Functional Characterization of Waterlogging and Heat Stresses Tolerance Gene Pyruvate decarboxylase 2 from Actinidia deliciosa
Abstract
1. Introduction
2. Results
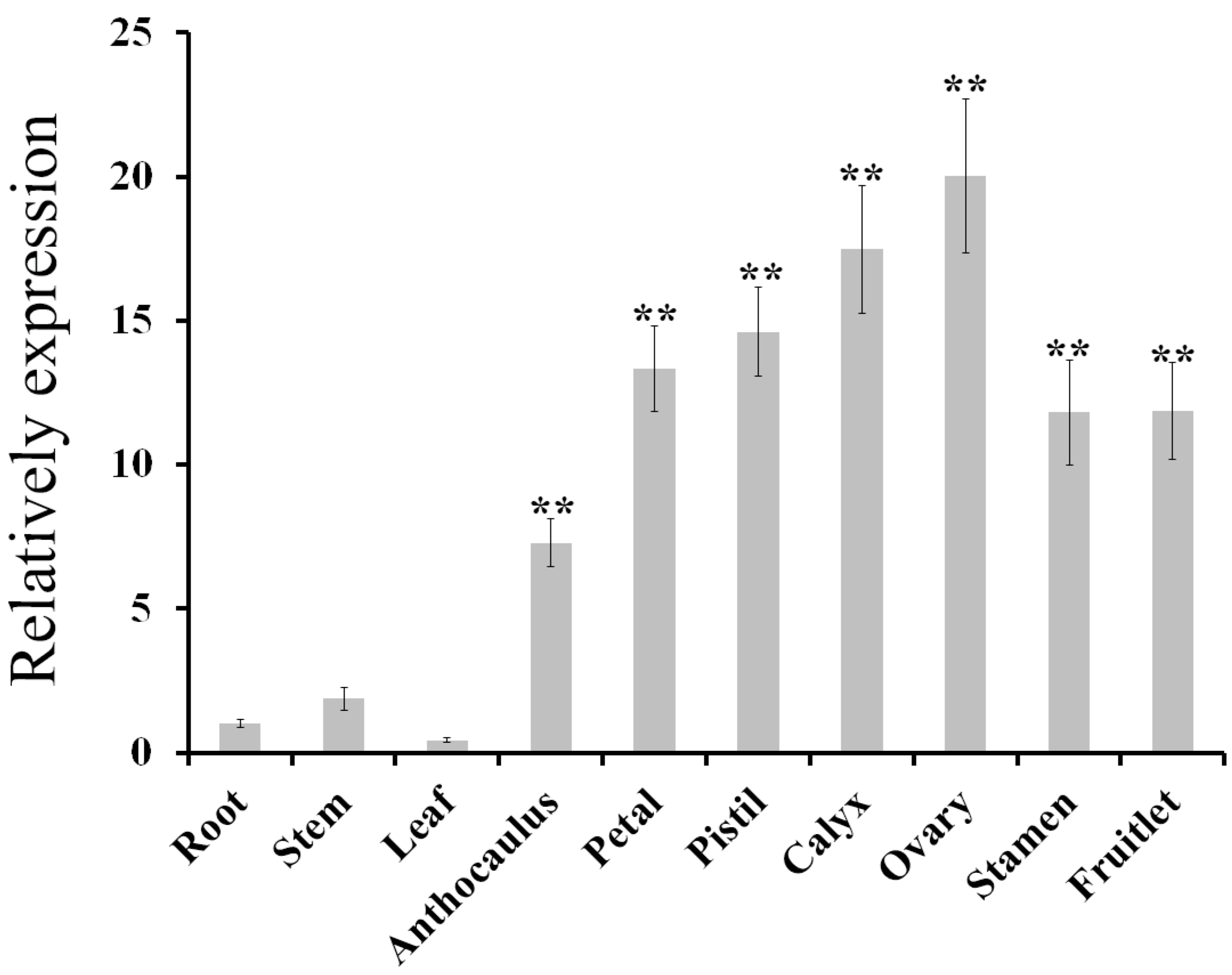
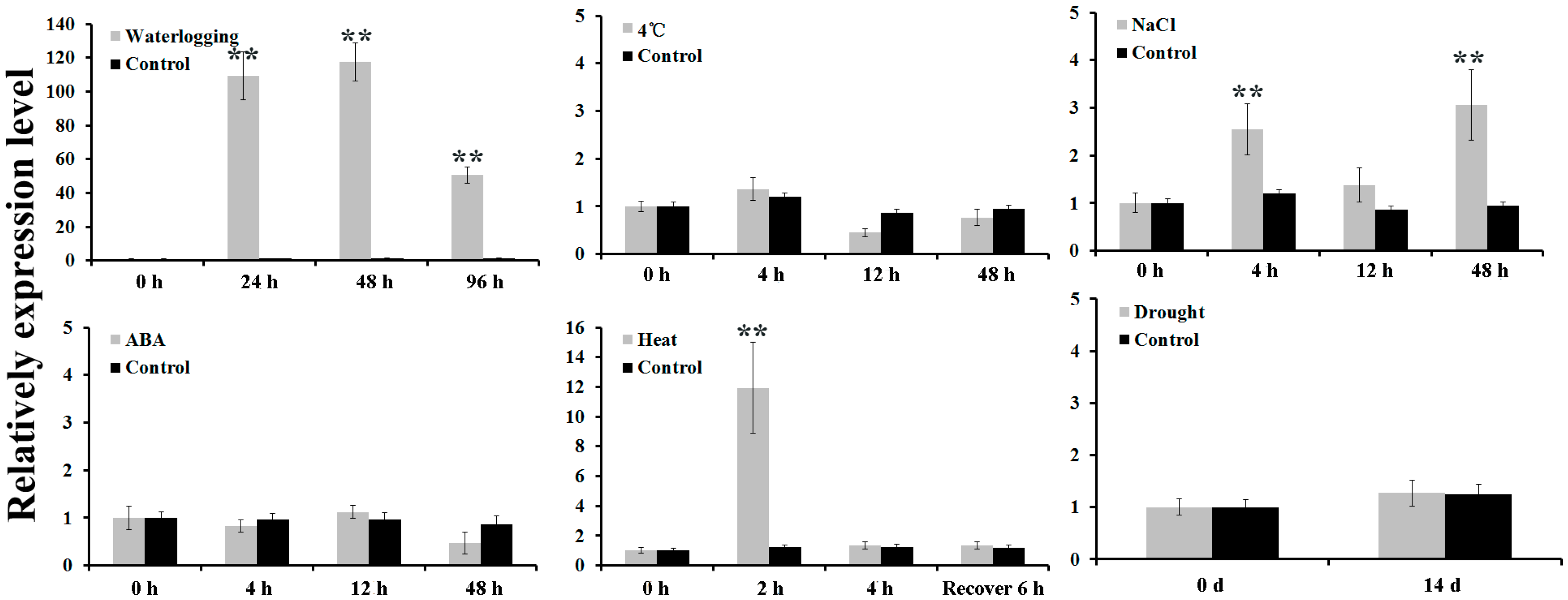
2.1. Isolation and Expression Analysis of AdPDC2
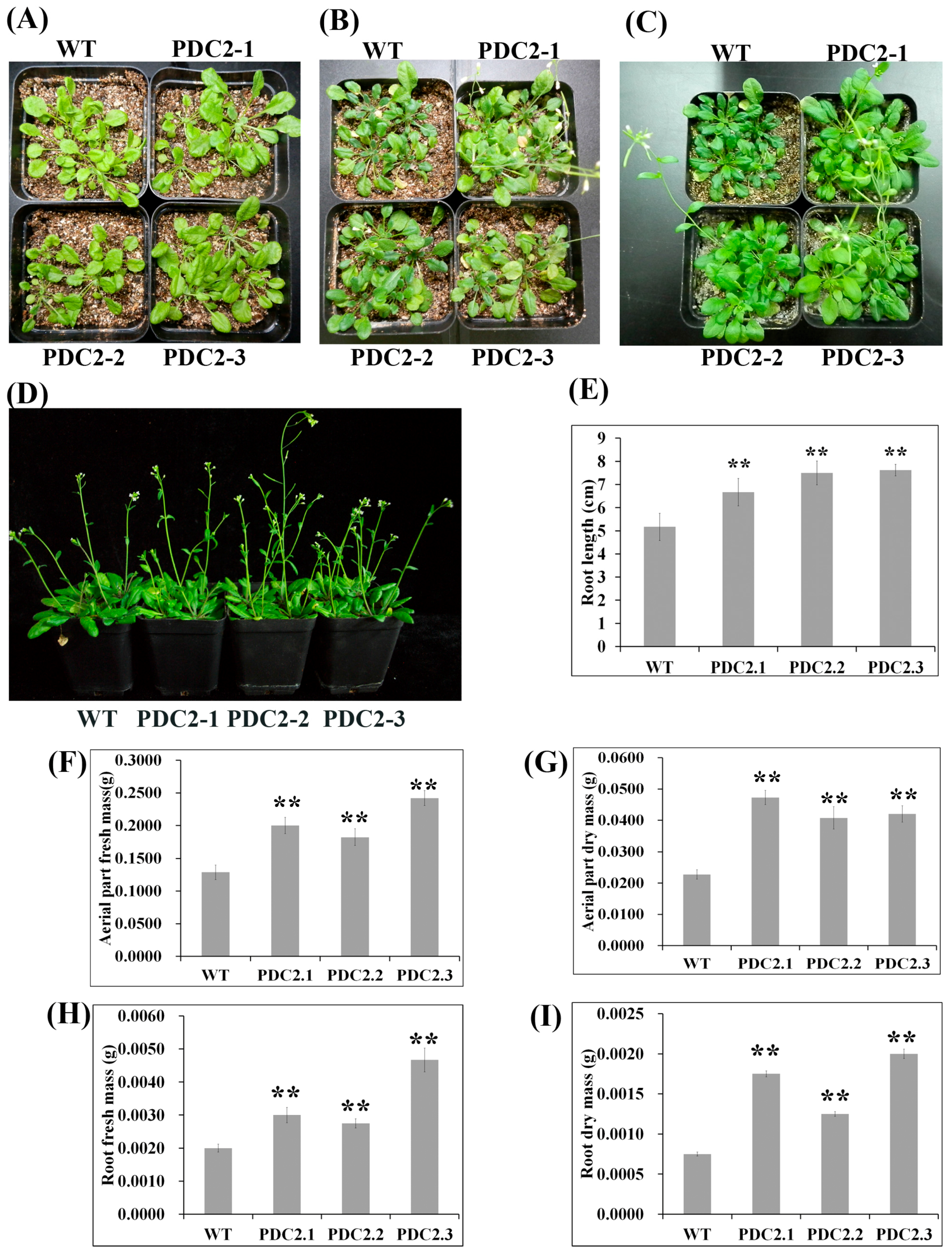
2.2. Overexpression of AdPDC2 Enhances Tolerance to Waterlogging Stress in Arabidopsis
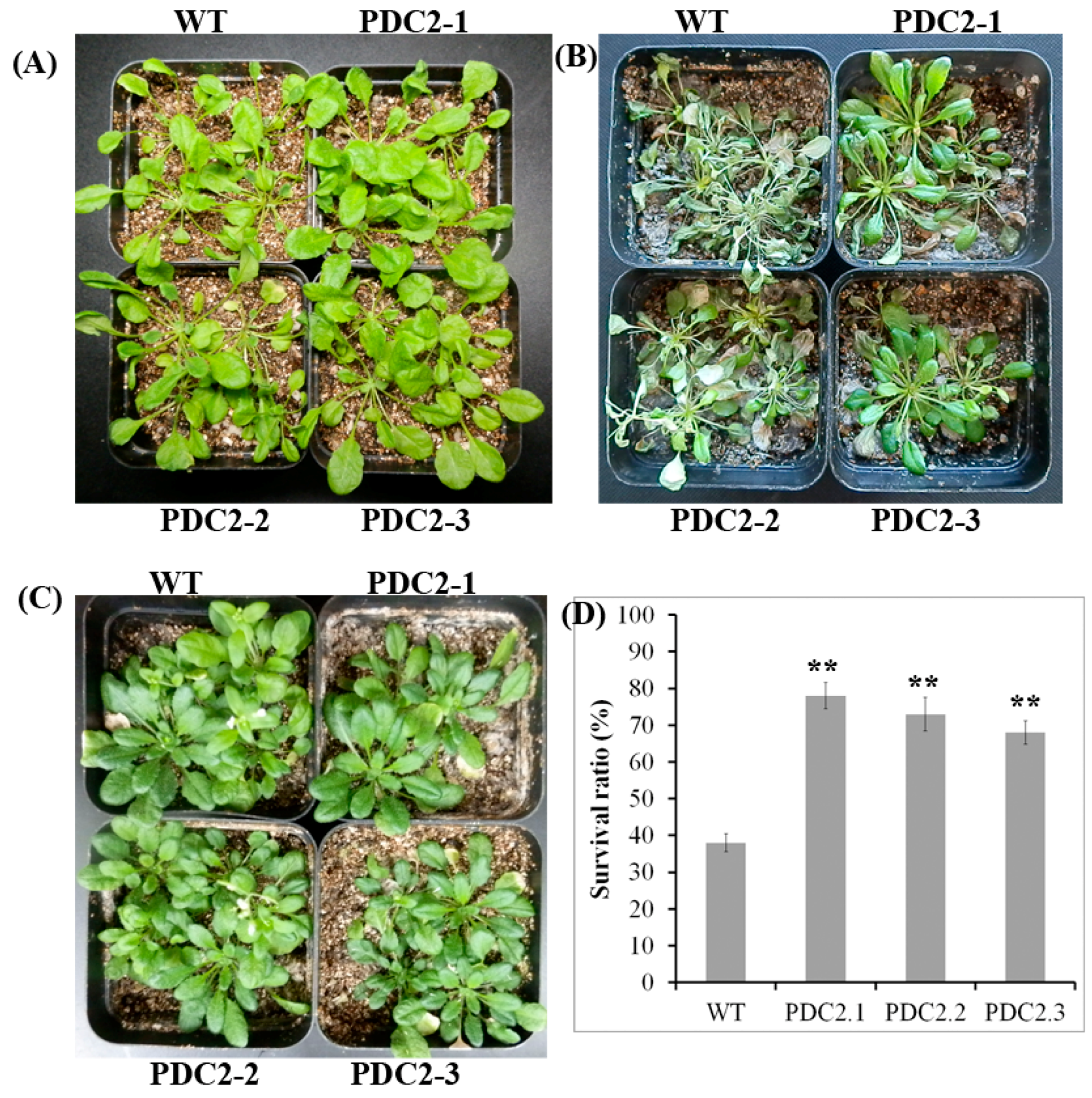
2.3. Overexpression of the AdPDC2 Gene Enhances Resistance to Heat Stress
2.4. Overexpression of the AdPDC2 Gene Could not Enhance Resistance to Salinity and Osmotic Stresses
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Treatment of A. deliciosa with Abiotic Stresses
4.3. Total Ribonucleic Acid (RNA) Isolation and cDNA Synthesis
4.4. Cloning and Sequence Analysis of AdPDC2
4.5. Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Assay for AdPDC2 Gene Expression
4.6. Generation of AdPDC2-Overexpressing Arabidopsis Plants
4.7. Analysis of Transgenic Lines for Tolerance to Waterlogging, NaCl, and Mannitol
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jackson, M.B.; Colmer, T.D. Response and adaptation by plants to flooding stress. Ann. Bot. 2005, 96, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Ismond, K.P.; Dolferus, R.; Pauw, M.D.; Dennis, E.S.; Good, A.G. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 2003, 132, 1292–1302. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.; Krishnan, S.P.; Swarup, S.; Bajic, V.B. Detection and preliminary analysis of motifs in promoters of anaerobically induced genes of different plant species. Ann. Bot. 2005, 96, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Huang, S.N.; Mo, Z.H.; Xuan, J.P.; Jia, X.D.; Wang, G.; Guo, Z.R. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Mol. Breed. 2015, 35, 208. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Huang, S.N.; Wang, G.; Xuan, J.P.; Guo, Z.R. Overexpression of Actinidia deliciosa Pyruvate decarboxylase 1 gene enhances waterlogging stress in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2016, 106, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Huang, S.N.; Chen, Y.H.; Wang, G.; Guo, Z.R. Identification and characterization of two waterlogging responsive alcohol dehydrogenase genes (AdADH1 and AdADH2) in Actinidia deliciosa. Mol. Breed. 2017, 37, 52. [Google Scholar] [CrossRef]
- Jung, K.H.; Gho, H.J.; Nguyen, M.X.; Kim, S.R.; An, G. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genom. 2013, 13, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Rajan, V.B.; D’Silva, P. Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genom. 2009, 9, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Ann. Rev. Plant Boil. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Bacardit, J.; Bachmair, A.; Holdsworth, M.J. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014, 24, 603. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Pucciariello, C.; Perata, P. New role for an old rule: N-end rule-mediated degradation of ethylene responsive factor proteins governs low oxygen response in plants. J. Integr. Plant Boil. 2013, 55, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Waters, I.; Morrell, S.; Greenway, H.; Colmer, T.D. Effects of anoxia on wheat seedlings: II. Influence of O2 supply prior to anoxia on tolerance to anoxia, alcoholic fermentation, and sugar levels. J. Exp. Bot. 1991, 42, 1437–1447. [Google Scholar] [CrossRef]
- Wignarajah, K.; Greenway, H.; John, C.D. Effect of waterlogging on growth and activity of alcohol dehydrogenase in barley and rice. New Phytol. 2006, 77, 585–592. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, X.; Yang, G.; Li, Z.; Lu, H.; Kong, X.; Eneji, A.; Dong, H. Physiological and molecular adjustment of cotton to waterlogging at peak-flowering in relation to growth and yield. Field Crop Res. 2015, 179, 164–172. [Google Scholar] [CrossRef]
- Dennis, E.; Dolferus, R.; Ellis, M.; Rahman, M.; Wu, Y.; Hoeren, F.; Grover, A.; Ismond, K.; Good, A.; Peacock, W. Molecular strategies for improving waterlogging tolerance in plants. J. Exp. Bot. 2000, 51, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Shiao, T.L.; Ellis, M.H.; Dolferus, R.; Dennis, E.S.; Doran, P.M. Overexpression of alcohol dehydrogenase or Pyruvate decarboxylase improves growth of hairy roots at reduced oxygen concentrations. Biotechnol. Bioeng. 2002, 77, 455. [Google Scholar] [CrossRef] [PubMed]
- Kürsteiner, O.; Dupuis, I.; Kuhlemeier, C. The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia, but not other environmental stresses. Plant Physiol. 2003, 132, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Mithran, M.; Paparelli, E.; Novi, G.; Perata, P.; Loreti, E. Analysis of the role of the Pyruvate decarboxylase gene family in Arabidopsis thaliana under low-oxygen conditions. Plant Biol. 2014, 16, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Huq, E.; Grover, A.; Dennis, E.S.; Peacock, W.J.; Hodges, T.K. Characterization of Pyruvate decarboxylase genes from rice. Plant Mol. Biol. 1996, 31, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Rivoal, J.; Thind, S.; Pradet, A.; Ricard, B. Differential induction of Pyruvate decarboxylase subunits and transcripts in anoxic rice seedlings. Plant Physiol. 1997, 114, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Coram, T.E.; Huang, X.; Zhan, G.; Settles, M.L.; Chen, X. Meta-analysis of transcripts associated with race-specific resistance to stripe rust in wheat demonstrates common induction of blue copper-binding protein, heat-stress transcription factor, pathogen-induced WIR1A protein, and ent-kaurene synthase transcripts. Funct. Integr. Genom. 2010, 10, 383–392. [Google Scholar]
- Guo, H.; Li, Z.; Zhou, M.; Cheng, H. cDNA-AFLP analysis reveals heat shock proteins play important roles in mediating cold, heat, and drought tolerance in Ammopiptanthus mongolicus. Funct. Integr. Genom. 2014, 14, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; He, B.; Wang, C.; Fang, Y.; Qi, J.; Tang, C. Molecular identification and characterization of the Pyruvate decarboxylase gene family associated with latex regeneration and stress response in rubber tree. Plant Physiol. Biochem. 2015, 87, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Pinhero, R.; Pazhekattu, R.; Marangoni, A.G.; Liu, Q.; Yada, R.Y. Alleviation of low temperature sweetening in potato by expressing Arabidopsis Pyruvate decarboxylase gene and stress-inducible rd29A: A preliminary study. Physiol. Mol. Biol. Plants 2011, 17, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Gass, N.; Glagotskaia, T.; Mellema, S.; Stuurman, J.; Barone, M.; Mandel, T.; Roessner-Tunali, U.; Kuhlemeier, C. Pyruvate decarboxylase provides growing pollen tubes with a competitive advantage in petunia. Plant Cell 2005, 17, 2355–2368. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Han, B. Primary function analysis of a pyruvate decarboxylase gene, OsPDC3, in rice. Chin. J. Rice Sci. 2011, 25, 567–574. [Google Scholar]
- Li, Y.; Ohtsu, K.; Nemoto, K.; Tsutsumi, N.; Hirai, A.; Nakazono, M. The rice pyruvate decarboxylase 3 gene, which lacks introns, is transcribed in mature pollen. J. Exp. Bot. 2004, 55, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Qu, S.C.; Dong, C.; Gao, Z.H.; Qiao, Y.S.; Zhang, Z. Utility and construction of full-length cDNA library of Malus hupehensis post-introduced with salicylic acid. Acta Bot. Boreali Occident. Sin. 2010, 30, 1527–1533. [Google Scholar]
- ExPASy Bioinformatics Resource Portal. Available online: http://web.expasy.org/ (accessed on 8 July 2017).
- Yin, X.R.; Allan, A.C.; Xu, Q.; Burdon, J.; Dejnoprat, S.; Chen, K.S.; Ferguson, I.B. Differential expression of kiwifruit ERF genes in response to postharvest abiotic stress. Postharvest Biol. Technol. 2012, 66, 1–7. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In planta agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar] [PubMed]
- Papdi, C.; Pérezsalamó, I.; Joseph, M.P.; Giuntoli, B.; Bögre, L.; Koncz, C. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor-vii genes rap2.12, rap2.2 and rap2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, H.-T.; Zhang, J.-Y.; Wang, G.; Jia, Z.-H.; Huang, S.-N.; Wang, T.; Guo, Z.-R. Functional Characterization of Waterlogging and Heat Stresses Tolerance Gene Pyruvate decarboxylase 2 from Actinidia deliciosa. Int. J. Mol. Sci. 2017, 18, 2377. https://doi.org/10.3390/ijms18112377
Luo H-T, Zhang J-Y, Wang G, Jia Z-H, Huang S-N, Wang T, Guo Z-R. Functional Characterization of Waterlogging and Heat Stresses Tolerance Gene Pyruvate decarboxylase 2 from Actinidia deliciosa. International Journal of Molecular Sciences. 2017; 18(11):2377. https://doi.org/10.3390/ijms18112377
Chicago/Turabian StyleLuo, Hui-Ting, Ji-Yu Zhang, Gang Wang, Zhan-Hui Jia, Sheng-Nan Huang, Tao Wang, and Zhong-Ren Guo. 2017. "Functional Characterization of Waterlogging and Heat Stresses Tolerance Gene Pyruvate decarboxylase 2 from Actinidia deliciosa" International Journal of Molecular Sciences 18, no. 11: 2377. https://doi.org/10.3390/ijms18112377
APA StyleLuo, H.-T., Zhang, J.-Y., Wang, G., Jia, Z.-H., Huang, S.-N., Wang, T., & Guo, Z.-R. (2017). Functional Characterization of Waterlogging and Heat Stresses Tolerance Gene Pyruvate decarboxylase 2 from Actinidia deliciosa. International Journal of Molecular Sciences, 18(11), 2377. https://doi.org/10.3390/ijms18112377






