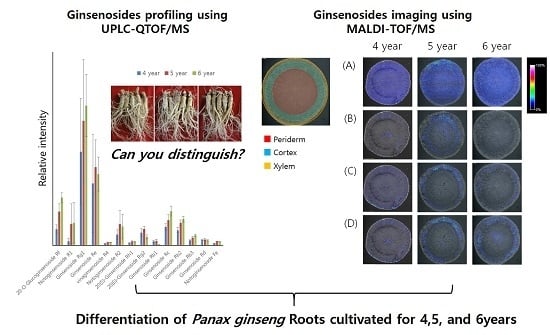
Mass Spectrometry Based Profiling and Imaging of Various Ginsenosides from Panax ginseng Roots at Different Ages
Abstract
:1. Introduction
2. Results
2.1. Optimal UPLC-QTOF/MS Method to Profile Various Ginsenosides
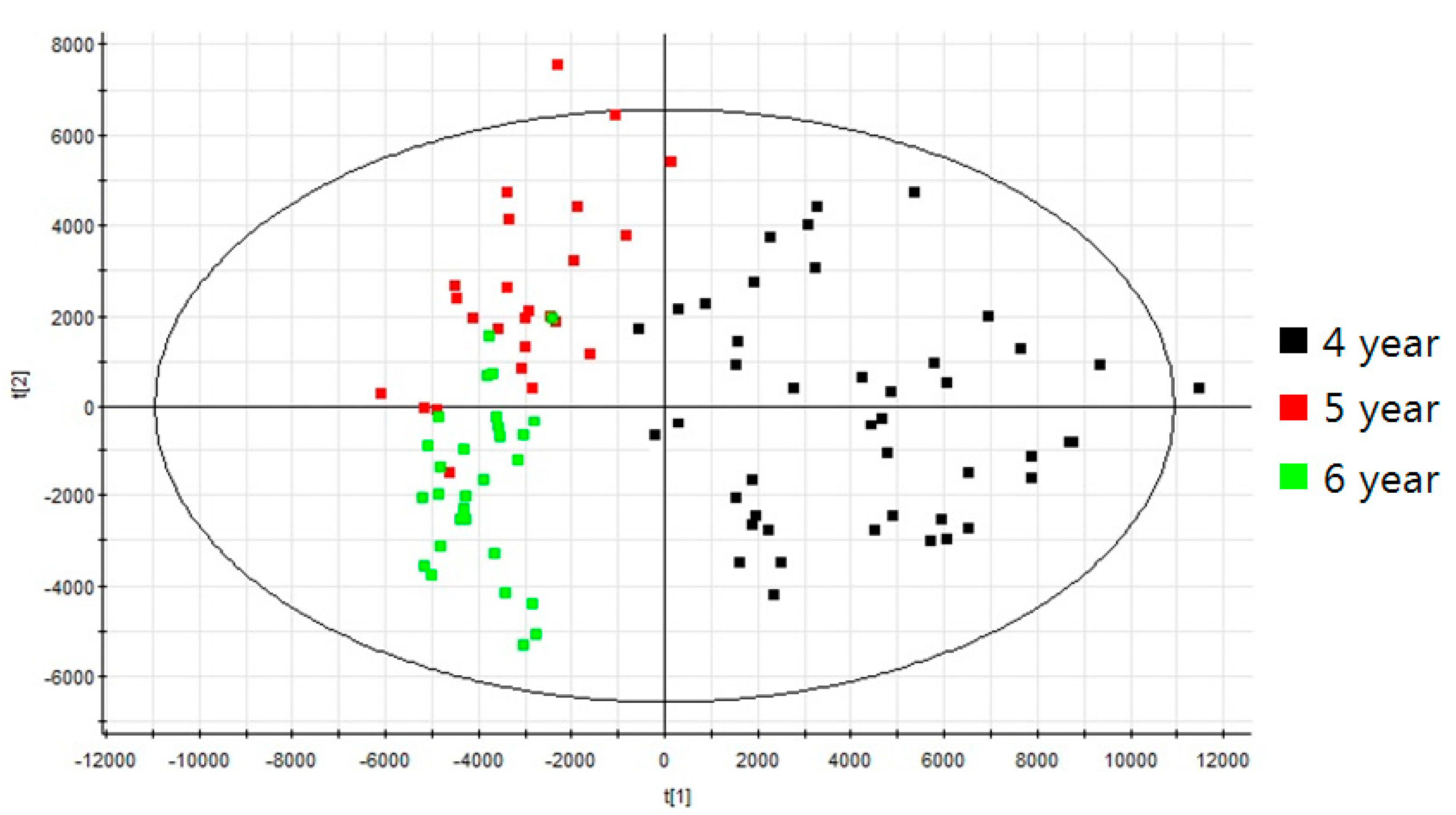
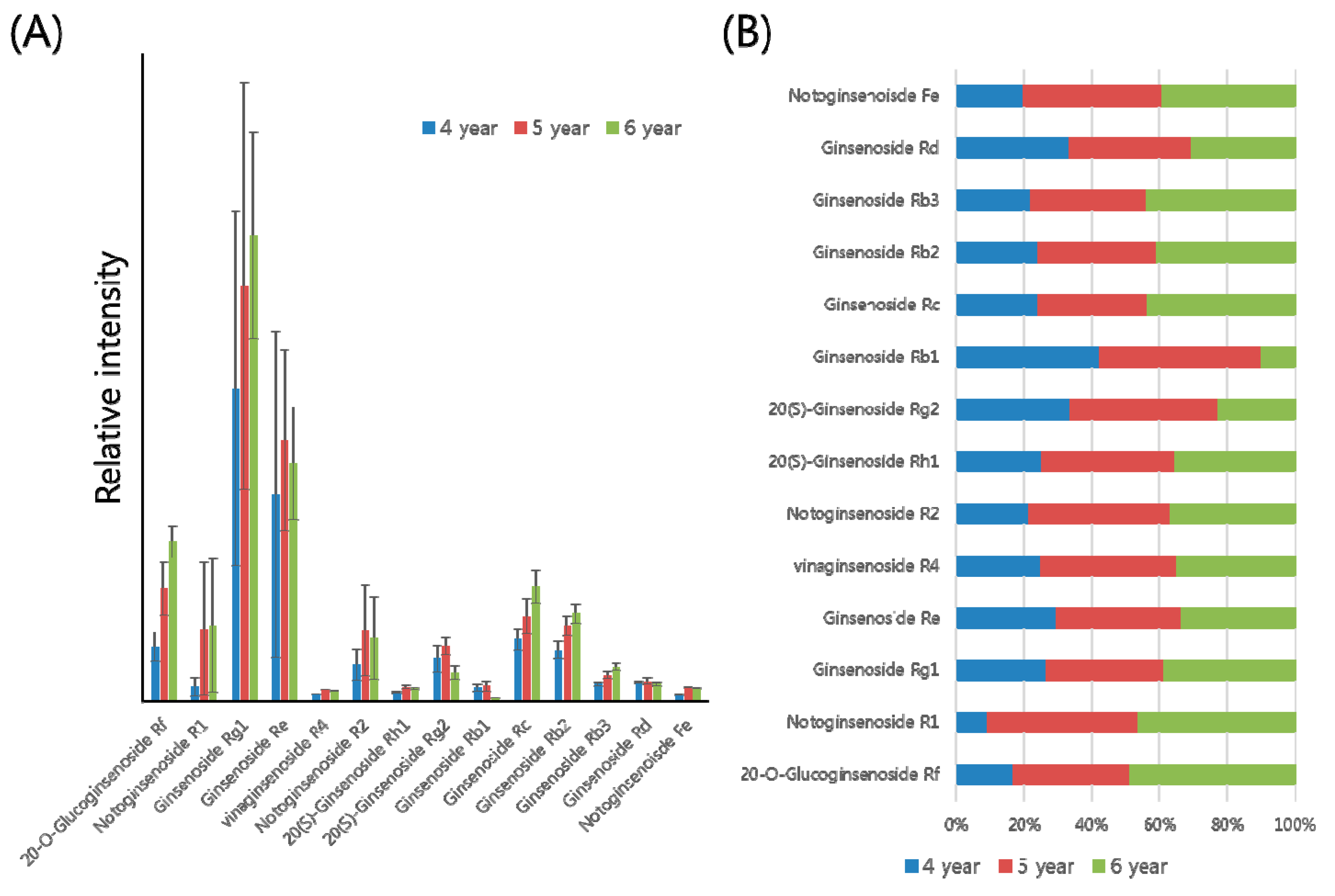
2.2. Profiling of Various Ginsenosides from P. ginseng Roots with Different Ages
2.3. Optimal MALDI-TOF/MS-Based Imaging of Ginsenosides
2.4. Localization of Ginsenosides in the P. ginseng Roots at Different Ages
3. Discussion
4. Materials and Methods
4.1. Standard Constituents and Reagents
4.2. Panax Ginseng Sample
4.3. Extraction of Ginsenosides
4.4. UPLC-QTOF/MS Conditions for Ginsenosides Profiling
4.5. Data Processing and Multivariate Analysis
4.6. Ginseng Root Tissue Sectioning
4.7. Sample Preparation and Matrix Application for MS Imaging
4.8. MALDI-IMS Conditions for Ginsenoside Imaging
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| UPLC | Ultra performance liquid chromatography |
| QTOF | Quadrupole time of flight |
| ESI | Electrospray ionization |
| MALDI | Matrix-assisted laser desorption ionization |
| MS | Mass spectrometry |
| IMS | Imaging mass spectrometry |
| LOD | Limits of detection |
| RT | Retention time |
| PCA | Principal component analysis |
References
- Yun, T.K. Brief introduction of Panax ginseng CA Meyer. J. Korean Med. Sci. 2001, 16, S3. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng CA Meyer. Acta Pharmacol. Sin. 2008, 29, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.Y.K.; Mak, N.K.; Cheng, Y.K.; Leung, K.W.; Ng, T.B.; Fan, D.T.P.; Yeung, H.W.; Wong, R.N.S. Pharmacogenomics and the Yin/Yang actions of ginseng: Anti-tumor, angiomodulating and steroid-like activities of ginsenosides. Chin. Med. 2007, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Kenarova, B.; Neychev, H.; Hadjiivanova, C.; Petkov, V.D. Immunomodulating activity of ginsenoside Rg1 from Panax ginseng. Jpn. J. Pharmacol. 1990, 54, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qu, S.; Sui, D.; Yu, X.; Lv, Z. Effects of ginsenoside-Rb on blood lipid metabolism and anti-oxidation in hyperlipidemia rats. Zhongguo Zhongyao Zazhi 2004, 29, 1085–1088. [Google Scholar] [PubMed]
- Cheng, Y.; Zhang, J.T. Anti-amnestic and anti-aging effects of ginsenoside Rg1 and Rb1 and its mechanism of action. Acta Pharmacol. Sin. 2005, 26, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Wang, J.; Geng, L.; Wei, Z.; Tian, X. Determination of ginseng with different ages using a taste-sensing system. Sens. Mater. 2013, 25, 241–255. [Google Scholar]
- Cui, S.; Wang, J.; Yang, L.; Wu, J.; Wang, X. Qualitative and quantitative analysis on aroma characteristics of ginseng at different ages using E-nose and GC–MS combined with chemometrics. J. Pharm. Biomed. Anal. 2015, 102, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.-M.; Luo, J.-G.; Huang, F.; Kong, L.-Y. Chemical characteristics combined with bioactivity for comprehensive evaluation of Panax ginseng CA Meyer in different ages and seasons based on HPLC-DAD and chemometric methods. J. Pharm. Biomed. Anal. 2014, 89, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.-S.; Bang, K.-H.; In, D.-S.; Kim, O.-T.; Hyun, D.-Y.; Ahn, I.-O.; Ku, B.C.; Kim, S.-W.; Seong, N.-S.; Cha, S.-W. Fingerprinting analysis of fresh ginseng roots of different ages using 1H-NMR spectroscopy and principal components analysis. Arch. Pharmacal Res. 2007, 30, 1625–1628. [Google Scholar] [CrossRef]
- Shi, W.; Wang, Y.; Li, J.; Zhang, H.; Ding, L. Investigation of ginsenosides in different parts and ages of Panax ginseng. Food Chem. 2007, 102, 664–668. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, J.Y.; Xiao, X.Y.; Lin, R.C.; Cheng, Y.Y. Simultaneous determination of ginsenosides in Panax ginseng with different growth ages using high-performance liquid chromatography–mass spectrometry. Phytochem. Anal. 2006, 17, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, J.-L.; Sheng, Y.-X.; Guo, D.-A.; Wang, Q.; Guo, H.-Z. Simultaneous quantification of six major active saponins of Panax notoginseng by high-performance liquid chromatography-UV method. J. Pharm. Biomed. Anal. 2005, 38, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Fitzloff, J.F. HPLC determination of ginsenosides content in ginseng dietary supplements using ultraviolet detection. J. Liquid Chromatogr. Relat. Technol. 2002, 25, 2485–2500. [Google Scholar] [CrossRef]
- Kim, S.N.; Ha, Y.W.; Shin, H.; Son, S.H.; Wu, S.J.; Kim, Y.S. Simultaneous quantification of 14 ginsenosides in Panax ginseng CA Meyer (Korean red ginseng) by HPLC-ELSD and its application to quality control. J. Pharm. Biomed. Anal. 2007, 45, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.-S.; Gu, L.-J.; Fang, Z.-M.; Wang, C.-Y.; Wang, Z.; Lee, M.-R.; Li, Z.; Li, J.-J.; Sung, C.-K. Simultaneous quantification of 19 ginsenosides in black ginseng developed from Panax ginseng by HPLC–ELSD. J. Pharm. Biomed. Anal. 2009, 50, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.X.; Ni, Y.; Su, M.M.; Zhang, Y.Y.; Zhao, A.H.; Gao, X.F.; Liu, Z.; Xiao, P.G.; Jia, W. Application of ultra-performance LC-TOF MS metabolite profiling techniques to the analysis of medicinal Panax herbs. Metabolomics 2008, 4, 248–260. [Google Scholar] [CrossRef]
- Ji, Q.C.; Harkey, M.R.; Henderson, G.L.; Eric Gershwin, M.; Stern, J.S.; Hackman, R.M. Quantitative determination of ginsenosides by high-performance liquid chromatography-tandem mass spectrometry. Phytochem. Anal. 2001, 12, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Z.; Chen, H.; Yi, T.; Qin, M.; Liang, Z. Chemical Differentiation and Quality Evaluation of Commercial Asian and American Ginsengs based on a UHPLC–QTOF/MS/MS Metabolomics Approach. Phytochem. Anal. 2015, 26, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lee, D.Y.; Kang, K.B.; Kim, J.Y.; Kim, S.O.; Yoo, Y.H.; Sung, S.H. Identification of ginsenoside markers from dry purified extract of Panax ginseng by a dereplication approach and UPLC–QTOF/MS analysis. J. Pharm. Biomed. Anal. 2015, 109, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-M.; Li, S.-L.; Zhang, H.; Wang, Y.; Zhao, Z.-L.; Chen, S.-L.; Xu, H.-X. Holistic quality evaluation of commercial white and red ginseng using a UPLC-QTOF-MS/MS-based metabolomics approach. J. Pharm. Biomed. Anal. 2012, 62, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Perdian, D.C.; Song, Z.; Yeung, E.S.; Nikolau, B.J. Use of mass spectrometry for imaging metabolites in plants. Plant J. 2012, 70, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Heyman, H.M.; Dubery, I.A. The potential of mass spectrometry imaging in plant metabolomics: A review. Phytochem. Rev. 2016, 15, 297–316. [Google Scholar] [CrossRef]
- Eriksson, C.; Masaki, N.; Yao, I.; Hayasaka, T.; Setou, M. MALDI imaging mass spectrometry—A mini review of methods and recent developments. Mass Spectrom. 2013, 2, S0022. [Google Scholar] [CrossRef] [PubMed]
- Kaspar, S.; Peukert, M.; Svatos, A.; Matros, A.; Mock, H.P. MALDI-imaging mass spectrometry–An emerging technique in plant biology. Proteomics 2011, 11, 1840–1850. [Google Scholar] [CrossRef] [PubMed]
- Shroff, R.; Schramm, K.; Jeschke, V.; Nemes, P.; Vertes, A.; Gershenzon, J.; Svatoš, A. Quantification of plant surface metabolites by matrix-assisted laser desorption–ionization mass spectrometry imaging: Glucosinolates on Arabidopsis thaliana leaves. Plant J. 2015, 81, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Taira, S.; Ikeda, R.; Yokota, N.; Osaka, I.; Sakamoto, M.; Kato, M.; Sahashi, Y. Mass spectrometric imaging of ginsenosides localization in Panax ginseng root. Am. J. Chin. Med. 2010, 38, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Wang, S.; Liu, J.; Gao, D.; Jiang, Y.; Liu, H.; Cai, Z. Localization of ginsenosides in Panax ginseng with different age by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry imaging. J. Chromatogr. B 2016, 1026, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Sanada, S.; Shoji, J. Studies on the saponins of ginseng. III. Structures of ginsenoside-Rb3 and 20-glucoginsenoside-Rf. Chem. Pharm. Bull. 1978, 26, 1694–1697. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, M.; Taniyasu, S.; Besso, H.; Tanaka, O.; Saruwatari, Y.; Fuwa, T. Dammarane-saponins of sanchi-ginseng, roots of Panax notoginseng (BURK.) FH Chen (Araliaceae): Structures of new saponins, notoginsenosides-R1 and-R2, and identification of ginsenosides-Rg2 and-Rh1. Chem. Pharm. Bull. 1981, 29, 2844–2850. [Google Scholar] [CrossRef]
- Yang, T.-R.; Kasai, R.; Zhou, J.; Tanaka, O. Dammarane saponins of leaves and seeds of Panax notoginseng. Phytochemistry 1983, 22, 1473–1478. [Google Scholar] [CrossRef]
- Qiu, L.; Jiao, Y.; Huang, G.K.; Xie, J.Z.; Miao, J.H.; Yao, X.S. New Dammarane-type saponins from the roots of Panax notoginseng. Helv. Chim. Acta 2014, 97, 102–111. [Google Scholar] [CrossRef]
- Li, M.; Wang, Z.; Zhang, P.; Xua, Y.; Ding, H.; Sui, D.; Xu, H.; Chen, Y. Two new triterpenoid saponins from the structural modification of pseudoginsenoside-F11 and their cytotoxic activities. Nat. Prod. Commun. 2016, 11, 601. [Google Scholar] [PubMed]
- Tung, N.H.; Song, G.Y.; Park, Y.J.; Kim, Y.H. Two new dammarane-type saponins from the leaves of Panax ginseng. Chem. Pharm. Bull. 2009, 57, 1412–1414. [Google Scholar] [CrossRef] [PubMed]
- Li, K.-K.; Xu, F.; Gong, X.-J. Isolation, purification and quantification of ginsenoside F5 and F3 isomeric compounds from crude extracts of flower buds of Panax ginseng. Molecules 2016, 21, 315. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.Y.; Li, Y.W.; Kwok-Po Hau, D.; Jiang, Z.H.; Yu, Z.L.; Fong, W.F. Acylated protopanaxadiol-type ginsenosides from the root of Panax ginseng. Chem. Biodivers. 2011, 8, 1853–1863. [Google Scholar] [CrossRef] [PubMed]
- Yahara, S.; Kasai, R.; Tanaka, O. New dammarane type saponins of leaves of Panax japonicus CA Meyer. (1). Chikusetsusaponins-L5, -L9a and -L10. Chem. Pharm. Bull. 1977, 25, 2041–2047. [Google Scholar] [CrossRef]
- Ko, S.-R.; Choi, K.-J.; Suzuki, K.; Suzuki, Y. Enzymatic preparation of ginsenosides Rg2, Rh1, and F1. Chem. Pharm. Bull. 2003, 51, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Takeda, T.; Zhu, T.; Chen, Y.; Yao, X.; Tanaka, O.; Ogihara, Y. A new minor saponin from the leaves of Panax ginseng. Planta Med. 1990, 56, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Park, J.H.; Ryu, J.-H.; Park, J.D.; Lee, Y.H.; Park, J.-H.; Kim, T.-H.; Kim, J.M.; Baek, N.-I. Ginsenoside Rg5, a genuine dammarane glycoside from Korean red ginseng. Arch. Pharmacal Res. 1996, 19, 551–553. [Google Scholar] [CrossRef]
- Kim, D.-S.; Lee, Y.-H.; Park, J.-D.; Jeong, S.-Y.; Lee, C.-B.; Kim, S.-I.; Baek, N.-I. Complete assignment of 1H and 13C-NMR signals for (20S)-and (20R)-ginsenoside Rh2 by 2D-NMR Techniques. J. Korean Soc. Appl. Biol. Chem. 1995, 38, 184–189. [Google Scholar]
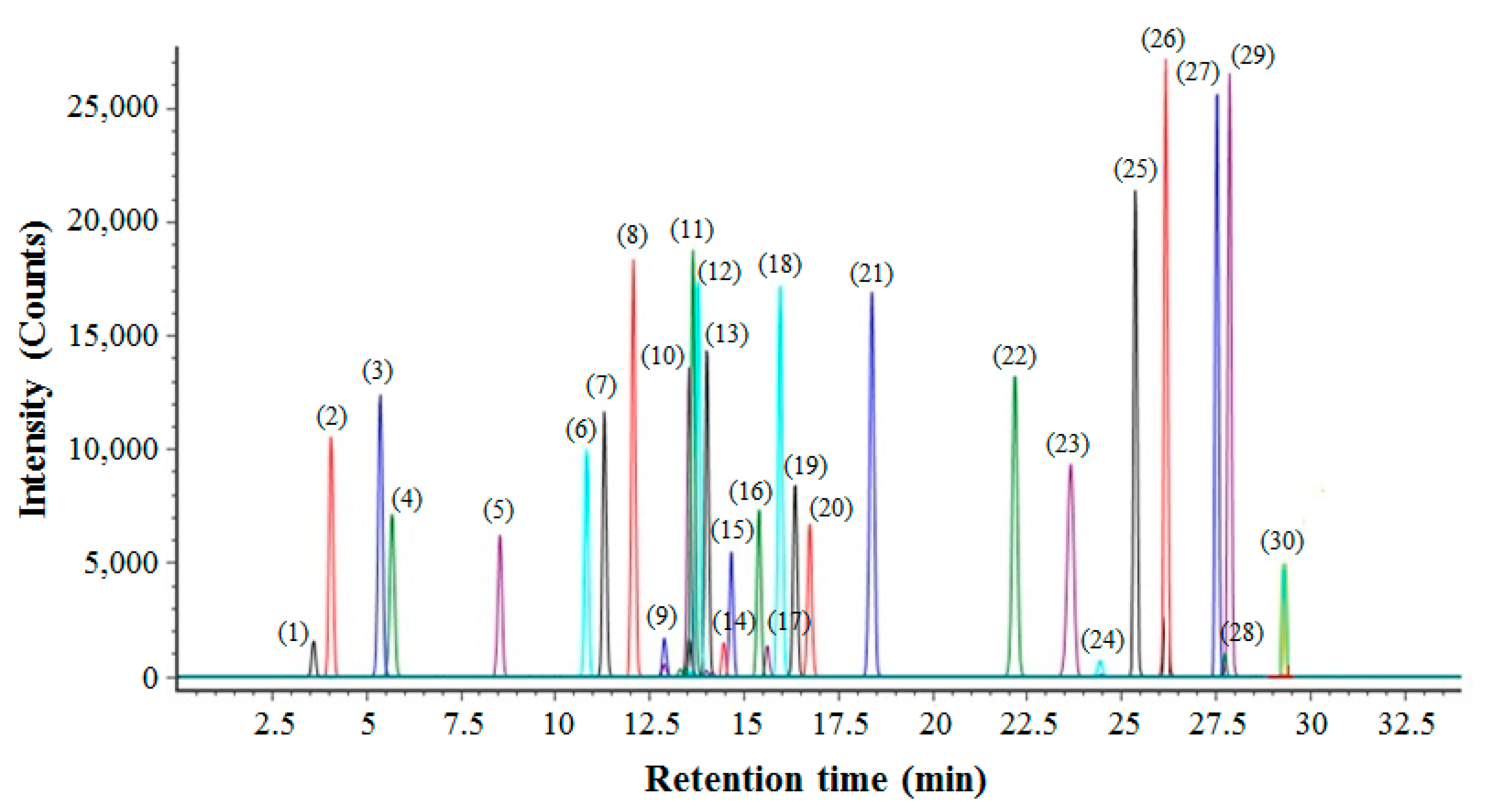


| No. | RT (min) | Ginsenosides | Molecular Formula | Expected Neutral Mass (Da) | Observed Neutral Mass (Da) | QTOF/MS (ESI−) m/z | Mass Accuracy (ppm) | Adducts |
|---|---|---|---|---|---|---|---|---|
| 1 | 3.61 | 20-O-glucoginsenoside Rf (Rf) | C48H82O19 | 962.545 | 962.5428 | 1007.541 | 2.2 | +HCOO |
| 2 | 4.07 | Notoginsenoside R1 (R1) | C47H80O18 | 932.5345 | 932.5336 | 977.5318 | 0.9 | +HCOO |
| 3 | 5.38 | Ginsenoside Rg1 (Rg1) | C42H72O14 | 800.4922 | 800.4905 | 845.4887 | 2 | +HCOO |
| 4 | 5.69 | Ginsenoside Re (Re) | C48H82O18 | 946.5501 | 946.5483 | 991.5465 | 1.9 | +HCOO |
| 5 | 8.54 | Ginsenoside Rh6 (Rh6) | C36H62O11 | 670.4292 | 670.4266 | 715.4248 | 3.6 | +HCOO |
| 6 | 10.82 | Ginsenoside Rh21 (Rh21) | C37H64O10 | 668.4499 | 668.4478 | 713.4460 | 3 | +HCOO |
| 7 | 11.29 | Vinaginsenoside R4 (R4) | C48H82O19 | 962.5450 | 962.5433 | 1007.5415 | 1.8 | +HCOO |
| 8 | 12.06 | Pseudo-ginsenoside F11 (F11) | C42H72O14 | 800.4922 | 800.4897 | 845.4879 | 2.9 | +HCOO |
| 9 | 12.9 | Notoginsenoside R2 (R2) | C41H70O13 | 770.4816 | 770.4783 | 815.4765 | 4.1 | +HCOO |
| 10 | 13.55 | Ginsenoside F5 (F5) | C41H70O13 | 770.4816 | 770.4809 | 815.4791 | 0.9 | +HCOO |
| 11 | 13.64 | 20(S)-ginsenoside Rh1 (Rh1) | C36H62O9 | 638.4394 | 638.4370 | 683.4352 | 3.4 | +HCOO |
| 12 | 13.77 | 20(S)-ginsenoside Rg2 (Rg2) | C42H72O13 | 784.4973 | 784.4967 | 829.4949 | 0.7 | +HCOO |
| 13 | 14.02 | Ginsenoside F3 (F3) | C41H70O13 | 770.4816 | 770.4806 | 815.4788 | 1.3 | +HCOO |
| 14 | 14.45 | Ginsenoside Ra2 (Ra2) | C58H98O26 | 1210.6346 | 1210.6347 | 1255.6329 | 0.1 | +HCOO |
| 15 | 14.63 | Ginsenoside Rb1 (Rb1) | C54H92O23 | 1108.6029 | 1108.6036 | 1153.6018 | 0.5 | +HCOO |
| 16 | 15.36 | Ginsenoside Rc (Rc) | C53H90O22 | 1078.5924 | 1078.5920 | 1123.5902 | 0.3 | +HCOO |
| 17 | 15.58 | Ginsenoside Ra1 (Ra1) | C58H98O26 | 1210.6346 | 1210.6332 | 1255.6314 | 1.1 | +HCOO |
| 18 | 15.94 | Ginsenoside F1 (F1) | C36H62O9 | 638.4394 | 638.4386 | 683.4368 | 1.2 | +HCOO |
| 19 | 16.36 | Ginsenoside Rb2 (Rb2) | C53H90O22 | 1078.5924 | 1078.5926 | 1123.5908 | 0.2 | +HCOO |
| 20 | 16.71 | Ginsenoside Rb3 (Rb3) | C53H90O22 | 1078.5924 | 1078.5915 | 1123.5897 | 0.8 | +HCOO |
| 21 | 18.38 | Ginsenoside Rd (Rd) | C48H82O18 | 946.5501 | 946.5499 | 991.5481 | 0.2 | +HCOO |
| 22 | 22.1 | Notoginsenoisde Fe (Fe) | C47H80O17 | 916.5396 | 916.5377 | 961.5359 | 1.9 | +HCOO |
| 23 | 23.63 | Ginsenoside Rg4 (Rg4) | C42H70O12 | 766.4867 | 766.4849 | 811.4831 | 2.3 | +HCOO |
| 24 | 24.47 | Ginsenoside F4 (F4) | C42H70O12 | 766.4867 | 766.4851 | 811.4833 | 2 | +HCOO |
| 25 | 25.37 | Ginsenoside F2 (F2) | C42H72O13 | 784.4973 | 784.4952 | 829.4934 | 2.5 | +HCOO |
| 26 | 26.17 | Ginsenoside Rg3 (Rg3) | C42H72O13 | 784.4973 | 784.4956 | 829.4938 | 2 | +HCOO |
| 27 | 27.53 | Compound K (CK) | C36H62O8 | 622.4445 | 622.4422 | 667.4404 | 3.3 | +HCOO |
| 28 | 27.72 | Ginsenoside Rg5 (Rg5) | C42H70O12 | 766.4867 | 766.4846 | 811.4828 | 2.6 | +HCOO |
| 29 | 27.85 | Ginsenoside Rh2 (Rh2) | C36H62O8 | 622.4445 | 622.4431 | 667.4413 | 2 | +HCOO |
| 30 | 29.3 | Ginsenoside Rh3 (Rh3) | C36H60O7 | 604.4339 | 604.4338 | 649.4320 | 0.1 | +HCOO |
| Ginsenosides | Correlation (R2) | Linear Range (pg) * | LOD (pg) * | Ginsenosides | Correlation (R2) | Linear Range (pg) * | LOD (pg) * |
|---|---|---|---|---|---|---|---|
| Rf | 0.9947 | 70–2000 | 70 | Rc | 0.9963 | 200–4000 | 200 |
| R1 | 0.9997 | 9–2000 | 9 | Ra1 | 0.9985 | 70–8000 | 70 |
| Rg1 | 0.9992 | 12–4000 | 12 | F1 | 0.9969 | 5–2000 | 5 |
| Re | 0.9840 | 10–12,000 | 10 | Rb2 | 0.9969 | 2–4000 | 2 |
| Rh6 | 0.9999 | 8–400 | 8 | Rb3 | 0.9999 | 4–2000 | 4 |
| Rh21 | 0.9824 | 2–160 | 2 | Rd | 0.9964 | 6–4000 | 6 |
| R4 | 0.9999 | 4–20 | 4 | Fe | 0.9900 | 10–200 | 10 |
| F11 | 0.9845 | 4–2000 | 4 | Rg4 | 0.9984 | 20–1000 | 20 |
| R2 | 0.9991 | 40–4000 | 40 | F4 | 0.9990 | 200–2000 | 200 |
| F5 | 0.9977 | 4–1000 | 4 | F2 | 0.9998 | 1–10 | 1 |
| Rh1 | 0.9998 | 4–2000 | 4 | Rg3 | 0.9964 | 2–120 | 2 |
| Rg2 | 0.9969 | 5–400 | 5 | CK | 0.9999 | 4–180 | 4 |
| F3 | 0.9923 | 6–2000 | 6 | Rg5 | 0.9987 | 20–2000 | 20 |
| Ra2 | 0.9995 | 200–7200 | 200 | Rh2 | 0.9997 | 4–1000 | 4 |
| Rb1 | 0.9906 | 3–7400 | 3 | Rh3 | 0.9983 | 16–100 | 16 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.W.; Ji, S.-H.; Lee, Y.-S.; Choi, D.J.; Choi, B.-R.; Kim, G.-S.; Baek, N.-I.; Lee, D.Y. Mass Spectrometry Based Profiling and Imaging of Various Ginsenosides from Panax ginseng Roots at Different Ages. Int. J. Mol. Sci. 2017, 18, 1114. https://doi.org/10.3390/ijms18061114
Lee JW, Ji S-H, Lee Y-S, Choi DJ, Choi B-R, Kim G-S, Baek N-I, Lee DY. Mass Spectrometry Based Profiling and Imaging of Various Ginsenosides from Panax ginseng Roots at Different Ages. International Journal of Molecular Sciences. 2017; 18(6):1114. https://doi.org/10.3390/ijms18061114
Chicago/Turabian StyleLee, Jae Won, Seung-Heon Ji, Young-Seob Lee, Doo Jin Choi, Bo-Ram Choi, Geum-Soog Kim, Nam-In Baek, and Dae Young Lee. 2017. "Mass Spectrometry Based Profiling and Imaging of Various Ginsenosides from Panax ginseng Roots at Different Ages" International Journal of Molecular Sciences 18, no. 6: 1114. https://doi.org/10.3390/ijms18061114
APA StyleLee, J. W., Ji, S.-H., Lee, Y.-S., Choi, D. J., Choi, B.-R., Kim, G.-S., Baek, N.-I., & Lee, D. Y. (2017). Mass Spectrometry Based Profiling and Imaging of Various Ginsenosides from Panax ginseng Roots at Different Ages. International Journal of Molecular Sciences, 18(6), 1114. https://doi.org/10.3390/ijms18061114