Abstract
The nucleolus is the subnuclear membrane-less organelle where rRNA is transcribed and processed and ribosomal assembly occurs. During the last 20 years, however, the nucleolus has emerged as a multifunctional organelle, regulating processes that go well beyond its traditional role. Moreover, the unique organization of rDNA in tandem arrays and its unusually high transcription rates make it prone to unscheduled DNA recombination events and frequent RNA:DNA hybrids leading to DNA double strand breaks (DSBs). If not properly repaired, rDNA damage may contribute to premature disease onset and aging. Deregulation of ribosomal synthesis at any level from transcription and processing to ribosomal subunit assembly elicits a stress response and is also associated with disease onset. Here, we discuss how genome integrity is maintained within nucleoli and how such structures are functionally linked to nuclear DNA damage response and repair giving an emphasis on the newly emerging roles of the nucleolus in mammalian physiology and disease.
1. Introduction
To sustain life, cells must be able to maintain and appropriately utilize their genome with the ultimate goal to produce the multitude of macromolecules ranging from small non-coding RNAs to large multisubunit proteins. Unlike RNAs, however, proteins require that cells allocate most of their energy reservoirs to generate functional ribosomes, the ribonucleoprotein factories where proteins are synthesized. It is, therefore, not surprising that ribosome synthesis represents a universal and continuous process for all living systems. A fundamental role in this process is held by the nucleolus, where ribosomal RNA is synthesized and ribosomal subunits are assembled. Though nucleoli are not visible during distinct phases of the cell cycle or specific differentiation stages, it is commonly accepted that any cell devoid of its nucleoli is either deceased or dying [1].
Under the microscope, the nucleolus is often the most prominent sub-nuclear formation of almost every eukaryotic cell during interphase. Consequently, it was described as early as 1781 by Fontana [2] and extensively studied for over a century [3]. By the end of the 19th century, researchers had ended up with conclusions that are still valid [4]. The number of visible nucleoli per cell varies enormously depending on cell cycle, cellular activity or differentiation status. For example, nucleoli are not visible in spermatozoa or in eukaryotic cells that undergo open mitosis (in open mitosis the nuclear envelope is completely disassembled). Moreover, nucleoli tend to be bigger in large cells and present in bigger numbers in growing cells [5]. The nucleolus forms at specific chromosomal loci [6,7,8] that are called “secondary constrictions” (the primary constriction is the centromere) or “nucleolar organizer regions” (NORs). NORs were found to consist of tandem arrays of ribosomal genes (rDNA) that encode 18S, 28S, and 5.8S ribosomal RNAs (rRNAs) [9,10,11,12] and the nucleolus was established as the site of ribosome biogenesis [13,14].
For three decades, research was focused on delineating the mechanisms that govern ribosomal synthesis and assembly. Recent evidence, however, suggests that the nucleolus is a multifunctional organelle. For instance, a large—if not the greatest—number of proteins in the nucleolus have functions that are not directly linked to ribosomal biogenesis [15,16,17]. Moreover, the presence of numerous RNA modifying enzymes in the nucleolus suggests that many non-ribosomal RNA post-transcriptional modifications take place in this organelle. In fact, all snRNAs localize transiently to the nucleolus to be modified by 2’-O-methylation and pseudouridylation [18]; interestingly, RNase P subunits and its RNA component are localized in the nucleolus suggesting that 5’ processing of tRNA might take place here [19,20,21]. Furthermore, the RNA component of Signal Recognition Particle and some of its subunits have a nucleolar biogenesis phase [22,23,24]. Other proteins, such as cell cycle regulatory factors, DNA repair enzymes or signaling kinases, are also known to reside in the nucleolus. Being the center of ribosomal biogenesis, the nucleolus provides a suitable territory to integrate environmental and intracellular signals, such as various stress stimuli ranging from nutrient deprivation and rapid pH fluctuations to genotoxic stress, in order to fine-tune complex biological processes.
In humans, rDNA represents only 0.4% of the genome and is found in low copy numbers within nucleoli, thus, any rDNA insults are expected to represent a minor fraction of total nuclear DNA damage. However, rDNA is considered one of the most unstable genomic sites [25]; the repetitive nature of rDNA renders it prone to improper recombination that may often lead to deletions or rearrangements. Moreover, the high transcription rate of rDNA is thought to cause a great amount of topological stress as well as R-loops, the three-stranded nucleic acid structures that are composed of a RNA:DNA hybrid and the associated non-template single-stranded DNA [26]; if not properly restored, R-loops are thought to generate DSBs [27]. Here, we discuss the processes that surveil genome integrity within nucleoli, how DNA damage affects nucleolar morphology and function and how a stressed nucleolus might affect disease onset and cell viability.
2. Nucleolus and the rDNA: Structure, Function and Organization
The nucleolus is a self-organizing membrane-less “organelle” which is “formed by the act of building a ribosome” [28]. Indeed, the level of ribosome production is reflected on the size of the nucleolus [29] and the molecular processes arising in this organelle determine its structural organization [30]. In higher eukaryotes, electron microscopy reveals that during interphase nucleoli sustain a tripartite structure: the fibrillar center (FC), the dense fibrillar compartment (DFC), and the granular center (GC). FCs contain inactive rDNA with the potential to be directly activated and proteins of the transcription machinery, such as RNA polymerase I, Topoisomerase I and Upstream Binding Factor (UBF) [31,32,33]. The DFC surrounds the FCs and consists of pre-rRNA and early processing factors [34], while at the GC, late processing factors and ribosomal proteins reside and ribosomal assembly takes place [35]. It is now broadly accepted that transcription takes place at the boundary of the FC and the DFC; instead, rRNA processing begins at the DFC and is completed in the GC.
Interestingly, in many eukaryotic cells, the nucleolus becomes disorganized when the cell enters mitosis and transcription is inhibited [36]. Despite nucleolar disorganization, UBF and DNA topoisomerase I remain associated with the NORs. In contrast, proteins from the DFC and the GC, such as nucleophosmin and fibrillarin, dissociate from the nucleolus, although these factors still remain associated with the perichromosomal layer during metaphase and anaphase; in doing so, these proteins form prenucleolar bodies during telophase, before being recruited to the newly formed nucleoli. Depending on the cell type, the nucleolus reappears when RNA polymerase I associates again with one NOR during late anaphase or telophase. In G1, multiple nucleoli may then fuse to form larger nucleoli containing several NORs (reviewed in [37]). The yeast uses a closed mitosis model (the nuclear envelope remains intact); as such, nucleoli are not disrupted during mitosis and are visible throughout the cell cycle [38].
There are four rRNA genes, namely the small subunit S-rRNA (18S), the 5.8S rRNA, the large subunit L-rRNA (25S/28S) and the 5S rRNA. The first three are transcribed as a single precursor rRNA molecule—in the order of S-rRNA, 5.8S rRNA and L-rRNA—by RNA polymerase I, which is then processed to remove the 5’ external transcribed spacer (5’ETS), the two internal transcribed spacers (IGS) flanking the 5.8S rRNA and the 3’ external transcribed spacer (3’ETS). The rDNA repeats are separated by non-transcribed intergenic spacer regions (ITS) [39,40] (Figure 1). Whereas the rRNA genes are highly conserved among species, the various intergenic and transcribed spacer regions are highly divergent [1]. The fourth rRNA (5S rRNA) is also organized in tandem arrays but it is transcribed by RNA polymerase III outside the nucleolus. In yeast, the 5S rDNA is found in the same transcription unit as the other rRNA genes but it is transcribed in the opposite direction by RNA polymerase III. The number of rDNA repeats varies enormously throughout phylogeny. Birds and mammals have 100–300 copies per haploid genome, while amphibians and plants may have thousands of copies [40]. Humans and mice bear ~200 rRNA copies per haploid genome. In humans, rRNA genes are located between the short arm and the satellite body of acrocentric chromosomes 13, 14, 15, 21, and 26 [41]. In mice, rDNA clusters are found within the centromeric regions of chromosomes 12, 15, 16, 18, and 19 [42,43]. In Saccharomyces cerevisiae, the rRNA genes are located on the right arm of chromosome XII in a tandem array of 150–200 copies [44]. The rDNA can also be differentially amplified as in polytene chromosomes in Drosophila salivary gland nuclei or even extrachromosomally as in amphibian oocytes [1].
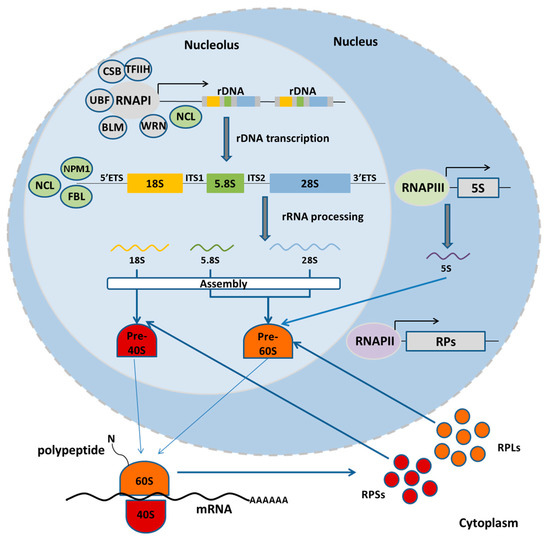
Figure 1.
The function of nucleolus. In normal state, the RNA Pol I machinery transcribes a polycistronic pre-rRNA molecule, which is subsequently processed to mature rRNAs (18S, 5.8S, and 28S) of the small and large ribosomal subunit. Outside the nucleolus, RNA Pol III transcribes the fourth rRNA (5S), while RNA Pol II transcribes the ribosomal protein genes to be translated in the cytoplasm. Ribosomal proteins of the large (RPLs) and the small (RPSs) subunit enter the nucleolus to associate with rRNAs and assemble the ribosomal subunits, which are then exported to the cytoplasm.
The structural and catalytic subunit of ribosomes is composed of rRNA, which, unlike its protein counterparts, cannot be amplified by translation. Instead of investing in higher transcription rates for rDNA or in maximizing ribosomal RNA stability, cells possess a large number of rRNA genes to sustain the high demand for ribosomes. Nevertheless, few hundred rRNA gene copies appear to be enough to meet this requirement, especially when one considers that only a fraction of ribosomal genes is routinely transcribed. Indeed, in yeast, flies or mammals, only 50% of the rRNA gene copies are transcribed; in line, mutant model organisms with less than half of the copies are viable [45,46,47]. Although, at present, such a highly conserved redundancy remains hard to explain, it is possible that distinct cell types or specific developmental stages require the rapid increase in protein synthesis. Moreover, Kobayashi has proposed that rDNA may act as a DNA damage sensor activating DNA repair or else triggering apoptosis [25] (further discussed in Section 6).
The formation and dissolution of nucleoli during cell cycle progression raised the question of what are the essential requirements for nucleolar formation. Subsequent studies revealed UBF, a transcription factor that recognizes the ribosomal RNA gene promoter, as a key factor in this process, as it binds excessively on human rDNA copies throughout the cell cycle. Indeed, a series of experiments with pseudoNORs, that contained megabase-long synthetic UBF-binding site repeats and were inserted into non-acrocentric human chromosomes, provided the first evidence that binding of UBF to specific DNA sites is required for the appearance of active NORs [48]. During metaphase, pseudoNORs form subnucleolar structures with a protein composition that is similar to that seen in endogenous active NORs. PseudoNORs, however, do not contain promoters or pre-rRNA-coding sequences and, thus, they do not form nucleoli during interphase. Further evidence for the pivotal role of UBF in nucleolar formation was given by neoNORs. Grob et al. constructed artificial NORs by combining UBF-binding site arrays with human promoters that drive the transcription of mouse pre-rRNAs. NeoNORs do form functional neonucleoli, organized in FCs, DFCs and GCs and produce functional ribosomal subunits that can be found in polysomes [49]. In essence, UBF binding to DNA, along with active rDNA transcription, is sufficient to form a nucleolus in human cells. In line, UBF marks the presence of active NORs throughout the cell cycle; in doing so, it remains attached to those NORs that were active during the previous interphase to ensure the rapid re-initiation of transcription. This strategy appears to be highly conserved, as UBF is present across all animal phyla.
3. The Nucleolus Associated Heterochromatin and Its Role in Safeguarding Genome Integrity
During interphase, each nucleolus is typically surrounded by heterochromatin. A subset of rDNA loci is transcribed by looping inside the nucleolus. Instead, inactive rDNA genes are associated with the nucleolar periphery and are characterized by repressive marks of constitutive heterochromatin [50,51]. Using next generation sequencing technologies, two independent studies have recently identified the nucleolus-associated chromatin domains (NADs), that is the DNA derived from isolated nucleoli [52,53]. Both studies identified similar NAD genomic regions containing apart from rDNA, also satellite, centromeric and pericentromeric DNA, silent chromatin and the absence of genes.
It is thought that the heterochromatin at rDNA repeats may play a role in regulating rRNA transcription. In mammalian cells, rDNA exists in an active, silent or stable silent chromatin state. Active rDNA genes are typically not CpG-methylated or associated with nucleosomes [54] and reside at the boundary of the FC and the DFC. The silent chromatin is organized in nucleosomes bearing histone repressive marks; it contains non-methylated DNA and is found at the center of FCs. Instead, stable silent rRNA genes acquire all heterochromatin marks, both histone repressive marks and DNA CpG methylation. This range of modifications likely allows for the fine-tuning of transcription regulation and for the control of rDNA stability. When rRNA synthesis needs to be minimized, rRNA genes acquire silent histone modifications but not CpG methylation; this is a silent chromatin state that can easily be reversed when necessary. In contrast, stable silent rDNA cannot easily be reverted owing to extensive CpG methylation [55,56,57].
The evolutionarily conserved organization of a fraction of rDNA into heterochromatin seems to be important not only for nucleolar structure and regulation of rDNA transcription, but also for safeguarding genomic stability by rendering heterochromatic rDNA less accessible to intrinsic genotoxic byproducts of metabolism, or to the cellular DNA recombination machinery. In mammals, switching rDNA between active and inactive state is mediated by the nucleolar remodeling complex (NoRC) that belongs to the ATP-dependent SNF2h family. Transient deletion of its large subunit TTF-1-interacting protein-5 (TIP5) [56,58,59] results in loss of heterochromatin, rDNA instability, nucleolar disorganization and cellular senescence [25,60], implying that rDNA silencing plays an important role in maintaining rDNA integrity. Loss of CpG methylation, by DNMT1 or DNMT3b inactivation or using the DNA methylation inhibitor aza-dCIn, reactivates silent rRNA genes and results in rDNA instability as evident by the formation of episomal rDNA circles [61]. In Drosophila, deletion of the histone methyltransferase Su(var)3-9, responsible for establishing H3K9me2 in constitutive heterochromatin, results in global heterochromatin disruption, rDNA instability and formation of extrachromosomal rDNA circles [60]. It cannot be distinguished, however, whether rDNA instability is caused directly by heterochromatin disruption or is the consequence of global genome instability.
4. Genome Maintenance inside the Nucleolus
rDNA is considered a highly fragile genomic entity. High transcription rates cause topological stress due to the extensive formation of RNA:DNA hybrids. If not readily dissolved in a timely manner, such structures may trigger cytotoxic DNA double strand breaks (DSBs). Topoisomerases, the enzymes that participate in the over- or un-winding of DNA, are known to relax this topological stress. In support, inhibition of topoisomerases is known to induce DSBs in rDNA or other highly transcribed sites [62,63,64]. Moreover, the repetitive nature of rDNA renders it particularly prone to homologous recombination events; i.e., DNA repeats within the same or between different clusters inappropriately recombined, which triggers either chromosomal translocations or the loss/amplification of rDNA. Interestingly, the striking copy number variation between individuals or even within cells of the same individual, suggests a high level of meiotic (>10%) and mitotic recombination frequency in the rDNA locus [65]. Moreover, increased mitotic instability of the rDNA locus is a trait of more than half of lung and colorectal adult cancer samples as detected by rDNA rearrangements, relative to the surrounding normal tissue or the peripheral blood of each patient [66]. Indeed, the recently identified presence of palindromic rDNA repeats in prematurely aging WRN patients [67], the abnormal nucleolar morphology of cancer cells as well as the cancer prone phenotype of certain ribosomopathies, suggest that rDNA instability may be, in several occasions, causal to premature disease onset and/or progression, including cancer. In this section, we discuss the mechanisms that maintain genome stability within the nucleolus, with a focus on DSB repair and Nucleotide Excision Repair. Repair of smaller non-helix distorting lesions by Base Excision Repair and Mismatch Repair is not included in this review, as the contribution of these pathways in nucleolar genome maintenance has not yet been evidenced.
4.1. DSB Repair and the Nucleolus
DNA DSBs are highly cytotoxic lesions that interfere with transcription and DNA replication, triggering large chromosomal rearrangements and threatening cell survival. Similar to every other genomic site, persistent R-loops, endogenously generated free radicals and exposure of rDNA to ionizing radiation (IR) or genotoxins e.g., bleomycin or zeocin may trigger DSBs. Eukaryotic cells have evolved two main pathways to resolve DSBs, the non-homologous end joining pathway (NHEJ) and the homologous recombination pathway (HR). In mammalian cells, NHEJ is active throughout the cell cycle; instead, HR is active in late S and G2 phases when the sister chromatid provides the homologous template required for repair DNA synthesis.
In mammalian cells, rDNA breaks cause nucleolar caps; these are unique structures at the nucleolar periphery that contain individual, most probably damaged, NORs [68]. Similar to the relocalization of rDNA DSBs to nucleolar caps, DSBs outside the nucleolus often cluster together [69,70], likely to facilitate HR repair through homologous pairing [71,72]. Accordingly, DSBs in heterochromatic DNA relocate outside heterochromatin for accessibility of repair factors [73,74]. Interestingly, approximately half of the rDNA repeats are silent and localized at the perinucleolar heterochromatin. rDNA DSBs relocate to nucleolar caps, probably independently of the chromatin state where the lesion occured [68], which could be considered a consensus mechanism of DSB relocation for repair in any genomic locus. In yeast, DNA DSBs also relocate to the periphery of the nucleolus, but unlike mammalian cells, the nucleolus does not segregate [75]. Nucleolar segregation is a consequence of DNA-dependent protein kinase (DNA-PK) and/or Ataxia-telangiectasia mutated (ATM) dependent transcription inhibition. In mouse cells, ATM inhibits RNA polymerase I by interfering with the formation of the pre-initiation complex, leading to the premature displacement of elongating RNA polymerases [76,77]. On the contrary, studies in human cells show that rDNA silencing upon exposure to IR occurs in a DNA-PK-dependent manner [78]; the latter recruits PARP1 to damaged chromatin sites to cease ongoing transcription. DNA-PK has also been shown to inhibit RNA polymerase I in vitro by blocking the formation of the pre-initiation complex [79,80] (Figure 2a). Even so, both kinases are able to contribute in post-DNA damage associated rDNA silencing through the (in)direct phosphorylation of nucleolar proteins involved in transcription and rRNA processing [81,82,83,84].
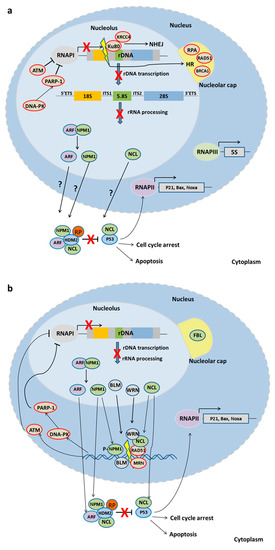
Figure 2.
The nucleolar response to DNA damage. (a) DSBs in the rDNA cause nucleolar segregation and nucleolar cap formation. While DSBs outside the nucleolus result in a global nuclear response, DNA damage inside the nucleolus elicits only a localized response. DNA damage kinases, ATM and DNA-PK, inhibit RNA Pol I to cease transcription and subsequent rRNA processing and ribosomal assembly. Ribosomal proteins and possibly nucleolar proteins activate the P53 pathway. Damaged DNA is predominantly repaired by NHEJ, while HR, when activated, contributes to rDNA instability. (b) DSBs outside the nucleolus also induce nucleolar segregation and cap formation. RNA Pol I is inhibited by ATM and DNA-PK kinases while nucleolar proteins translocate to the damaged site in the nucleoplasm to participate in DNA repair as well as to the cytoplasm to activate the stress response.
To determine whether rDNA DSBs are repaired by NHEJ or HR, three recent studies have employed the Physarum polycephalum endonuclease I-Ppol that recognizes a 15-base pair site within the 28S coding sequence to induce DSBs within rDNA. Harding et al. identified NHEJ as the predominant DSB repair pathway in the nucleolus [85]. Unlike the depletion of HR factors, inhibition of DNA-PK or the transient depletion of NHEJ factors triggered the accumulation of DSBs upon I-Ppol induction. The exact site of NHEJ repair has not yet been clarified; Ku80, DNA-PK and XRCC4 are detected in the nucleoli but are absent in nucleolar caps, suggesting that these factors associate with DNA lesions within nucleoli [86]. Instead, van Sluis et al. showed that HR is also involved in the DSBs repair of rDNA [68]. HR takes place in nucleolar caps that stain positive for rDNA, γH2AX and distinct HR proteins, such as BRCA1, RPA2 and Rad51. In addition, unscheduled DNA synthesis was detected in nucleolar caps, indicating repair by DNA synthesis. HR as well as unscheduled DNA synthesis in nucleolar caps, occurred also in G1, suggesting that HR in rDNA happens in the absence of a sister chromatid, possibly by using other rDNA repeats as a template. A drawback of the methodology used, however, may be that the I-Ppol recognition site is found in every rDNA repeat. Thus, both studies were likely performed under an unusually high number of rDNA DSBs. Warderdam et al. investigated the time course of rDNA breaks resolution and concluded that DNA damage is repaired by NHEJ independently of the break source, I-Ppol or IR; in fact, HR appears to impede proper repair and when the latter DNA repair mechanism is active, it results in the reduction of rDNA repeats [87]. Indeed, rDNA HR mediates an error-prone repair process that causes rDNA instability. In essence, the presence of DSBs in nucleoli triggers an ATM- and/or DNA-PK-dependent rDNA silencing leading to nucleolar segregation and cap formation. NHEJ is likely the predominant repair pathway, while HR takes place in a cell cycle independent manner and possibly results in rDNA repeat reduction. Interestingly, the response to rDNA DSB in nucleoli is also unique; unlike the pan-nuclear response of cells to DSBs, in nucleoli ATM is activated locally and transcription inhibition is restricted only to specific rDNA sites [68,76].
4.2. Nucleotide Excision Repair and the Nucleolus
Nucleotide excision repair (NER) is a versatile DNA repair pathway that resolves a variety of helix distorting lesions, such as bulky adducts and UV-induced cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts, which impede transcription and DNA replication. NER is divided into two sub-pathways: the global genome repair (GG-NER), which resolves DNA lesions throughout genome, and the transcription-coupled repair (TC-NER) that is directed on the transcribed strand of active genes. The two NER subpathways differ in how the lesion is recognized, with XPC-RAD23 being specific for GG-NER whereas, during TC-NER, CSA and CSB factors are recruited at the DNA damage-stalled RNA Pol II; both subpathways then merge to a common repair machinery [88].
Although TC-NER factors are localized in nucleoli, a number of studies have shown that TC-NER is not active in rDNA loci whereas GG-NER may be hampered by the structural compartmentalization of rDNA, evident by the absence of XPC in the nucleolus [89]. In hamster cells, CPDs remained unresolved in the rDNA locus even 24 h post UV irradiation [90,91]. Both NER subpathways are also inactive in mouse rDNA [92]. In human cells, CPDs are removed from rDNA at a very slow rate, both in growing and quiescent cells, implying that CPD repair occurs independently of transcription. On the other hand, in the absence of functional TC-NER (CSA or CSB), CPD removal was substantially reduced compared to repair-proficient cells, whereas cells deficient in GG-NER showed no repair of CPDs [93], implying a role for both GG- and TC-NER in repairing the rDNA locus. Conclusively, how and to what extent NER factors contribute to the resolution of UV-induced damage remains unclear, though it has been proposed that rDNA is likely maintained by recombination instead of TC-NER [91]. This may be further supported by recent findings that implicate CSB in Transcription-Coupled Homologous Recombination of oxidatively induced DSBs [94], though such a mechanism has not yet been described, for neither the nucleolus nor UV-induced lesions. Recently, CSB has also been shown to participate in repair of nucleolar oxidative lesions [89]. Interestingly, unlike DSB formation that can be targeted to the nucleolus by employing sequence-specific nucleases such as I-Ppol, exposure to UV irradiation introduces lesions to the whole nucleus and elicits the DNA damage response that may potentially impede rDNA repair due to nucleolar re-organization upon stress.
5. The Nucleolus upon Stress
Being the hub integrating environmental and intracellular signals, the nucleolus undergoes dramatic structural changes upon DNA damage. When cells are exposed to damage, for example UV or IR, the FC and the GC condense and separate from the nucleolar body resulting in the so-called nucleolar segregation. This is accompanied by proteins moving from nucleoli to the nucleoplasm and vice versa.
It is now established that any kind of stress that impairs any step of ribosomal biogenesis, i.e., rDNA transcription, rRNA processing, ribosomal protein synthesis and the ribosomal subunits assembly, causes nucleolar disruption and subsequently activates P53-dependent response [95,96,97,98], resulting in cell cycle arrest or apoptosis. P53-independent nucleolar stress response has also been described in yeast, that lacks P53, and in Drosophila, that lacks a Double minute 2 (MDM2) homologue [98]. In mammals though, such a pathway remains either silent or not yet uncovered. In the nucleus, P53 upregulates transcription of cell cycle (e.g., p21) and apoptotic regulators (e.g., Bax, Noxa) transcribed by RNA pol II, whereas in the cytoplasm P53 activates the mitochondrial apoptotic pathway [99,100,101]. P53 may also inhibit RNA Pol I transcription by disrupting the interaction between the basal transcription factors SL1-UBF, which results in a decrease in ribosome subunit biogenesis [102]. Under normal physiological conditions, the P53 levels are kept low: the interaction with E3 ubiquitin ligases, such as HDM2 for human and MDM2 for mouse, ensures the P53 export from the nucleus (monoubiquitinylation) and degradation (polyubiquitinylation) or inhibition of its transactivation domain. Upon cellular stress (e.g., DNA damage, pH fluctuation, heat shock or nutrient deprivation), the HDM2-P53 interaction is disrupted, thus activating P53. Importantly, the P53-mediated stress response is elicited only if the nucleolar function is affected [97] (Figure 2b).
Nucleolar and ribosomal proteins play a central role in the regulation of the P53-mediated stress response pathway. Nucleophosmin (NPM1 or B23) is a nucleolar protein with diverse functions, including ribosome biogenesis, histone chaperoning and centrosome duplication (reviewed in [103]). NPM1 can regulate the P53-HDM2 axis by its interaction with ARF. ARF (p19ARF for human, p14ARF for mice) is a tumor suppressor transcribed from an alternative reading frame (ARF) of the p16INK4a cyclin-dependent kinase inhibitor; ARF disrupts P53-HDM2 complex by its interaction with HDM2. Under physiological conditions, NPM1 interacts constitutively with ARF, which is then sequestered in the nucleolus. Upon stress, activation of NPM1 releases ARF which in turn translocates to the nucleoplasm, where it interacts with HDM2 to destabilize the P53-HDM2 complex and eventually activate P53 [104]. NPM1 itself has been reported to directly bind either HDM2 or p53 to regulate the HDM2-p53 pathway [105].
Nucleolin (NCL) also modulates the HDM2-P53 axis. NCL is a multifunctional nucleolar protein which participates in rDNA chromatin remodeling through its activity as histone chaperone of H2A-H2B, is essential for basal transcription factors loading and RNA Pol I progression and contributes to pre-rRNA processing (reviewed in [103]). The multifunctionality of NCL is due to its tripartite structure (contains three distinct functional domains) and its ability to shuttle between the nucleus and the cytoplasm. NCL interacts with both HDM2 and P53 [106,107] to regulate P53 stability. Upon stress, NCL translocates to the nucleoplasm where it interacts with HDM2, thus promoting HDM2 ubiquitination and degradation and disrupting its interaction with P53. Moreover, upon DNA damage, NCL forms a ternary complex with P53 and HDM2, which recruits the ubiquitin protease HAUSP. HAUSP regulates NCL stability by deubiquination [108], but how these interactions contribute to P53 regulation remain unclear. NCL has also been reported to regulate P53 on the translational level. NCL binds both 5’ and 3’UTR of p53 mRNA to repress its translation. DNA damage releases ribosomal proteins from the ribosomes, resulting in increased levels of ribosomal protein RPL26 which disrupts the NCL interaction with p53 mRNA, allowing for its translation [109].
Nucleolar Proteins in DNA Repair
Besides their role in the stress response, NCL and NPM1 emerge also as players in repair of damaged DNA outside the nucleolus. NCL interacts with Rad51, the key recombinase of HR-mediated DSB repair; NCL inhibition sensitizes cells to DSBs induced by the topoisomerase II inhibitor amsacrine [110]. Moreover, both NCL and NPM1 are part of the switch recombinase complex SWAP of B-lymphocytes, suggesting a contribution of these nucleolar proteins in recombination repair [111]. DNA DSBs generated by exposure of cells to IR or to genotoxins, e.g., camptothecin, induce NCL translocation to the nucleoplasm [106,112], where it colocalizes with DSB repair proteins [113]. When NCL is depleted from cells treated with genotoxic agents, DSB signaling and repair factors, such as the MRN complex, H2Ax, ATM and MDC1, are recruited at sites of damage but no DSB resolution occurs, indicating a defect at later stages of repair [113,114]. During DSB repair, the nucleosome at the damaged site is disrupted by eviction of histones H2A-H2B and H3-H4, to enable access of the repair proteins to DNA. In NCL depleted cells, H2A-H2B remains in place, indicating that NCL mediates H2A-H2B mobilization from chromatin to the nucleoplasm [113,114]. In support, NCL facilitates H2A-H2B removal from transcriptional sites by the chromatin remodeling complexes SWI/SNF and ACF, both of which are also involved in DSB repair [115,116]. Moreover, NCL interacts with WRN inside the nucleolus, whereas upon IR- or camptothecin-induced DSBs, both translocate at damaged sites in the nucleoplasm where they re-associate. Their interaction inhibits the helicase but not exonuclease activity of WRN in vitro, but how this may contribute to repair remains elusive [112].
Besides NCL, several lines of evidence support an emerging role for NPM1 in DSB repair. Unlike NCL, NPM1 does not massively translocate to the nucleoplasm upon induction of DNA DSBs. A minor population though is phosphorylated at residue T199 and subsequently recruited to ubiquitin conjugates present at DSBs [117]. In vivo expression of a T199A non-phosphorylatable mutant of NPM1 did not compromise DSB foci formation or the recruitment of DNA repair factors, but similar to NCL, resulted in failure of clearance of DSBs at 6 h post IR [117]. NPM1 has been reported to hold chaperone activity with histones H3–H4 as well as H1 and H2 [118,119,120], suggesting that, together with NCL, it could contribute to nucleosome eviction at DSB sites.
By participating in DNA repair outside the nucleolus, nucleolar proteins might as well serve as signals for switching on (nucleolar disruption and protein translocation) and off (sequestration back to the nucleolus, which is then reformed) the stress response.
6. rDNA Damage and Disease Onset
The emerging role of rDNA damage in disease onset supports the notion that rDNA damage accumulation may be causal to accelerated aging (progeria) and/or age-related diseases. At present, the impact of rDNA damage replicative senescence and aging has been extensively studied in yeast. For instance, the ribosomal gene copy number is critical for yeast lifespan and is tightly maintained by a mechanism that involves Fob1p, Sir2p and the replication fork barriers (RFBs). RFBs are found downstream of the pre-rRNA coding sequences [121] and are considered rDNA recombination hot spots [122]. Fob1 binding to an RFB [123] mediates a DSB at the stalled fork [124,125] which is repaired by HR [126] and is regulated by the histone deacetylase Sir2p and its target, non-coding promoter E-pro at the intergenic spacer [127]. When the ribosomal gene copy number is below the wild type dosage, Sir2p is inactivated allowing E-pro to be transcribed. E-pro transcripts remove cohesins that hold sister chromatids together, thus promoting unequal pairing and resulting in copy number increase. In case the rDNA copy number is close to wild type levels, Sir2p represses E-pro transcription, cohesins stay in place and equal sister chromatid recombination takes place without altering the copy number. Aberrant intra-chromosomal recombination results in extra-chromosomal rDNA circles (ERCs). Fob1p, Sir2p and ERCs have been shown to determine yeast lifespan: deletion of FOB1 increases rDNA stability and lifespan [45,128], SIR2 mutants accumulate ERCs and show reduced life span [45] and ERCs accumulate preferentially in the aging mother cell [129,130]. Thus, it has been suggested that ERCs are toxic byproducts that act as pro-senescence effectors. Recently, Kobayashi has proposed that inactive rDNA copies are essential for suppressing DNA damage response [131] and that rDNA loss itself activates DDR which in turn causes senescence in a ribosomal biogenesis-independent manner [132,133]. Interestingly, yeast strains with low copy rDNA, where most of the repeats are transcriptionally active, show increased sensitivity to DNA damage, e.g., UV irradiation [132].
It is widely accepted that DNA damage accumulation is associated with aging, evident by the accelerated aging syndromes, many of which are caused by inborn DNA repair defects. Thus, the notion that rDNA instability may causally trigger progeria gains scientific interest. Recently, RFBs have been identified in mouse and human rRNA genes [134]. Mammalian SIR2 homologue is not known to be involved in nucleolar DNA integrity, though Sirtuin 7 resides in the nucleolus and activates RNA polymerase I [135,136]. Almost 30% of rDNA repeats in normal individuals exist as palindromic rather than tandem repeats and are probably non-functional. Their frequency goes up to 50% in individuals with Werner syndrome (WRN) progeria; WRN patients carry inborn defects in the WRN RecQ helicase; the latter implies that altering of the fine balance between canonical and palindromic repeats may lead to premature aging phenotypes [67]. It remains, however, to be found whether such palindromic repeats are equivalent to the yeast ERCs. BLM and ATM are also candidates for linking aging to rDNA maintenance. Cells derived from Bloom syndrome (mutated BLM) or Ataxia Telangiectasia (mutated ATM) patients show high variability of the rDNA copy number due to mitotic recombination in relation to their wild type counterparts [137,138]. Moreover, recent evidence suggests reduction of ribosomal DNA repeats as a trait of human aging [139,140]. Age-related neurological disorders, such as Hodgkin’s disease [141] and Parkinson’s disease [142], show deregulated rDNA transcription and nucleolar dysfunction. rDNA copy number variability has been associated with neurodegeneration: increased number was detected in patients with dementia with Lewy bodies [138] and elevated repeats of the 18S rDNA locus combined with increased silent chromatin marks in Alzheimer’s disease patients [143].
The association of nucleolar function with cancer was established over a hundred years ago, when it was observed that cancer cells have large and abnormal nucleoli [144]. Abnormal morphology is in large attributed to hyperactivation of rDNA transcription required to sustain the high metabolic and proliferation rate of cancer cells [145]. Cancer cells are also characterized by rDNA instability, as manifested by increased mitotic instability observed in more than half of lung and colorectal adult cancer samples relative to surrounding normal tissue or peripheral blood of each patient [66]. Bloom syndrome and Ataxia telangiectasia patients also present with high risk of cancer incidence, though this could be acquired to generalized genomic instability.
DNA Repair Factors Regulating rDNA Transcription and Disease
Deregulation of rDNA transcription is associated with disease: hyperactivation is manifested in cardiovascular disease [146] and cancer, while downregulation is a trait of premature aging syndromes and age-related neurological diseases, e.g., Hodgkin’s disease and Parkinson’s disease. Besides their role in genome maintenance, BLM, WRN and NER factors all interact with RNA Pol I and are all causal to progeria and growth retardation. BLM and WRN both belong to the RecQ subfamily of ATP dependent 3’–5’ DNA helicases, participate in replication and repair of DSBs and positively regulate rDNA transcription. WRN localization in the nucleolus is tightly connected to active rDNA transcription: in quiescent cells, or cells treated with transcription blocking agents, WRN localizes to the nucleoplasm, while upon RNA Pol I reinitiation WRN shuttles back to the nucleolus [147,148]. WRN deficient fibroblasts show decreased levels of rRNA transcription. Exogenously expressed wild type WRN recovers rRNA transcription through interaction of WRN with RNA Pol I [147]. Moreover, WRN mediates promoter clearance and vascular endothelial growth factor (VEGF), fibroblast GF-β (FGF-β) and epidermal GF (EGF) stimulation of RNA Pol I [149]. BLM localization to the nucleolus is also positively associated with ongoing rDNA transcription. BLM deficient cells also show reduced rRNA transcription. BLM interacts with the RNA Pol I subunit RPA194 and possibly participates together with DNA topoisomerase I in unwinding the RNA:DNA hybrids that form in the rDNA and inhibit ongoing transcription [150,151].
Patients with defects in the GG-NER or TC-NER sub-pathway, manifest with strikingly heterogeneous clinical outcomes, ranging from cancer to progeria and developmental or metabolic abnormalities. The latter argues for NER factors having additional roles beyond DNA repair. Indeed, NER factors are now known to function in the regulation of gene expression in human cells [152], the transcriptional reprogramming of pluripotent stem cells [153] and the fine regulation of growth genes during murine development [154,155]. Patients with Cockayne syndrome (defective in Csb or Csa) and Trichothiodystrophy (defective in Xpb or Xpd) manifest with progeria and mental retardation. Cells isolated from patients baring mutations in Csb, Xpb or Xpd or cells transiently knocked down for CSB show reduced rRNA synthesis [156,157]. Inside the nucleolus, CSB interacts with RNA Pol I, TFIIH subunits XPB and XPD, XPG endonuclease and basal RNA Pol I-associated transcription factors to activate rRNA transcription [156]. Moreover CSB, XPB and XPD are implicated in positive regulation of RNA Pol I elongation [158,159]. A recent study suggests a possible role for CSB in rDNA transcription. rDNA tends to form G quadruplexes (G4) that block ongoing transcription. In CSB deficient cells, nucleolar transcription is hampered, leading to mitochondrial dysfunction. CSB can resolve unimolecular G4 structures in vitro and recombinant CSB rescues the mitochondrial dysfunction of CSB deficient cells [160].
7. Conclusions
The nucleolus is well recognized as a central cellular hub for sensing stress stimuli and coordinating stress response. The functional interplay of genome maintenance and ribosomal biogenesis pathways is just emerging as a new player in age-related diseases and premature aging syndromes (Figure 3). DNA damage causes dramatic changes in nucleolar architecture, which is accompanied by protein translocation from nucleoli to the cytoplasm and vice versa. Shuttling of nucleolar proteins between the nucleolus and the cytoplasm might also serve as another way of nucleolar sensing of DNA damage and, subsequently orchestrating a response. Moreover, DNA repair proteins that reside in nucleoli to maintain rDNA stability and regulate rRNA transcription offer another level of ribosomal control and disease onset. Premature aging syndromes present with segmental progeria instead of the global functional decline observed in aging. Progeria and a number of age-related diseases show tissue specific aberrant nucleolar function, implying cell type dependent requirements for proper nucleolar function. We do not know though whether nucleolar dysfunction is the direct cause or an effect of disease. The use of in vivo tagging technologies and tissue specific knock out animal models as well as targeting or excluding proteins from the nucleolus, if possible, may contribute to understanding this intertwined network and suggest new therapeutic strategies targeting the nucleolus.
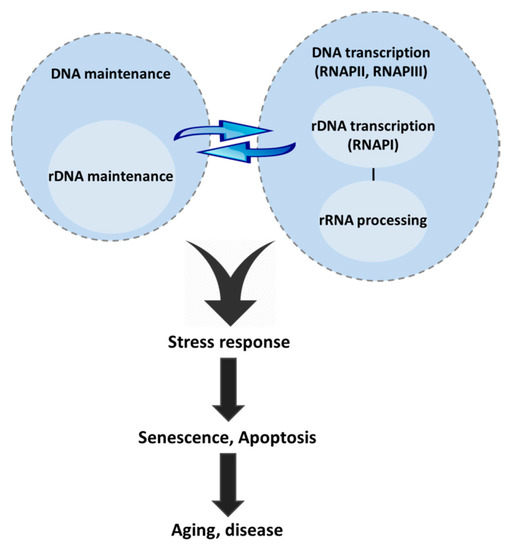
Figure 3.
The crosstalk between the DNA and rDNA maintenance pathways and rRNA biogenesis regulates the stress response. DNA damage dramatically affects the nucleolar architecture and function through a network of interactions and translocations where rRNA biogenesis factors participate in DNA repair. Vice versa, DNA maintenance factors participate in and regulate rDNA transcription. In addition, nucleolar factors directly regulate the stress response. Genome maintenance pathways, nucleolar transcription and rRNA processing form a complex network that converges to the stress response, activation of which ultimately leads to aging and/or age-related disease or progeria, whereas evasion leads to cancer.
Acknowledgments
We would like to thank George Garinis for his valuable comments. The THALIS “miREG” (MIS380247), the ARISTEIA “TagNER” (45) and the Horizon 2020 ERC Consolidator grant “DeFiNER” (GA64663) supported this work.
Author Contributions
Maria Tsekrekou conceived and wrote the manuscript; Kalliopi Stratigi wrote and edited; and Georgia Chatzinikolaou wrote, edited and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hadjiolov, A.A. The Nucleolus and Ribosome Biogenesis Cell Biology Monographs; Springer: New York, NY, USA, 1985. [Google Scholar]
- Fontana, F. Traite sur le Venin de la Viper, sur les Poisons Americains, sur le Laurier-Cerise et sur Quelques Autres Poisons Vegetaux; Gibelin: Florence, Italy, 1781. [Google Scholar]
- Montgomery, T. Comparative cytological studies, with especial regard to the morphology of the nucleolus. J. Morphol. 1898, 15, 265–582. [Google Scholar] [CrossRef]
- Maggi, L.B., Jr.; Weber, J.D. Nucleolar adaptation in human cancer. Cancer Investig. 2005, 23, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Rudra, D.; Warner, J.R. What better measure than ribosome synthesis? Genes Dev. 2004, 18, 2431–2436. [Google Scholar] [CrossRef] [PubMed]
- Heitz, E. Nukleolen und chromosomen in der Gattung Vicia. Planta 1931, 15, 495–505. [Google Scholar] [CrossRef]
- McClintock, B. The relationship of a particular chrmosomal element to the development of the nucleoli in Zea mays. Cell Tissue Res. 1934, 21, 294–326. [Google Scholar]
- Pederson, T. The nucleolus. Cold Spring Harbor Perspect. Biol. 2011, 3, a000638. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.D.; Gurdon, J.B. Absence of Ribosomal Rna Synthesis in the Anucleolate Mutant of Xenopus Laevis. Proc. Natl. Acad. Sci. USA 1964, 51, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Ritossa, F.M.; Spiegelman, S. Localization of DNA Complementary to Ribosomal RNA in the Nucleolus Organizer Region of Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 1965, 53, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Birnstiel, M.L.; Wallace, H.; Sirlin, J.L.; Fischberg, M. Localization of the ribosomal DNA complements in the nucleolar organizer region of Xenopus laevis. Natl. Cancer Inst. Monogr. 1966, 23, 431–447. [Google Scholar] [PubMed]
- Ritossa, F.M.; Atwood, K.C.; Lindsley, D.L.; Spiegelman, S. On the chromosomal distribution of DNA complementary to ribosomal and soluble RNA. Natl. Cancer Inst. Monogr. 1966, 23, 449–472. [Google Scholar] [PubMed]
- Edstrom, J.E.; Grampp, W.; Schor, N. The intracellular distribution and heterogeneity of ribonucleic acid in starfish oocytes. J. Biophys. Biochem. Cytol. 1961, 11, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.P.; Errera, M. The role of the nucleolus in ribonucleic acid-and protein synthesis: I. Incorporation of cytidine into normal and nucleolar inactivated HeLa cells. Biochim. Biophys. Acta 1961, 49, 47–57. [Google Scholar] [CrossRef]
- Pendle, A.F.; Clark, G.P.; Boon, R.; Lewandowska, D.; Lam, Y.W.; Andersen, J.; Mann, M.; Lamond, A.I.; Brown, J.W.; Shaw, P.J. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 2005, 16, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lam, Y.W.; Leung, A.K.; Ong, S.E.; Lyon, C.E.; Lamond, A.I.; Mann, M. Nucleolar proteome dynamics. Nature 2005, 433, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.J.; Eggleton, P.; Young, P.J. Joining the dots: Production, processing and targeting of U snRNP to nuclear bodies. Biochim. Biophys. Acta 2008, 1783, 2137–2144. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.R.; Cao, L.G.; Taneja, K.; Singer, R.H.; Wang, Y.L.; Pederson, T. Nuclear domains of the RNA subunit of RNase P. J. Cell Sci. 1997, 110, 829–837. [Google Scholar] [PubMed]
- Bertrand, E.; Houser-Scott, F.; Kendall, A.; Singer, R.H.; Engelke, D.R. Nucleolar localization of early tRNA processing. Genes Dev. 1998, 12, 2463–2468. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Wolenski, J.S.; Wesolowski, D.; Lee, C.; Altman, S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J. Cell Biol. 1999, 146, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.R.; Pederson, T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. USA 1998, 95, 7981–7986. [Google Scholar] [CrossRef] [PubMed]
- Politz, J.C.; Yarovoi, S.; Kilroy, S.M.; Gowda, K.; Zwieb, C.; Pederson, T. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA 2000, 97, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Sommerville, J.; Brumwell, C.L.; Politz, J.C.; Pederson, T. Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J. Cell Sci. 2005, 118, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. Bioessays 2008, 30, 267–272. [Google Scholar] [CrossRef] [PubMed]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Hamperl, S.; Cimprich, K.A. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair 2014, 19, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Melese, T.; Xue, Z. The nucleolus: An organelle formed by the act of building a ribosome. Curr. Opin. Cell Biol. 1995, 7, 319–324. [Google Scholar] [CrossRef]
- Smetana, K.; Busch, H. The Nucleolus and Nucleolar DNA. In The Cell Nucleus; Busch, H., Ed.; Academic Press: New York, NY, USA, 1974; pp. 73–147. [Google Scholar]
- Stoykova, A.S.; Dabeva, M.D.; Dimova, R.N.; Hadjiolov, A.A. Ribosome biogenesis and nucleolar ultrastructure in neuronal and oligodendroglial rat brain cells. J. Neurochem. 1985, 45, 1667–1676. [Google Scholar] [CrossRef] [PubMed]
- Casafont, I.; Navascues, J.; Pena, E.; Lafarga, M.; Berciano, M.T. Nuclear organization and dynamics of transcription sites in rat sensory ganglia neurons detected by incorporation of 5-fluorouridine into nascent RNA. Neuroscience 2006, 140, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.O.; Barthelmes, H.U.; Boege, F.; Mielke, C. The N-terminal domain anchors human topoisomerase I at fibrillar centers of nucleoli and nucleolar organizer regions of mitotic chromosomes. J. Biol. Chem. 2002, 277, 35932–35938. [Google Scholar] [CrossRef] [PubMed]
- Roussel, P.; Andre, C.; Masson, C.; Geraud, G.; Hernandez-Verdun, D. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 1993, 104, 327–337. [Google Scholar] [PubMed]
- Puvion-Dutilleul, F.; Puvion, E.; Bachellerie, J.P. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with a 5’ETS leader probe. Chromosoma 1997, 105, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Sirri, V.; Urcuqui-Inchima, S.; Roussel, P.; Hernandez-Verdun, D. Nucleolus: The fascinating nuclear body. Histochem. Cell Biol. 2008, 129, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Grummt, I. Cell cycle-dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA 1999, 96, 6096–6101. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Verdun, D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011, 2, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Carmo-Fonseca, M.; Mendes-Soares, L.; Campos, I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000, 2, e107–e112. [Google Scholar] [CrossRef] [PubMed]
- Miller, O.L., Jr.; Beatty, B.R. Visualization of nucleolar genes. Science 1969, 164, 955–957. [Google Scholar] [CrossRef] [PubMed]
- Long, E.O.; Dawid, I.B. Repeated genes in eukaryotes. Annu. Rev. Biochem. 1980, 49, 727–764. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.S.; Warburton, D.; Atwood, K.C. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. USA 1972, 69, 3394–3398. [Google Scholar] [CrossRef] [PubMed]
- Dev, V.G.; Tantravahi, R.; Miller, D.A.; Miller, O.J. Nucleolus organizers in Mus musculus subspecies and in the RAG mouse cell line. Genetics 1977, 86, 389–398. [Google Scholar] [PubMed]
- Kurihara, Y.; Suh, D.S.; Suzuki, H.; Moriwaki, K. Chromosomal locations of Ag-NORs and clusters of ribosomal DNA in laboratory strains of mice. Mamm. Genome 1994, 5, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Petes, T.D. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA 1979, 76, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Horiuchi, T.; Kobayashi, T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003, 17, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Kwan, E.X.; Wang, X.S.; Amemiya, H.M.; Brewer, B.J.; Raghuraman, M.K. rDNA Copy Number Variants Are Frequent Passenger Mutations in Saccharomyces cerevisiae Deletion Collections and de Novo Transformants. G3 (Bethesda) 2016, 6, 2829–2838. [Google Scholar] [CrossRef] [PubMed]
- Paredes, S.; Maggert, K.A. Expression of I-CreI endonuclease generates deletions within the rDNA of Drosophila. Genetics 2009, 181, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Mais, C.; Wright, J.E.; Prieto, J.L.; Raggett, S.L.; McStay, B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005, 19, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Grob, A.; Colleran, C.; McStay, B. Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev. 2014, 28, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Pontvianne, F.; Blevins, T.; Chandrasekhara, C.; Mozgova, I.; Hassel, C.; Pontes, O.M.; Tucker, S.; Mokros, P.; Muchova, V.; Fajkus, J.; et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013, 27, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, A.; Guibert, S.; Tiwari, V.K.; Ohlsson, R.; Langst, G. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J. 2008, 27, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, A.; Conesa, A.; Santoyo-Lopez, J.; Medina, I.; Montaner, D.; Peterfia, B.; Solovei, I.; Cremer, T.; Dopazo, J.; Langst, G. Initial genomics of the human nucleolus. PLoS Genet. 2010, 6, e1000889. [Google Scholar] [CrossRef] [PubMed]
- Van Koningsbruggen, S.; Gierlinski, M.; Schofield, P.; Martin, D.; Barton, G.J.; Ariyurek, Y.; den Dunnen, J.T.; Lamond, A.I. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 2010, 21, 3735–3748. [Google Scholar] [CrossRef] [PubMed]
- Zentner, G.E.; Saiakhova, A.; Manaenkov, P.; Adams, M.D.; Scacheri, P.C. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011, 39, 4949–4960. [Google Scholar] [CrossRef] [PubMed]
- Santoro, R.; Li, J.; Grummt, I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002, 32, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Santoro, R.; Grummt, I. Epigenetic mechanism of rRNA gene silencing: Temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell Biol. 2005, 25, 2539–2546. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Santoro, R.; Grummt, I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002, 21, 4632–4640. [Google Scholar] [CrossRef] [PubMed]
- Strohner, R.; Nemeth, A.; Jansa, P.; Hofmann-Rohrer, U.; Santoro, R.; Langst, G.; Grummt, I. NoRC—A novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001, 20, 4892–4900. [Google Scholar] [CrossRef] [PubMed]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Karpen, G.H. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007, 9, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Gagnon-Kugler, T.; Langlois, F.; Stefanovsky, V.; Lessard, F.; Moss, T. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell 2009, 35, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Govoni, M.; Farabegoli, F.; Pession, A.; Novello, F. Inhibition of topoisomerase II activity and its effect on nucleolar structure and function. Exp. Cell Res. 1994, 211, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.O.; Krokowski, R.M.; Barthelmes, H.U.; Hock, R.; Boege, F.; Mielke, C. Distinct effects of topoisomerase I and RNA polymerase I inhibitors suggest a dual mechanism of nucleolar/nucleoplasmic partitioning of topoisomerase I. J. Biol. Chem. 2004, 279, 21873–21882. [Google Scholar] [CrossRef] [PubMed]
- Leppard, J.B.; Champoux, J.J. Human DNA topoisomerase I: Relaxation, roles, and damage control. Chromosoma 2005, 114, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Stults, D.M.; Killen, M.W.; Pierce, H.H.; Pierce, A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008, 18, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Stults, D.M.; Killen, M.W.; Williamson, E.P.; Hourigan, J.S.; Vargas, H.D.; Arnold, S.M.; Moscow, J.A.; Pierce, A.J. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009, 69, 9096–9104. [Google Scholar] [CrossRef] [PubMed]
- Caburet, S.; Conti, C.; Schurra, C.; Lebofsky, R.; Edelstein, S.J.; Bensimon, A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005, 15, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Van Sluis, M.; McStay, B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015, 29, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Aten, J.A.; Stap, J.; Krawczyk, P.M.; van Oven, C.H.; Hoebe, R.A.; Essers, J.; Kanaar, R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 2004, 303, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Neumaier, T.; Swenson, J.; Pham, C.; Polyzos, A.; Lo, A.T.; Yang, P.; Dyball, J.; Asaithamby, A.; Chen, D.J.; Bissell, M.J.; et al. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc. Natl. Acad. Sci. USA 2012, 109, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Dion, V.; Kalck, V.; Horigome, C.; Towbin, B.D.; Gasser, S.M. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 2012, 14, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Evdokimova, V.N.; Cuenco, K.T.; Nikiforova, M.N.; Kelly, L.M.; Stringer, J.R.; Bakkenist, C.J.; Nikiforov, Y.E. Homologous chromosomes make contact at the sites of double-strand breaks in genes in somatic G0/G1-phase human cells. Proc. Natl. Acad. Sci. USA 2012, 109, 9454–9459. [Google Scholar] [CrossRef] [PubMed]
- Chiolo, I.; Minoda, A.; Colmenares, S.U.; Polyzos, A.; Costes, S.V.; Karpen, G.H. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 2011, 144, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Jakob, B.; Splinter, J.; Conrad, S.; Voss, K.O.; Zink, D.; Durante, M.; Lobrich, M.; Taucher-Scholz, G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011, 39, 6489–6499. [Google Scholar] [CrossRef] [PubMed]
- Torres-Rosell, J.; Sunjevaric, I.; de Piccoli, G.; Sacher, M.; Eckert-Boulet, N.; Reid, R.; Jentsch, S.; Rothstein, R.; Aragon, L.; Lisby, M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007, 9, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Kruhlak, M.; Crouch, E.E.; Orlov, M.; Montano, C.; Gorski, S.A.; Nussenzweig, A.; Misteli, T.; Phair, R.D.; Casellas, R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 2007, 447, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Oka, Y.; Suzuki, K.; Yamauchi, M.; Mitsutake, N.; Yamashita, S. Recruitment of the cohesin loading factor NIPBL to DNA double-strand breaks depends on MDC1, RNF168 and HP1γ in human cells. Biochem. Biophys. Res. Commun. 2011, 411, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Calkins, A.S.; Iglehart, J.D.; Lazaro, J.B. DNA damage-induced inhibition of rRNA synthesis by DNA-PK and PARP-1. Nucleic Acids Res. 2013, 41, 7378–7386. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Gottlieb, T.M.; Jackson, S.P.; Grummt, I. DNA-dependent protein kinase: A potent inhibitor of transcription by RNA polymerase I. Genes Dev. 1995, 9, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Michaelidis, T.M.; Grummt, I. Mechanism of inhibition of RNA polymerase I transcription by DNA-dependent protein kinase. Biol. Chem. 2002, 383, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Beli, P.; Lukashchuk, N.; Wagner, S.A.; Weinert, B.T.; Olsen, J.V.; Baskcomb, L.; Mann, M.; Jackson, S.P.; Choudhary, C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 2012, 46, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, M.V.; Larsen, D.H.; Bunkenborg, J.; Bartek, J.; Lukas, J.; Andersen, J.S. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell Proteomics. 2010, 9, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Bensimon, A.; Schmidt, A.; Ziv, Y.; Elkon, R.; Wang, S.Y.; Chen, D.J.; Aebersold, R.; Shiloh, Y. ATM-dependent and-independent dynamics of the nuclear phosphoproteome after DNA damage. Sci. Signal 2010, 3, rs3. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.M.; Boiarsky, J.A.; Greenberg, R.A. ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell Rep. 2015, 13, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Moore, H.M.; Bai, B.; Boisvert, F.M.; Latonen, L.; Rantanen, V.; Simpson, J.C.; Pepperkok, R.; Lamond, A.I.; Laiho, M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol. Cell Proteomics. 2011, 10, M111009241. [Google Scholar] [CrossRef] [PubMed]
- Warmerdam, D.O.; van den Berg, J.; Medema, R.H. Breaks in the 45S rDNA Lead to Recombination-Mediated Loss of Repeats. Cell Rep. 2016, 14, 2519–2527. [Google Scholar] [CrossRef] [PubMed]
- Kamileri, I.; Karakasilioti, I.; Garinis, G.A. Nucleotide excision repair: New tricks with old bricks. Trends Genet. 2012, 28, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Menoni, H.; Hoeijmakers, J.H.; Vermeulen, W. Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J. Cell Biol. 2012, 199, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Stevnsner, T.; May, A.; Petersen, L.N.; Larminat, F.; Pirsel, M.; Bohr, V.A. Repair of ribosomal RNA genes in hamster cells after UV irradiation, or treatment with cisplatin or alkylating agents. Carcinogenesis 1993, 14, 1591–1596. [Google Scholar] [CrossRef] [PubMed]
- Christians, F.C.; Hanawalt, P.C. Lack of transcription-coupled repair in mammalian ribosomal RNA genes. Biochemistry 1993, 32, 10512–10518. [Google Scholar] [CrossRef] [PubMed]
- Fritz, L.K.; Smerdon, M.J. Repair of UV damage in actively transcribed ribosomal genes. Biochemistry 1995, 34, 13117–13124. [Google Scholar] [CrossRef] [PubMed]
- Christians, F.C.; Hanawalt, P.C. Repair in ribosomal RNA genes is deficient in xeroderma pigmentosum group C and in Cockayne’s syndrome cells. Mutat. Res. 1994, 323, 179–187. [Google Scholar] [CrossRef]
- Wei, L.; Levine, A.S.; Lan, L. Transcription-coupled homologous recombination after oxidative damage. DNA Repair 2016, 44, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.O. Sensing cellular stress: Another new function for the nucleolus? Sci. STKE 2004, 224, e10. [Google Scholar] [CrossRef] [PubMed]
- Pederson, T.; Tsai, R.Y. In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 2009, 184, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Rubbi, C.P.; Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003, 22, 6068–6077. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Wang, Y.; Raje, H.; Rosby, R.; DiMario, P. Nucleolar stress with and without p53. Nucleus 2014, 5, 402–426. [Google Scholar] [CrossRef] [PubMed]
- Mihara, M.; Erster, S.; Zaika, A.; Petrenko, O.; Chittenden, T.; Pancoska, P.; Moll, U.M. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 2003, 11, 577–590. [Google Scholar] [CrossRef]
- Leu, J.I.; Dumont, P.; Hafey, M.; Murphy, M.E.; George, D.L. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat. Cell Biol. 2004, 6, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Chipuk, J.E.; Kuwana, T.; Bouchier-Hayes, L.; Droin, N.M.; Newmeyer, D.D.; Schuler, M.; Green, D.R. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 2004, 303, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Comai, L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell Biol. 2000, 20, 5930–5938. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.D.; Oeffinger, M. Nucleolin and nucleophosmin: Nucleolar proteins with multiple functions in DNA repair. Biochem. Cell Biol. 2016, 94, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Gjerset, R.A.; Bandyopadhyay, K. Regulation of p14ARF through subnuclear compartmentalization. Cell Cycle 2006, 5, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.; Marine, J.C.; Danovi, D.; Falini, B.; Pelicci, P.G. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002, 4, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Daniely, Y.; Dimitrova, D.D.; Borowiec, J.A. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol. Cell Biol. 2002, 22, 6014–6022. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Rorie, C.J.; Dimitrova, D.; Daniely, Y.; Borowiec, J.A. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene 2006, 25, 7274–7288. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.H.; Park, J.J.; Gu, B.H.; Kim, J.O.; Park, S.G.; Baek, K.H. HAUSP-nucleolin interaction is regulated by p53-Mdm2 complex in response to DNA damage response. Sci. Rep. 2015, 5, 12793. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guo, K.; Kastan, M.B. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J. Biol. Chem. 2012, 287, 16467–16476. [Google Scholar] [CrossRef] [PubMed]
- De, A.; Donahue, S.L.; Tabah, A.; Castro, N.E.; Mraz, N.; Cruise, J.L.; Campbell, C. A novel interaction [corrected] of nucleolin with Rad51. Biochem. Biophys. Res. Commun. 2006, 344, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Borggrefe, T.; Wabl, M.; Akhmedov, A.T.; Jessberger, R. A B-cell-specific DNA recombination complex. J. Biol. Chem. 1998, 273, 17025–17035. [Google Scholar] [CrossRef] [PubMed]
- Indig, F.E.; Rybanska, I.; Karmakar, P.; Devulapalli, C.; Fu, H.; Carrier, F.; Bohr, V.A. Nucleolin inhibits G4 oligonucleotide unwinding by Werner helicase. PLoS ONE 2012, 7, e35229. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Fujimoto, H.; Sato, J.; Hayashi, I.; Burma, S.; Matsuura, S.; Chen, D.J.; Komatsu, K. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS ONE 2012, 7, e49245. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, M.; Derheimer, F.A.; Tait-Mulder, J.; Kastan, M.B. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2013, 110, 16874–16879. [Google Scholar] [CrossRef] [PubMed]
- Angelov, D.; Bondarenko, V.A.; Almagro, S.; Menoni, H.; Mongelard, F.; Hans, F.; Mietton, F.; Studitsky, V.M.; Hamiche, A.; Dimitrov, S.; et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006, 25, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Ui, A.; Nakajima, S.; Hatakeyama, K.; Hoshi, M.; Watanabe, R.; Janicki, S.M.; Ogiwara, H.; Kohno, T.; Kanno, S.; et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell 2010, 40, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Koike, A.; Nishikawa, H.; Wu, W.; Okada, Y.; Venkitaraman, A.R.; Ohta, T. Recruitment of phosphorylated NPM1 to sites of DNA damage through RNF8-dependent ubiquitin conjugates. Cancer Res. 2010, 70, 6746–6756. [Google Scholar] [CrossRef] [PubMed]
- Okuwaki, M.; Matsumoto, K.; Tsujimoto, M.; Nagata, K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001, 506, 272–276. [Google Scholar] [CrossRef]
- Holmberg Olausson, K.; Nister, M.; Lindstrom, M.S. Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription. J. Biol. Chem. 2014, 289, 34601–34619. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, V.; Kishore, A.H.; Febitha, K.K.; Kundu, T.K. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol. Cell Biol. 2005, 25, 7534–7545. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J.; Lockshon, D.; Fangman, W.L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 1992, 71, 267–276. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nomura, M.; Horiuchi, T. Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol. Cell Biol. 2001, 21, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell Biol. 2003, 23, 9178–9188. [Google Scholar] [CrossRef] [PubMed]
- Weitao, T.; Budd, M.; Campbell, J.L. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat. Res. 2003, 532, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Burkhalter, M.D.; Sogo, J.M. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol. Cell 2004, 15, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Horiuchi, T.; Tongaonkar, P.; Vu, L.; Nomura, M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 2004, 117, 441–453. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ganley, A.R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 2005, 309, 1581–1584. [Google Scholar] [CrossRef] [PubMed]
- Defossez, P.A.; Prusty, R.; Kaeberlein, M.; Lin, S.J.; Ferrigno, P.; Silver, P.A.; Keil, R.L.; Guarente, L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 1999, 3, 447–455. [Google Scholar] [CrossRef]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Park, P.U.; Defossez, P.A.; Guarente, L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell Biol. 1999, 19, 3848–3856. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. How does genome instability affect lifespan?: Roles of rDNA and telomeres. Genes Cells 2011, 16, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Ide, S.; Miyazaki, T.; Maki, H.; Kobayashi, T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 2010, 327, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Ganley, A.R.; Ide, S.; Saka, K.; Kobayashi, T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol. Cell 2009, 35, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, Y.; Kobayashi, T. The Human RNA Polymerase I Transcription terminator complex acts as a replication fork barrier that coordinates the progress of replication with rRNA transcription activity. Mol. Cell Biol. 2015, 35, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006, 20, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Grob, A.; Roussel, P.; Wright, J.E.; McStay, B.; Hernandez-Verdun, D.; Sirri, V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 2009, 122, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Killen, M.W.; Stults, D.M.; Adachi, N.; Hanakahi, L.; Pierce, A.J. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 2009, 18, 3417–3428. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Pietrzak, M.; Rempala, G.; Nelson, P.T.; Hetman, M. Neurodegeneration-associated instability of ribosomal DNA. Biochim. Biophys. Acta 2014, 1842, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Deng, L.; Xue, Y.; Suzuki, K.; Zhang, W.; Yu, Y.; Wu, J.; Sun, L.; Gong, X.; Luan, H.; et al. Visualization of aging-associated chromatin alterations with an engineered TALE system. Cell Res. 2017, 27, 483–504. [Google Scholar] [CrossRef] [PubMed]
- Zafiropoulos, A.; Tsentelierou, E.; Linardakis, M.; Kafatos, A.; Spandidos, D.A. Preferential loss of 5S and 28S rDNA genes in human adipose tissue during ageing. Int. J. Biochem. Cell Biol. 2005, 37, 409–415. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, R.A.; Spitzer, D.; Bar-Am, I.; Sylvester, J.E.; Kaufmann, M.; Wernich, A.; Drexler, H.G. Karyotypic dissection of Hodgkin’s disease cell lines reveals ectopic subtelomeres and ribosomal DNA at sites of multiple jumping translocations and genomic amplification. Leukemia 2000, 14, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Rieker, C.; Engblom, D.; Kreiner, G.; Domanskyi, A.; Schober, A.; Stotz, S.; Neumann, M.; Yuan, X.; Grummt, I.; Schutz, G.; et al. Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. J. Neurosci. 2011, 31, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, M.; Rempala, G.; Nelson, P.T.; Zheng, J.J.; Hetman, M. Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS ONE 2011, 6, e22585. [Google Scholar] [CrossRef] [PubMed]
- Quin, J.E.; Devlin, J.R.; Cameron, D.; Hannan, K.M.; Pearson, R.B.; Hannan, R.D. Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta 2014, 1842, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Rice, W.G.; Grummt, I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, N.; Sussman, M.A. Stressing on the nucleolus in cardiovascular disease. Biochim. Biophys. Acta 2014, 1842, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, M.; Suzuki, T.; Itoh, C.; Goto, M.; Furuichi, Y.; Matsumoto, T. WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene 2002, 21, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.D.; Wang, L.; Youssoufian, H.; Martin, G.M.; Oshima, J. Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp. Cell Res. 1998, 242, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Lutomska, A.; Lebedev, A.; Scharffetter-Kochanek, K.; Iben, S. The transcriptional response to distinct growth factors is impaired in Werner syndrome cells. Exp. Gerontol. 2008, 43, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Grierson, P.M.; Lillard, K.; Behbehani, G.K.; Combs, K.A.; Bhattacharyya, S.; Acharya, S.; Groden, J. BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum. Mol. Genet. 2012, 21, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Grierson, P.M.; Acharya, S.; Groden, J. Collaborating functions of BLM and DNA topoisomerase I in regulating human rDNA transcription. Mutat. Res. 2013, 743–744, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Le May, N.; Mota-Fernandes, D.; Velez-Cruz, R.; Iltis, I.; Biard, D.; Egly, J.M. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell 2010, 38, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Fong, Y.W.; Inouye, C.; Yamaguchi, T.; Cattoglio, C.; Grubisic, I.; Tjian, R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 2011, 147, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Kamileri, I.; Karakasilioti, I.; Sideri, A.; Kosteas, T.; Tatarakis, A.; Talianidis, I.; Garinis, G.A. Defective transcription initiation causes postnatal growth failure in a mouse model of nucleotide excision repair (NER) progeria. Proc. Natl. Acad. Sci. USA 2012, 109, 2995–3000. [Google Scholar] [CrossRef] [PubMed]
- Chatzinikolaou, G.; Apostolou, Z.; Aid-Pavlidis, T.; Ioannidou, A.; Karakasilioti, I.; Papadopoulos, G.L.; Aivaliotis, M.; Tsekrekou, M.; Strouboulis, J.; Kosteas, T.; et al. ERCC1-XPF cooperates with CTCF and cohesin to facilitate the developmental silencing of imprinted genes. Nat. Cell Biol. 2017, 19, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Bradsher, J.; Auriol, J.; Proietti de Santis, L.; Iben, S.; Vonesch, J.L.; Grummt, I.; Egly, J.M. CSB is a component of RNA pol I transcription. Mol. Cell 2002, 10, 819–829. [Google Scholar] [CrossRef]
- Yuan, X.; Feng, W.; Imhof, A.; Grummt, I.; Zhou, Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell 2007, 27, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Lebedev, A.; Scharffetter-Kochanek, K.; Iben, S. Truncated Cockayne syndrome B protein represses elongation by RNA polymerase I. J. Mol. Biol. 2008, 382, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Assfalg, R.; Lebedev, A.; Gonzalez, O.G.; Schelling, A.; Koch, S.; Iben, S. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2012, 40, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Scheibye-Knudsen, M.; Tseng, A.; Borch Jensen, M.; Scheibye-Alsing, K.; Fang, E.F.; Iyama, T.; Bharti, S.K.; Marosi, K.; Froetscher, L.; Kassahun, H.; et al. Cockayne syndrome group A and B proteins converge on transcription-linked resolution of non-B DNA. Proc. Natl. Acad. Sci. USA 2016, 113, 12502–12507. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).