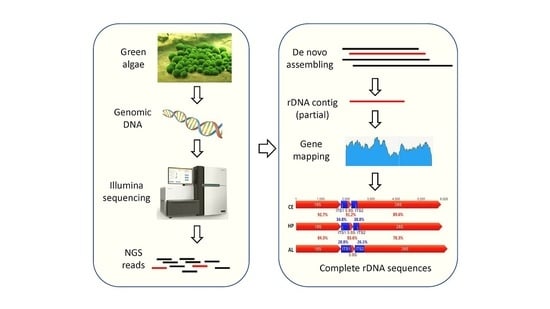
A Simple Method to Decode the Complete 18-5.8-28S rRNA Repeated Units of Green Algae by Genome Skimming
Abstract
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guiry, M.D. How many species of algae are there? J. Phycol. 2012, 48, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Fondriest Environmental, I. “Algae, Phytoplankton and Chlorophyll.” Fundamentals of Environmental Measurements. Available online: http://www.fondriest.com/environmental-measurements/parameters/water-quality/algae-phytoplankton-chlorophyll/ (accessed on 2 November 2017).
- Cho, H.U.; Kim, H.G.; Kim, Y.M.; Park, J.M. Volatile fatty acid recovery by anaerobic fermentation from blue-green algae: Effect of pretreatment. Bioresour. Technol. 2017, 244, 1433–1438. [Google Scholar] [CrossRef] [PubMed]
- Oheocha, C.; Haxo, F.T. Some atypical algal chromoproteins. Biochim. Biophys. Acta 1960, 41, 516–520. [Google Scholar] [CrossRef]
- Wells, M.L.; Potin, P.; Craigie, J.S.; Raven, J.A.; Merchant, S.S.; Helliwell, K.E.; Smith, A.G.; Camire, M.E.; Brawley, S.H. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol. 2017, 29, 949–982. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Koo, S.Y.; Lee, D.U. Antiproliferative effects of carotenoids extracted from chlorella ellipsoidea and chlorella vulgaris on human colon cancer cells. J. Agric. Food Chem. 2008, 56, 10521–10526. [Google Scholar] [CrossRef] [PubMed]
- Posten, C.; Chen, S.F. Microalgae Biotechnology; Springer International Publishing: Basel, Switzerland, 2015; Vol. 153, p. 188. [Google Scholar]
- Markou, G.; Nerantzis, E. Microalgae for high-value compounds and biofuels production: A review with focus on cultivation under stress conditions. Biotechnol. Adv. 2013, 31, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Hannon, M.; Gimpel, J.; Tran, M.; Rasala, B.; Mayfield, S. Biofuels from algae: Challenges and potential. Biofuels 2010, 1, 763–784. [Google Scholar] [CrossRef] [PubMed]
- Fučíková, K.; Lewis, P.O.; Lewis, L.A. Chloroplast phylogenomic data from the green algal order sphaeropleales (chlorophyceae, chlorophyta) reveal complex patterns of sequence evolution. Mol. Phylogenet. Evol. 2016, 98, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Leliaert, F.; Verbruggen, H.; Vanormelingen, P.; Steen, F.; López-Bautista, J.M.; Zuccarello, G.C.; De Clerck, O. DNA-based species delimitation in algae. Eur. J. Phycol. 2014, 49, 179–196. [Google Scholar] [CrossRef]
- Seckbach, J. Life as We Know It; Springer Netherlands: Amsterdam, The Netherlands, 2006; p. 765. [Google Scholar]
- Smith, A.M.C.G.; Dolan, L.; Harberd, N.; Jones, J.; Martin, C.; Sablowski, R.; Amey, A. Plant Biology; Garland Science: New York, NY, USA, 2009; p. 679. [Google Scholar]
- Boedeker, C.; Leliaert, F.; Zuccarello, G.C. Molecular phylogeny of the cladophoraceae (cladophorales, ulvophyceae), with the resurrection of acrocladus nageli and willeella borgesen, and the description of lurbica gen. nov. And pseudorhizoclonium gen. nov. J. Phycol. 2016, 52, 905–928. [Google Scholar] [CrossRef] [PubMed]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Ripma, L.A.; Simpson, M.G.; Hasenstab-Lehman, K. Geneious! Simplified genome skimming methods for phylogenetic systematic studies: A case study in oreocarya (boraginaceae). Appl. Plant Sci. 2014, 2, 1400062. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, K.R. Frontiers in Fungal Ecology, Diversity and Metabolites; IK International Publishing House: Delhi, India, 2008; p. 352. [Google Scholar]
- Bleidorn, C. Phylogenomics: An Introduction; Springer: Basel, Switzerland, 2017; p. XIII, 222. [Google Scholar]
- Webster, M.S. The Extended Specimen: Emerging Frontiers in Collections-Based Ornithological Research; CRC Press: Boca Raton, FL, USA, 2017; p. 240. [Google Scholar]
- Lorenz, R.T.; Cysewski, G.R. Commercial potential for haematococcus microalgae as a natural source of astaxanthin. Trends Biotechnol. 2000, 18, 160–167. [Google Scholar] [CrossRef]
- Cardozo, K.H.; Guaratini, T.; Barros, M.P.; Falcao, V.R.; Tonon, A.P.; Lopes, N.P.; Campos, S.; Torres, M.A.; Souza, A.O.; Colepicolo, P.; et al. Metabolites from algae with economical impact. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 146, 60–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pan, L.; Wei, X.; Gao, H.; Liu, J. Impact of astaxanthin-enriched algal powder of haematococcus pluvialis on memory improvement in balb/c mice. Environ. Geochem. Health 2007, 29, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Boussiba, S. Carotenogenesis in the green alga haematococcus pluvialis: Cellular physiology and stress response. Physiol. Plant. 2000, 108, 111–117. [Google Scholar] [CrossRef]
- Alasalvar, C.; Miyashita, K.; Shahidi, F.; Wanasundara, U. Handbook of Seafood Quality, Safety and Health Applications; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Herrero, M.; Ibanez, E.; Fanali, S.; Cifuentes, A. Quantitation of chiral amino acids from microalgae by mekc and lif detection. Electrophoresis 2007, 28, 2701–2709. [Google Scholar] [CrossRef] [PubMed]
- Soontornchaiboon, W.; Joo, S.S.; Kim, S.M. Anti-inflammatory effects of violaxanthin isolated from microalga chlorella ellipsoidea in raw 264.7 macrophages. Biol. Pharm. Bull. 2012, 35, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.C.; Kang, N.; Kim, E.A.; Kang, M.C.; Lee, S.H.; Kang, S.M.; Lee, J.B.; Jeon, B.T.; Kim, S.K.; Park, S.J. A novel angiotensin i-converting enzyme (ace) inhibitory peptide from a marine chlorella ellipsoidea and its antihypertensive effect in spontaneously hypertensive rats. Process Biochem. 2012, 47, 2005–2011. [Google Scholar] [CrossRef]
- Acton, Q.A. Issues in Earth Sciences, Geology, and Geophysics: 2011 Edition; ScholarlyEditions: Atlanta, GA, USA, 2012. [Google Scholar]
- Heibel, T.; De Give, T. Rooted in Design: Sprout Home’s Guide to Creative Indoor Planting; Ten Speed Press: Emeryville, CA, USA, 2015; p. 224. [Google Scholar]
- Boedeker, C.; Sviridenko, B.F. Cladophora koktschetavensis from kazakhstan is a synonym of aegagropila linnaei (cladophorales, chlorophyta) and fills the gap in the disjunct distribution of a widespread genotype. Aquat. Bot. 2012, 101, 64–68. [Google Scholar] [CrossRef]
- Hanyuda, T.; Wakana, I.; Arai, S.; Miyaji, K.; Watano, Y.; Ueda, K. Phylogenetic relationships within cladophorales (ulvophyceae, chlorophyta) inferred from 18 s rrna gene sequences, with special reference to aegagropila linnaei 1. J. Phycol. 2002, 38, 564–571. [Google Scholar] [CrossRef]
- Tiwari, K.; Jadhav, S.; Gupta, S. Modified ctab technique for isolation of DNA from some medicinal plants. Res. J. Med. Plant 2011, 201, 1. [Google Scholar]
- Magoč, T.; Salzberg, S.L. Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, S.; Madden, T.L. Blast: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Orr, R.J.; Stüken, A.; Rundberget, T.; Eikrem, W.; Jakobsen, K.S. Improved phylogenetic resolution of toxic and non-toxic alexandrium strains using a concatenated rdna approach. Harmful Algae 2011, 10, 676–688. [Google Scholar] [CrossRef]
- Heeg, J.S.; Wolf, M. Its2 and 18s rdna sequence-structure phylogeny of chlorella and allies (chlorophyta, trebouxiophyceae, chlorellaceae). Plant Gene 2015, 4, 20–28. [Google Scholar] [CrossRef]
- Buchheim, M.A.; Sutherland, D.M.; Buchheim, J.A.; Wolf, M. The blood alga: Phylogeny of haematococcus (chlorophyceae) inferred from ribosomal rna gene sequence data. Eur. J. Phycol. 2013, 48, 318–329. [Google Scholar] [CrossRef]
- Ali, A.B.; De Baere, R.; Van der Auwera, G.; De Wachter, R.; Van de Peer, Y. Phylogenetic relationships among algae based on complete large-subunit rrna sequences. Int. J. Syst. Evol. Microbiol. 2001, 51, 737–749. [Google Scholar] [PubMed]
- Daugbjerg, N.; Andersen, R.A. A molecular phylogeny of the heterokont algae based on analyses of chloroplast-encoded rbcl sequence data. J. Phycol. 1997, 33, 1031–1041. [Google Scholar] [CrossRef]
- Daugbjerg, N.; Andersen, R.A. Phylogenetic analyses of the rbcl sequences from haptophytes and heterokont algae suggest their chloroplasts are unrelated. Mol. Biol. Evol. 1997, 14, 1242–1251. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Friedl, T.; Hegewald, E. Phylogenetic relationships of scenedesmus and scenedesmus-like coccoid green algae as inferred from its-2 rdna sequence comparisons. Plant Biol. 1999, 1, 418–428. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. Bold: The barcode of life data system (http://www.Barcodinglife.Org). Mol. Ecol. Resour. 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Li, F.W.; Kuo, L.Y.; Rothfels, C.J.; Ebihara, A.; Chiou, W.L.; Windham, M.D.; Pryer, K.M. Rbcl and matk earn two thumbs up as the core DNA barcode for ferns. PLoS ONE 2011, 6, e26597. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Ben Ali, A.; Meyer, A. Microsporidia: Accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene 2000, 246, 1–8. [Google Scholar] [CrossRef]
- Van de Peer, Y.; De Wachter, R. Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18s rrna. J. Mol. Evol. 1997, 45, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Robba, L.; Russell, S.J.; Barker, G.L.; Brodie, J. Assessing the use of the mitochondrial cox1 marker for use in DNA barcoding of red algae (rhodophyta). Am. J. Bot. 2006, 93, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Shearer, T.L.; Van Oppen, M.J.; Romano, S.L.; Worheide, G. Slow mitochondrial DNA sequence evolution in the anthozoa (cnidaria). Mol. Ecol. 2002, 11, 2475–2487. [Google Scholar] [CrossRef] [PubMed]
- Vences, M.; Thomas, M.; Bonett, R.M.; Vieites, D.R. Deciphering amphibian diversity through DNA barcoding: Chances and challenges. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, M.; Backeljau, T.; Nevado, B.; De Meyer, M. Comparative performances of DNA barcoding across insect orders. BMC Bioinform. 2010, 11, 206. [Google Scholar] [CrossRef] [PubMed]
- Meusnier, I.; Singer, G.A.; Landry, J.F.; Hickey, D.A.; Hebert, P.D.; Hajibabaei, M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (gbs) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Torres-Machorro, A.L.; Hernandez, R.; Alderete, J.F.; Lopez-Villasenor, I. Comparative analyses among the trichomonas vaginalis, trichomonas tenax, and tritrichomonas foetus 5s ribosomal rna genes. Curr. Genet. 2009, 55, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Eickbush, T.H.; Eickbush, D.G. Finely orchestrated movements: Evolution of the ribosomal rna genes. Genetics 2007, 175, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Ganley, A.R.; Kobayashi, T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rdna repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007, 17, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, D.J.; Lajeunesse, T.C.; Santos, S.R. Measuring rdna diversity in eukaryotic microbial systems: How intragenomic variation, pseudogenes, and pcr artifacts confound biodiversity estimates. Mol. Ecol. 2007, 16, 5326–5340. [Google Scholar] [CrossRef] [PubMed]
- Leo, N.; Barker, S. Intragenomic variation in its2 rdna in the louse of humans, pediculus humanus: Its2 is not a suitable marker for population studies in this species. Insect Mol. Biol. 2002, 11, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Alanagreh, L.; Pegg, C.; Harikumar, A.; Buchheim, M. Assessing intragenomic variation of the internal transcribed spacer two: Adapting the illumina metagenomics protocol. PLoS ONE 2017, 12, e0181491. [Google Scholar] [CrossRef] [PubMed]
- Simon, U.K.; Weiß, M. Intragenomic variation of fungal ribosomal genes is higher than previously thought. Mol. Biol. Evol. 2008, 25, 2251–2254. [Google Scholar] [CrossRef] [PubMed]
- Pereira, T.J.; Baldwin, J.G. Contrasting evolutionary patterns of 28 s and its rrna genes reveal high intragenomic variation in cephalenchus (nematoda): Implications for species delimitation. Mol. Phylogenet. Evol. 2016, 98, 244–260. [Google Scholar] [CrossRef] [PubMed]
- Shapoval, N.A.; Lukhtanov, V.A. Intragenomic variations of multicopy its2 marker in agrodiaetus blue butterflies (lepidoptera, lycaenidae). Comp. Cytogenet. 2015, 9, 483. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; McLellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding, L. Varscan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009, 25, 2283–2285. [Google Scholar] [CrossRef] [PubMed]


| Species | Chlorella ellipsoidea (CE) | Haematococcus pluvialis (HP) | Aegagropila linnaei (AL) |
|---|---|---|---|
| Total reads | 67,016,212 | 68,697,604 | 7,502,824 |
| 18-5.8-28S reads | 159,445 | 74,748 | 1648 |
| 18-5.8-28S reads % * | 0.24 | 0.11 | 0.02 |
| Coverage (fold) | 2736 | 825 | 26 |
| Sequence (bp) | 5785 | 5817 | 6028 |
| NCBI accession number | KY364701 | KY364700 | KY364699 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, G.-M.; Lai, Y.-H.; Audira, G.; Hsiao, C.-D. A Simple Method to Decode the Complete 18-5.8-28S rRNA Repeated Units of Green Algae by Genome Skimming. Int. J. Mol. Sci. 2017, 18, 2341. https://doi.org/10.3390/ijms18112341
Lin G-M, Lai Y-H, Audira G, Hsiao C-D. A Simple Method to Decode the Complete 18-5.8-28S rRNA Repeated Units of Green Algae by Genome Skimming. International Journal of Molecular Sciences. 2017; 18(11):2341. https://doi.org/10.3390/ijms18112341
Chicago/Turabian StyleLin, Geng-Ming, Yu-Heng Lai, Gilbert Audira, and Chung-Der Hsiao. 2017. "A Simple Method to Decode the Complete 18-5.8-28S rRNA Repeated Units of Green Algae by Genome Skimming" International Journal of Molecular Sciences 18, no. 11: 2341. https://doi.org/10.3390/ijms18112341
APA StyleLin, G.-M., Lai, Y.-H., Audira, G., & Hsiao, C.-D. (2017). A Simple Method to Decode the Complete 18-5.8-28S rRNA Repeated Units of Green Algae by Genome Skimming. International Journal of Molecular Sciences, 18(11), 2341. https://doi.org/10.3390/ijms18112341