Assay
4.6. RNA Isolation, cDNA Synthesis and Quantitative Real-Time PCR
4.7. Western Blot Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| c-MYC | V-Myc avian myelocytomatosis viral oncogene homolog |
| GCN5 | General control nonderepressible 5 acetyltransferase |
| GNAT | GCN5-related N-acetyltransferases |
| MDM2 | Mouse double minute 2 |
| PCAF | P300/CBP-associated factor acetyltransferase |
| UC | Urothelial carcinoma |
| UCC | Urothelial carcinoma cell line |
| HDAC | Histone deacetylase |
| HAT | Histone acetyltransferase |
| UP | Uroepithel cells, cultured normal urothelial cells |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Kamat, A.M.; Hahn, N.M.; Efstathiou, J.A.; Lerner, S.P.; Malmstrom, P.U.; Choi, W.; Guo, C.C.; Lotan, Y.; Kassouf, W. Bladder cancer. Lancet 2016, 388, 2796–2810. [Google Scholar] [CrossRef]
- Volanis, D.; Papadopoulos, G.; Doumas, K.; Gkialas, I.; Delakas, D. Molecular mechanisms in urinary bladder carcinogenesis. J. BUON 2011, 16, 589–601. [Google Scholar] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar]
- Gui, Y.; Guo, G.; Huang, Y.; Hu, X.; Tang, A.; Gao, S.; Wu, R.; Chen, C.; Li, X.; Zhou, L.; et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011, 43, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, W.; Shi, L.; Sun, L.; Liang, J.; Yi, X.; Li, Q.; Zhang, Y.; Yang, F.; Han, X.; et al. HAT4, a golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol. Cell 2011, 44, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Z.; Tora, L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 2007, 26, 5341–5357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Faiola, F.; Xu, M.; Pan, S.; Martinez, E. Human ATAC is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 2008, 283, 33808–33815. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wei, T.; Si, X.; Wang, Q.; Li, Y.; Leng, Y.; Deng, A.; Chen, J.; Wang, G.; Zhu, S.; et al. Lysine acetyltransferase GCN5 potentiates the growth of non-small cell lung cancer via promotion of E2F1, Cyclin D1, and Cyclin E1 expression. J. Biol. Chem. 2013, 288, 14510–14521. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhang, Q.; Lan, H.; Wang, L.; Mou, P.; Shao, W.; Liu, D.; Yang, W.; Lin, Z.; Lin, Q.; et al. GCN5 potentiates glioma proliferation and invasion via STAT3 and Akt signaling pathways. Int. J. Mol. Sci. 2015, 16, 21897–21910. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.W.; Jin, H.J.; Zhao, W.; Gao, B.; Fang, J.; Wei, J.; Zhang, D.D.; Zhang, J.; Fang, D. The histone acetyltransferase GCN5 expression is elevated and regulated by c-MYC and E2F1 transcription factors in human colon cancer. Gene Expr. 2015, 16, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Majaz, S.; Tong, Z.; Peng, K.; Wang, W.; Ren, W.; Li, M.; Liu, K.; Mo, P.; Li, W.; Yu, C. Histone acetyl transferase GCN5 promotes human hepatocellular carcinoma progression by enhancing AIB1 expression. Cell Biosci. 2016, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Kenneth, N.S.; Ramsbottom, B.A.; Gomez-Roman, N.; Marshall, L.; Cole, P.A.; White, R.J. TRRAP and GCN5 are used by c-MYC to activate RNA polymerase III transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 14917–14922. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Vorontchikhina, M.; Wang, Y.L.; Faiola, F.; Martinez, E. STAGA recruits mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol. Cell Biol. 2008, 28, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Ichikawa, W.; Faiola, F.; Lo, S.Y.; Liu, X.; Martinez, E. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim. Biophys. Acta 2014, 1839, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Malatesta, M.; Steinhauer, C.; Mohammad, F.; Pandey, D.P.; Squatrito, M.; Helin, K. Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 2013, 73, 6323–6333. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Gai, X.; Ding, F.; Lu, Z.; Tu, K.; Yao, Y.; Liu, Q. Histone acetyltransferase PCAF up-regulated cell apoptosis in hepatocellular carcinoma via acetylating histone H4 and inactivating Akt signaling. Mol. Cancer 2013, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Gai, X.; Tu, K.; Li, C.; Lu, Z.; Roberts, L.R.; Zheng, X. Histone acetyltransferase PCAF accelerates apoptosis by repressing a GLI1/BCL2/BAX axis in hepatocellular carcinoma. Cell Death Dis. 2015, 6, e1712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, G.; Wang, Z.; Wan, Y.; Guo, J.; Shi, L. PCAF-mediated Akt1 acetylation enhances the proliferation of human glioblastoma cells. Tumour. Biol. 2015, 36, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Fei, H.J.; Zu, L.D.; Wu, J.; Jiang, X.S.; Wang, J.L.; Chin, Y.E.; Fu, G.H. PCAF acts as a gastric cancer suppressor through a novel PCAF-p16-CDK4 axis. Am. J. Cancer Res. 2016, 6, 2772–2786. [Google Scholar] [PubMed]
- Wan, J.; Xu, W.; Zhan, J.; Ma, J.; Li, X.; Xie, Y.; Wang, J.; Zhu, W.G.; Luo, J.; Zhang, H. PCAF-mediated acetylation of transcriptional factor HOXB9 suppresses lung adenocarcinoma progression by targeting oncogenic protein JMJD6. Nucleic Acids Res. 2016, 44, 10662–10675. [Google Scholar] [CrossRef] [PubMed]
- Linares, L.K.; Kiernan, R.; Triboulet, R.; Chable-Bessia, C.; Latreille, D.; Cuvier, O.; Lacroix, M.; Le Cam, L.; Coux, O.; Benkirane, M. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat. Cell Biol. 2007, 9, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zeng, S.X.; Dai, M.S.; Yang, X.J.; Lu, H. MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J. Biol. Chem. 2002, 277, 30838–30843. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zeng, S.X.; Lee, H.; Lu, H. MDM2 mediates p300/CREB-binding protein-associated factor ubiquitination and degradation. J. Biol. Chem. 2004, 279, 20035–20043. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBIO cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Gunasekaran, P.; Leung, K.; Bindal, N.; Boutselakis, H.; Ding, M.; Bamford, S.; Cole, C.; Ward, S.; et al. Cosmic: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015, 43, D805–D811. [Google Scholar] [CrossRef] [PubMed]
- Pinkerneil, M.; Hoffmann, M.J.; Deenen, R.; Kohrer, K.; Arent, T.; Schulz, W.A.; Niegisch, G. Inhibition of class I histone deacetylases 1 and 2 promotes urothelial carcinoma cell death by various mechanisms. Mol. Cancer Ther. 2016, 15, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Van der Meulen, J.; Sanghvi, V.; Mavrakis, K.; Durinck, K.; Fang, F.; Matthijssens, F.; Rondou, P.; Rosen, M.; Pieters, T.; Vandenberghe, P.; et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 2015, 125, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Love, I.M.; Sekaric, P.; Shi, D.; Grossman, S.R.; Androphy, E.J. The histone acetyltransferase PCAF regulates p21 transcription through stress-induced acetylation of histone H3. Cell Cycle 2012, 11, 2458–2466. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, H.; Takami, Y.; Nakayama, T. GCN5: A supervisor in all-inclusive control of vertebrate cell cycle progression through transcription regulation of various cell cycle-related genes. Gene 2005, 347, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Schulz, W.A. Understanding urothelial carcinoma through cancer pathways. Int. J. Cancer 2006, 119, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Helming, K.C.; Wang, X.; Wilson, B.G.; Vazquez, F.; Haswell, J.R.; Manchester, H.E.; Kim, Y.; Kryukov, G.V.; Ghandi, M.; Aguirre, A.J.; et al. ARID1b is a specific vulnerability in ARID1a-mutant cancers. Nat. Med. 2014, 20, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, H.; Sasaki, M.; Mitachi, T.; Oike, T.; Higuchi, S.; Tominaga, Y.; Kohno, T. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016, 6, 430–445. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.P.; Robaa, D.; Alhalabi, Z.; Sippl, W.; Jung, M. Katching-up on small molecule modulators of lysine acetyltransferases. J. Med. Chem. 2016, 59, 1249–1270. [Google Scholar] [CrossRef] [PubMed]
- Richters, A.; Koehler, A.N. Epigenetic modulation using small molecules-targeting histone acetyltransferases in disease. Curr. Med. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gajer, J.M.; Furdas, S.D.; Grunder, A.; Gothwal, M.; Heinicke, U.; Keller, K.; Colland, F.; Fulda, S.; Pahl, H.L.; Fichtner, I.; et al. Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo. Oncogenesis 2015, 4, e137. [Google Scholar] [CrossRef] [PubMed]
- Swiatkowski, S.; Seifert, H.H.; Steinhoff, C.; Prior, A.; Thievessen, I.; Schliess, F.; Schulz, W.A. Activities of MAP-kinase pathways in normal uroepithelial cells and urothelial carcinoma cell lines. Exp. Cell Res. 2003, 282, 48–57. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koutsogiannouli, E.A.; Wagner, N.; Hader, C.; Pinkerneil, M.; Hoffmann, M.J.; Schulz, W.A. Differential Effects of Histone Acetyltransferase GCN5 or PCAF Knockdown on Urothelial Carcinoma Cells. Int. J. Mol. Sci. 2017, 18, 1449. https://doi.org/10.3390/ijms18071449
Koutsogiannouli EA, Wagner N, Hader C, Pinkerneil M, Hoffmann MJ, Schulz WA. Differential Effects of Histone Acetyltransferase GCN5 or PCAF Knockdown on Urothelial Carcinoma Cells. International Journal of Molecular Sciences. 2017; 18(7):1449. https://doi.org/10.3390/ijms18071449
Chicago/Turabian StyleKoutsogiannouli, Evangelia A., Nicholas Wagner, Christiane Hader, Maria Pinkerneil, Michèle J. Hoffmann, and Wolfgang A. Schulz. 2017. "Differential Effects of Histone Acetyltransferase GCN5 or PCAF Knockdown on Urothelial Carcinoma Cells" International Journal of Molecular Sciences 18, no. 7: 1449. https://doi.org/10.3390/ijms18071449
APA StyleKoutsogiannouli, E. A., Wagner, N., Hader, C., Pinkerneil, M., Hoffmann, M. J., & Schulz, W. A. (2017). Differential Effects of Histone Acetyltransferase GCN5 or PCAF Knockdown on Urothelial Carcinoma Cells. International Journal of Molecular Sciences, 18(7), 1449. https://doi.org/10.3390/ijms18071449