The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling
Abstract
1. Innate Immunity Response in Higher Organisms to Pathogen-Associated Molecular Patterns
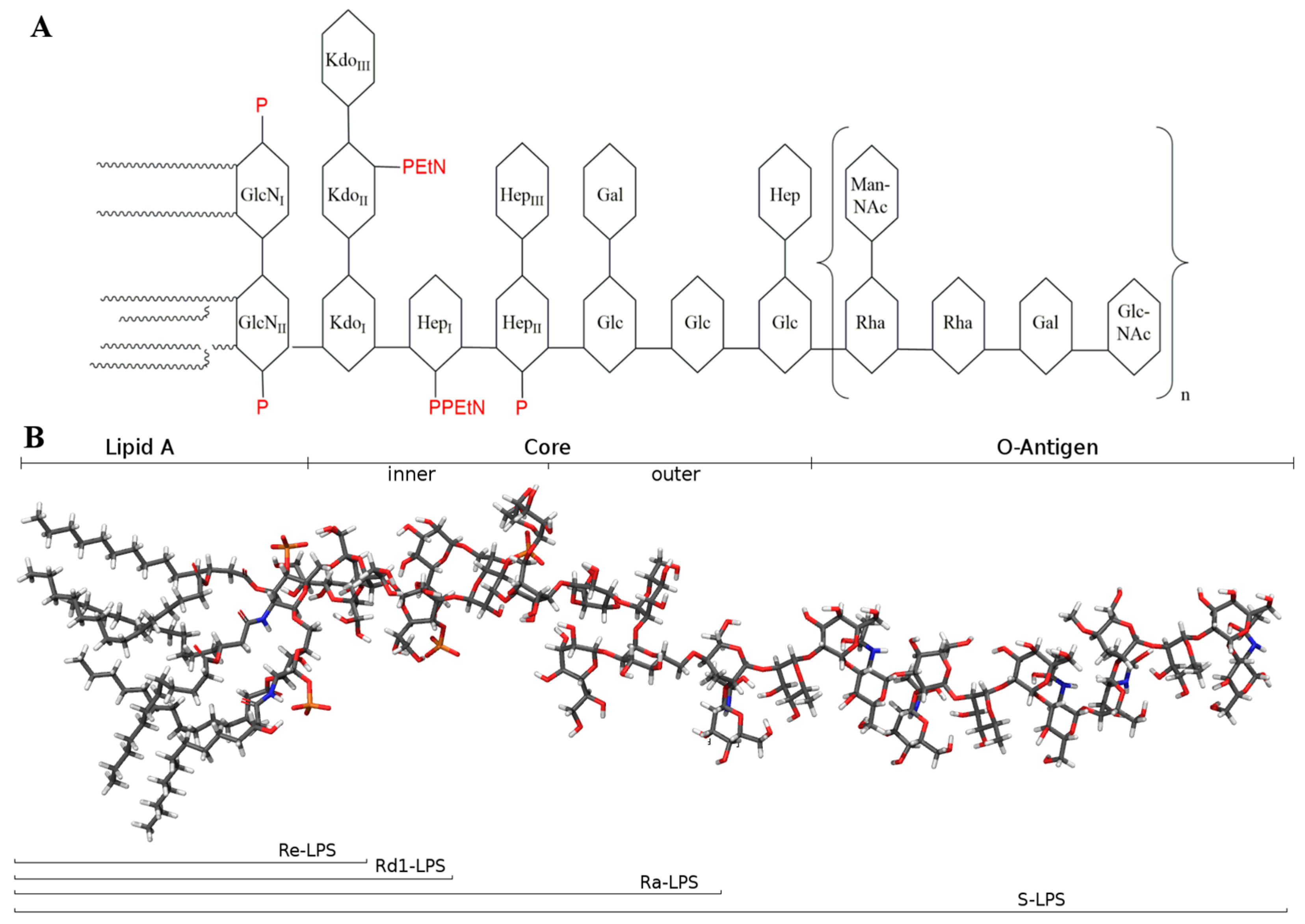
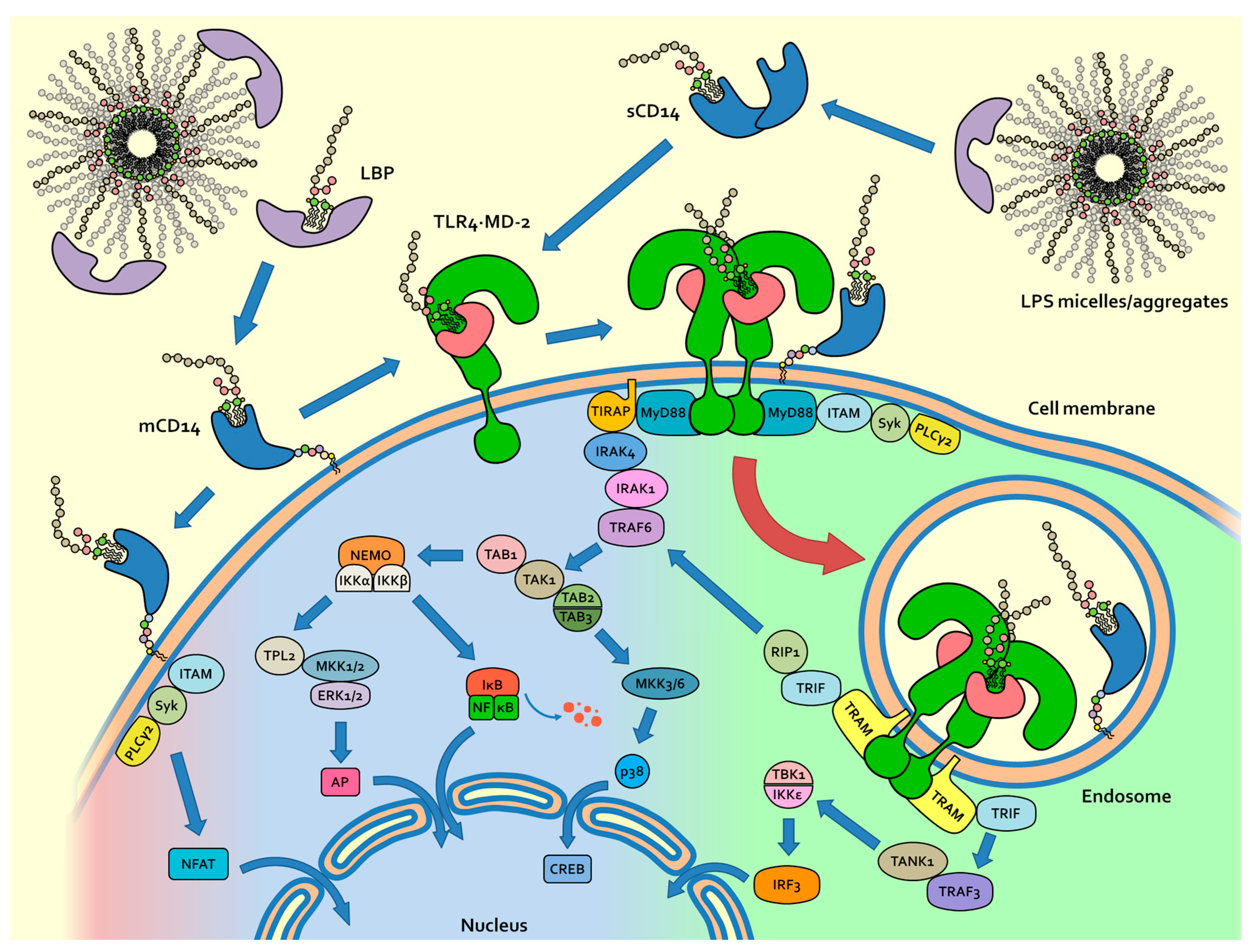
2. Physicochemical Properties of LPS and Molecular Mechanism of LPS/TLR4 Signalling
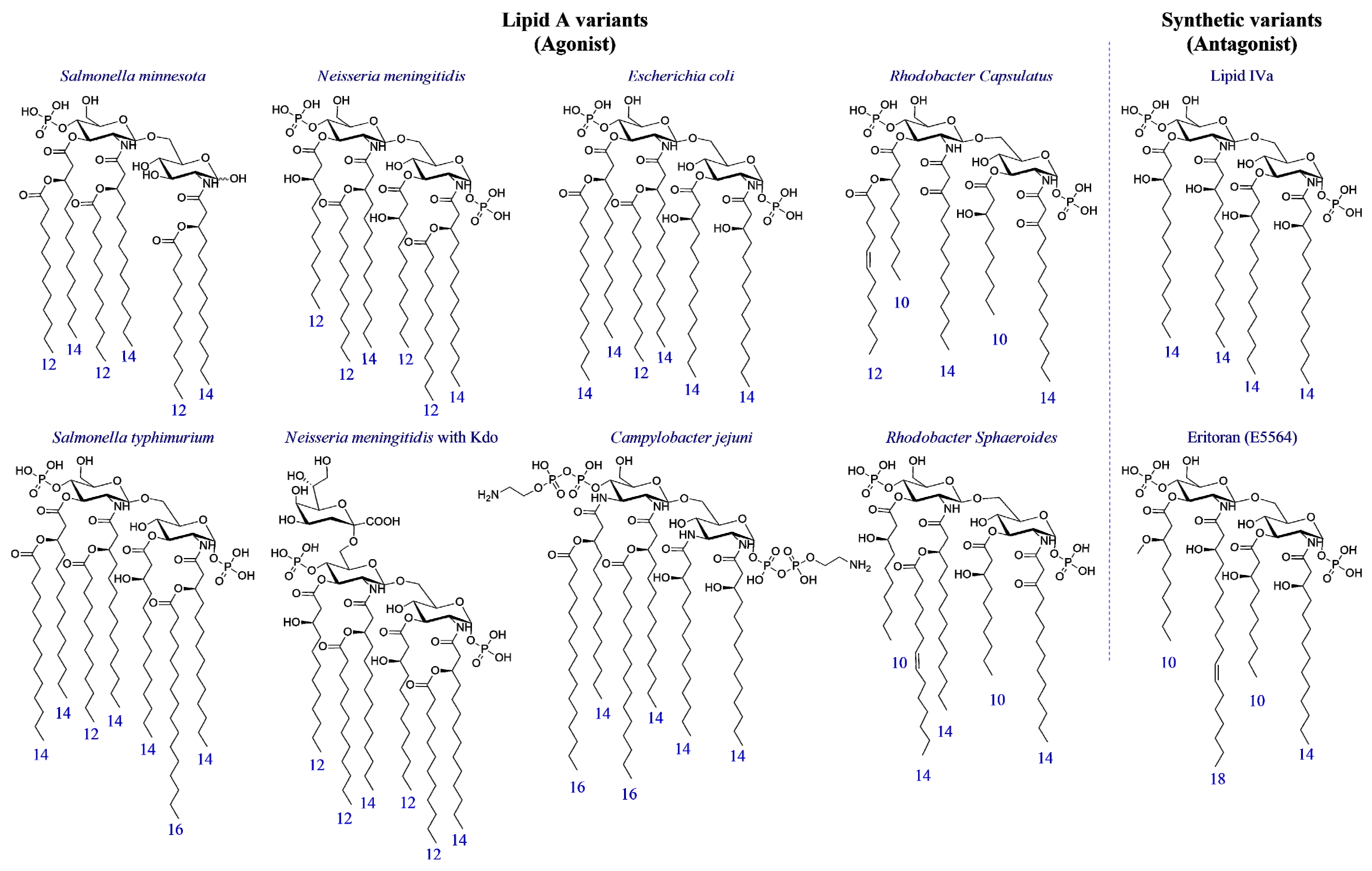
3. Structure-Activity Relationship in Natural and Synthetic Lipid A Variants
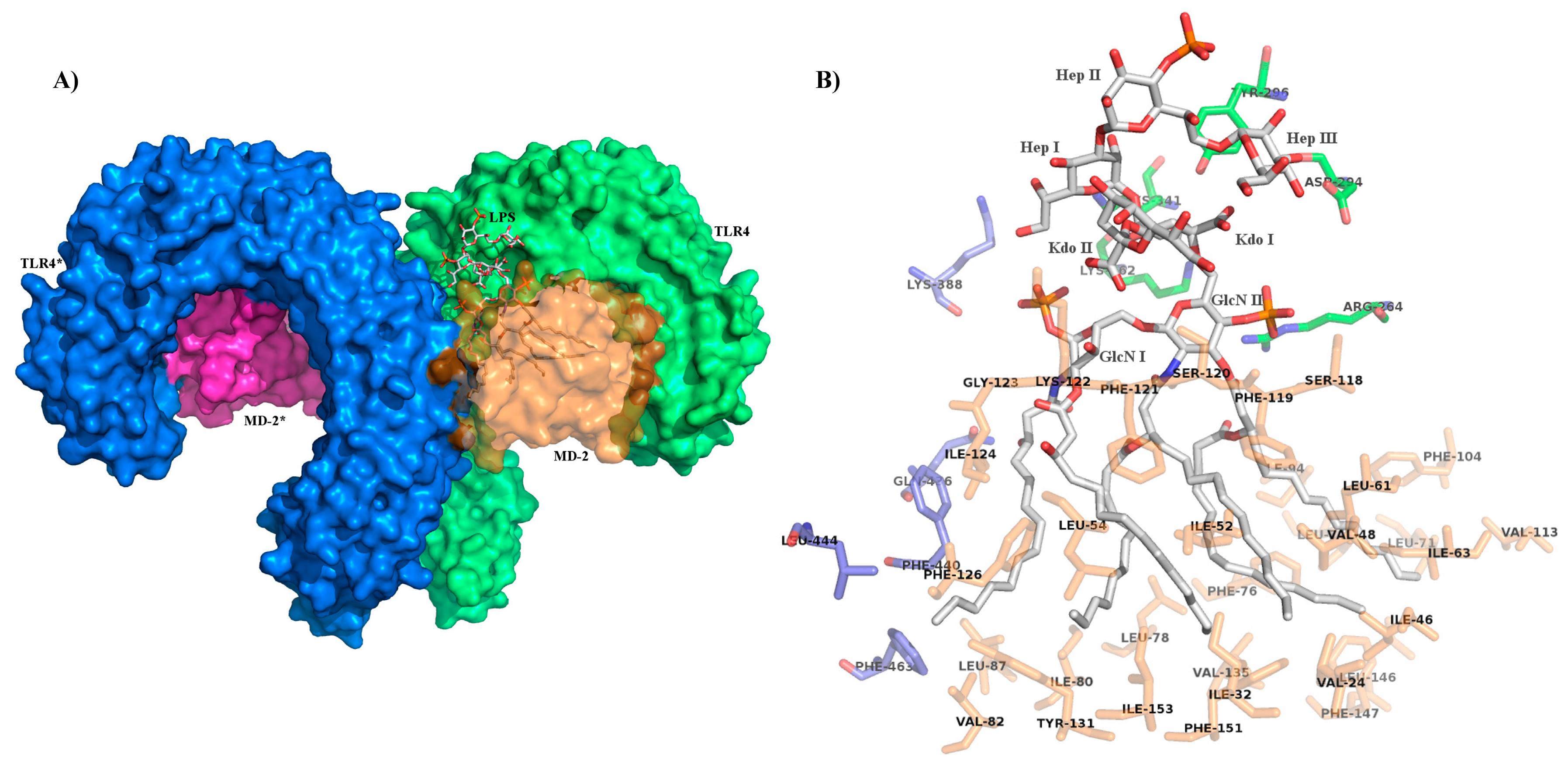
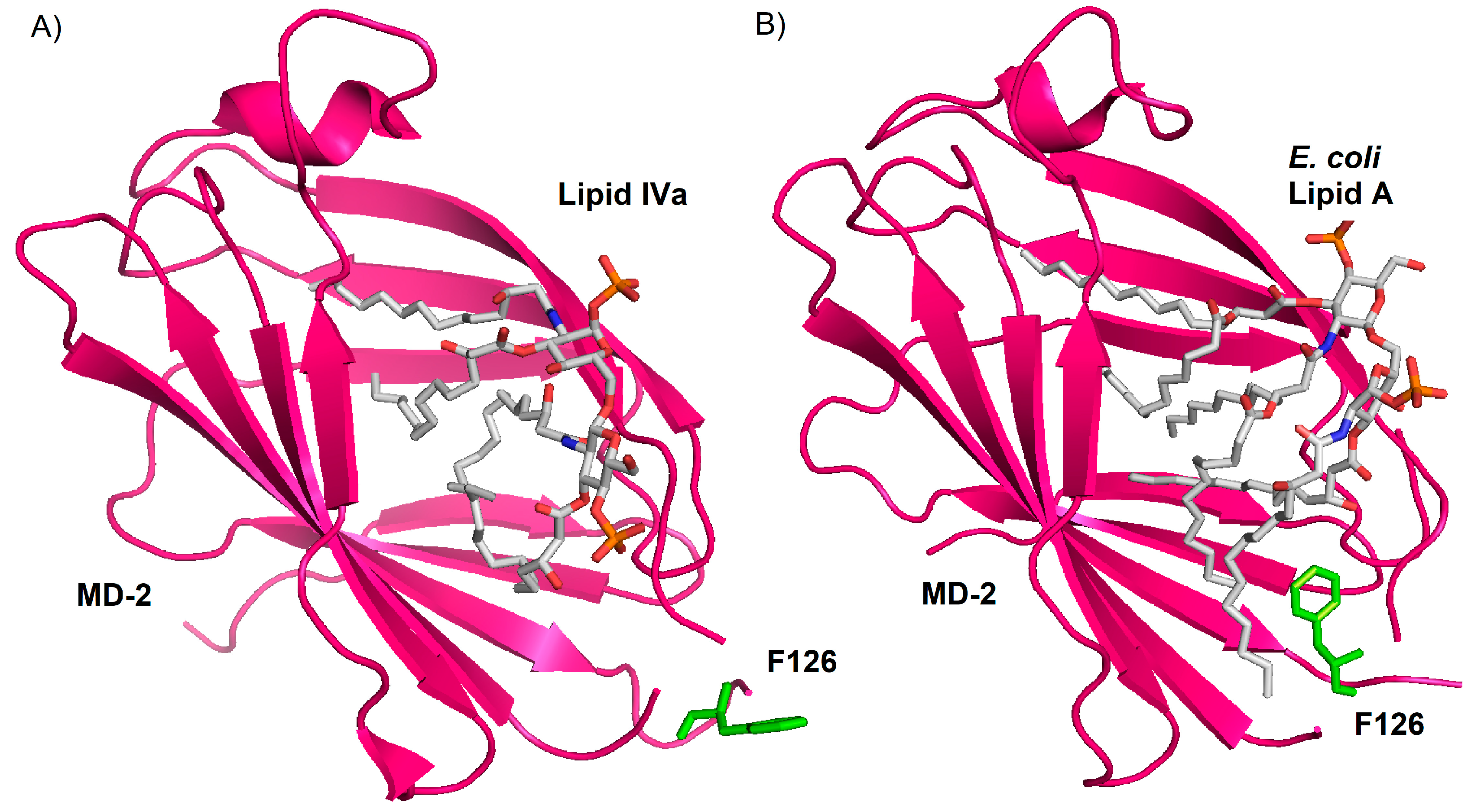
4. Role of Sugar/Protein and Lipid/Protein Interactions in the Formation and Stability of TLR4/MD-2/Endotoxin Complex
5. Role of the Sugar Kdo in the TLR4 Interaction of Natural and Synthetic LPS Variants
6. Conclusions and Future Perspectives
Acknowledgments
Conflicts of Interest
Abbreviations
| AIM2 | Interferon-inducible protein (Absent In Melanoma 2) |
| ALR | AIM2-Like Receptor |
| CD14 | Cluster of Differentiation 14 |
| CD80 | Cluster of Differentiation 80 |
| CD83 | Cluster of Differentiation 83 |
| CD86 | Cluster of Differentiation 86 |
| CLR | C-type Lectin Receptor |
| CMC | Critical Micellar Concentration |
| FA | Fatty acid |
| Gal | Galactose |
| Glc | Glucose |
| GlcN | Glucosamine |
| GlcN3N | 2,3-diamino-2,3-dideoxy-d-glucose |
| GlcNAc | N-Acetyl-Glucosamine |
| GPI | Glycosylphosphatidylinositol |
| HEK | Human Embryonic Kidney Cell 293 |
| Hep | heptosyl-2-keto-3-deoxy-octulosonate |
| HLADR | Human Leukocyte Antigen D Related |
| IL | Interleukin |
| INF | Interferon |
| IRF | Interferon Regulatory Factor |
| IRF3 | Interferon Regulatory Factor 3 |
| Kdo | 3-Deoxy-d-manno-oct-2-ulosonic acid |
| LBP | Lipid Binding Protein |
| LOS | Lipooligosaccharide (Hep3-Kdo2-lipid A) |
| LPS | Lipopolysaccharide |
| LRR | Leucine-Rich Repeat |
| MC | Mast Cells |
| MD-2 | Myeloid Differentiation factor 2 |
| MDDC | Human Monocyte Derived Dendritic Cell |
| MurNAc | N-Acetyl-Muramic acid |
| MyD88 | Myeloid differentiation primary response gene 88 |
| NBD | Nucleotide-Binding Domain |
| NF-κB | Nuclear Factor-kappa B |
| NLR | NOD-Like Receptors |
| NMR | Nuclear Magnetic Resonance |
| NOD 1 & 2 | Nucleotide-binding Oligomerization Domain receptor 1 & 2 |
| PAMP | Pathogen Associated Molecular Pattern |
| PGN | Peptidoglycan |
| PRR | Pattern Recognition Receptor |
| Ra-LPS | Lipid A + core |
| Re-LPS | (Kdo)2-Lipid A |
| RIG-1 | Retinoic Acid Inducible Gene 1 |
| R-LPS | Rough LPS |
| RLR | RIG-I-Like Receptor |
| SAR | Structure Activity Relationship |
| S-LPS | Smooth LPS |
| TANK | TRAF family member-associated NF-κ-B activator |
| TBK1 | TANK-Binding Kinase 1 (Serine/Threonine-protein Kinase TBK1) |
| TEM | Transmission Electron Microscopy |
| THP-1 | Human Leukemic Monocyte Cell Line |
| TIR | Toll/interleukin-1 receptor |
| TLR4 | Toll Like Receptor 4 |
| TNF | Tumor Necrosis Factor |
| TRAM | TNF Receptor Associated Modulator |
| TRIF | TIR-domain-containing adapter-inducing interferon-β |
References
- Janeway, C.A.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Shaw, M.H.; Kim, Y.-G.; Nuñez, G. NOD-Like Receptors: Role in Innate Immunity and Inflammatory Disease. Annu. Rev. Pathol. 2009, 4, 365–398. [Google Scholar] [CrossRef] [PubMed]
- Song, D.H.; Lee, J.-O. Sensing of microbial molecular patterns by Toll-like receptors. Immunol. Rev. 2012, 250, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Kieser, K.J.; Kagan, J.C. Multi-receptor detection of individual bacterial products by the innate immune system. Nat. Rev. Immunol. 2017, 17, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Imoto, M.; Yoshimura, H.; Sakaguchi, N.; Kusumoto, S.; Shiba, T. Total synthesis of lipid A. Tetrahedron Lett. 1985, 26, 1545–1548. [Google Scholar] [CrossRef]
- Yoshizaki, H.; Fukuda, N.; Sato, K.; Oikawa, M.; Fukase, K.; Suda, Y.; Kusumoto, S. First Total Synthesis of the Re-Type Lipopolysaccharide. Angew. Chem. Int. Ed. 2001, 40, 1475–1480. [Google Scholar] [CrossRef]
- Uehara, A.; Fujimoto, Y.; Kawasaki, A.; Kusumoto, S.; Fukase, K.; Takada, H. Meso-Diaminopimelic Acid and Meso-Lanthionine, Amino Acids Specific to Bacterial Peptidoglycans, Activate Human Epithelial Cells through NOD1. J. Immunol. 2006, 177, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Homma, J.Y.; Matsuura, M.; Kanegasaki, S.; Kawakubo, Y.; Kojima, Y.; Shibukawa, N.; Kumazawa, Y.; Yamamoto, A.; Tanamoto, K.; Yasuda, T.; et al. Structural Requirements of Lipid A Responsible for the Functions: A Study with Chemically Synthesized Lipid A and Its Analogues. J. Biochem. 1985, 98, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Kalis, C.; Keck, S.; Jiang, Z.; Georgel, P.; Du, X.; Shamel, L.; Sovath, S.; Mudd, S.; Beutler, B.; et al. R-form LPS, the master key to the activation of TLR4/MD-2-positive cells. Eur. J. Immunol. 2006, 36, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, Y.; Shao, F. Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr. Opin. Immunol. 2015, 32, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Gutsmann, T.; Schromm, A.B.; Brandenburg, K. The physicochemistry of endotoxins in relation to bioactivity. Int. J. Med. Microbiol. 2007, 297, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Din, Z.Z.; Mukerjee, P.; Cooke, P.H.; Kirkland, T.N. Physicochemical properties of the lipopolysaccharide unit that activisit. J. Biol. Chem. 1990, 265, 14023–14029. [Google Scholar] [PubMed]
- Takayama, K.; Mitchell, D.H.; Din, Z.Z.; Mukerjee, P.; Li, C.; Coleman, D.L. Monomeric Re lipopolysaccharide from Escherichia coli is more active than the aggregated form in the Limulus amebocyte lysate assay and in inducing Egr-1 mRNA in murine peritoneal macrophages. J. Biol. Chem. 1994, 269, 2241–2244. [Google Scholar] [PubMed]
- Yu, L.; Tan, M.; Ho, B.; Ding, J.L.; Wohland, T. Determination of critical micelle concentrations and aggregation numbers by fluorescence correlation spectroscopy: Aggregation of a lipopolysaccharide. Anal. Chim. Acta 2006, 556, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Giardina, P.C.; Gioannini, T.; Buscher, B.A.; Zaleski, A.; Zheng, D.S.; Stoll, L.; Teghanemt, A.; Apicella, M.A.; Weiss, J. Construction of Acetate Auxotrophs of Neisseria meningitidis to Study Host-Meningococcal Endotoxin Interactions. J. Biol. Chem. 2001, 276, 5883–5891. [Google Scholar] [CrossRef] [PubMed]
- Iovine, N.; Eastvold, J.; Elsbach, P.; Weiss, J.P.; Gioannini, T.L. The carboxyl-terminal domain of closely related endotoxin-binding proteins determines the target of protein-lipopolysaccharide complexes. J. Biol. Chem. 2002, 277, 7970–7978. [Google Scholar] [CrossRef] [PubMed]
- Post, D.M.B.; Zhang, D.; Eastvold, J.S.; Teghanemt, A.; Gibson, B.W.; Weiss, J.P. Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J. Biol. Chem. 2005, 280, 38383–38394. [Google Scholar] [CrossRef] [PubMed]
- Schromm, A.B.; Brandenburg, K.; Rietschel, E.T.; Flad, H.D.; Carroll, S.F.; Seydel, U. Lipopolysaccharide-binding protein mediates CD14-independent intercalation of lipopolysaccharide into phospholipid membranes. FEBS Lett. 1996, 399, 267–271. [Google Scholar] [CrossRef]
- Schumann, R.R. Function of lipopolysaccharide (LPS)-binding protein (LBP) and CD14, the receptor for LPS/LBP complexes: A short review. Res. Immunol. 1992, 143, 11–15. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.-S.; Lee, H.; Lee, J.-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.D.; Ramos, R.A.; Tobias, P.S.; Ulevitch, R.J.; Mathison, J.C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 1990, 249, 1431–1433. [Google Scholar] [CrossRef] [PubMed]
- Ulevitch, R.J.; Tobias, P.S. Recognition of Gram-negative bacteria and endotoxin by the innate immune system. Curr. Opin. Immunol. 1999, 11, 19–22. [Google Scholar] [CrossRef]
- Jiang, Q.; Akashi, S.; Miyake, K.; Petty, H.R. Cutting Edge: Lipopolysaccharide Induces Physical Proximity Between CD14 and Toll-Like Receptor 4 (TLR4) Prior to Nuclear Translocation of NF-κB. J. Immunol. 2000, 165, 3541–3544. [Google Scholar] [CrossRef] [PubMed]
- Gioannini, T.L.; Teghanemt, A.; Zhang, D.; Coussens, N.P.; Dockstader, W.; Ramaswamy, S.; Weiss, J.P. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc. Natl. Acad. Sci. USA 2004, 101, 4186–4191. [Google Scholar] [CrossRef] [PubMed]
- Teghanemt, A.; Prohinar, P.; Gioannini, T.L.; Weiss, J.P. Transfer of monomeric endotoxin from MD-2 to CD14: Characterization and functional consequences. J. Biol. Chem. 2007, 282, 36250–36256. [Google Scholar] [CrossRef] [PubMed]
- Deguine, J.; Barton, G.M. MyD88: A central player in innate immune signaling. F1000Prime Rep. 2014, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Kagan, J.C.; Su, T.; Horng, T.; Chow, A.; Akira, S.; Medzhitov, R. TRAM couples endocytosis of TLR4 to the induction of interferon beta. Nat. Immunol. 2008, 9, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Rowe, D.C.; Barnes, B.J.; Caffrey, D.R.; Visintin, A.; Latz, E.; Monks, B.; Pitha, P.M.; Golenbock, D.T. LPS-TLR4 Signaling to IRF-3/7 and NF-κB Involves the Toll Adapters TRAM and TRIF. J. Exp. Med. 2003, 198, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Brieger, A.; Rink, L.; Haase, H. Differential Regulation of TLR-Dependent MyD88 and TRIF Signaling Pathways by Free Zinc Ions. J. Immunol. 2013, 191, 1808–1817. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, I.; Granucci, F. Role of CD14 in host protection against infections and in metabolism regulation. Front. Cell. Infect. Microbiol. 2013, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed]
- Mata-Haro, V.; Cekic, C.; Martin, M.; Chilton, P.M.; Casella, C.R.; Mitchell, T.C. The Vaccine Adjuvant Monophosphoryl Lipid A as a TRIF-Biased Agonist of TLR4. Science 2007, 316, 1628–1632. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D. A Synthetic TLR4-active glycolipids as vaccine adjuvants and stand-alone immunotherapeutics. Curr. Top. Med. Chem. 2008, 8, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Scior, T.; Alexander, C.; Zaehringer, U. Reviewing and identifying amino acids of human, murine, canine and equine TLR4/MD-2 receptor complexes conferring endotoxic innate immunity activation by LPS/lipid A, or antagonistic effects by Eritoran, in contrast to species-dependent modulation. Comput. Struct. Biotechnol. J. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Seeley, J.J.; Ghosh, S. Molecular mechanisms of innate memory and tolerance to LPS. J. Leukoc. Biol. 2017, 101, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Shiozaki, M.; Doi, H.; Tanaka, D.; Shimozato, T.; Kurakata, S.I. Syntheses of glucose analogues of E5564 as a highly potent anti-sepsis drug candidate. Bioorg. Med. Chem. 2006, 14, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Gaekwad, J.; Zhang, Y.; Zhang, W.; Reeves, J.; Wolfert, M.A.; Boons, G.J. Differential induction of innate immune responses by synthetic lipid a derivatives. J. Biol. Chem. 2010, 285, 29375–29386. [Google Scholar] [CrossRef] [PubMed]
- Moran, A.P. Biological and serological characterization of Campylobacter jejuni lipopolysaccharides with deviating core and lipid A structures. FEMS Immunol. Med. Microbiol. 1995, 11, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; Billod, J.-M.; Martín-Santamaría, S.; Silipo, A.; Molinaro, A. Gram-Negative Extremophile Lipopolysaccharides: Promising Source of Inspiration for a New Generation of Endotoxin Antagonists. Eur. J. Org. Chem. 2017, 2017, 4055–4073. [Google Scholar] [CrossRef]
- Kusumoto, S.; Fukase, K.; Fukase, Y.; Kataoka, M.; Yoshizaki, H.; Sato, K.; Oikawa, M.; Suda, Y. Structural basis for endotoxic and antagonistic activities: Investigation with novel synthetic lipid A analogs. J. Endotoxin Res. 2003, 9, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Tamai, R.; Asai, Y.; Hashimoto, M.; Fukase, K.; Kusumoto, S.; Ishida, H.; Kiso, M.; Ogawa, T. Cell activation by monosaccharide lipid A analogues utilizing Toll-like receptor 4. Immunology 2003, 110, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Cighetti, R.; Ciaramelli, C.; Sestito, S.E.; Zanoni, I.; Kubik, Ł; Ardá-Freire, A.; Calabrese, V.; Granucci, F.; Jerala, R.; Martín-Santamaría, S.; et al. Modulation of CD14 and TLR4/MD-2 activities by a synthetic lipid A mimetic. ChemBioChem 2014, 15, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Perrin-Cocon, L.; Aublin-Gex, A.; Sestito, S.E.; Shirey, K.A.; Patel, M.C.; André, P.; Blanco, J.C.; Vogel, S.N.; Peri, F.; Lotteau, V. TLR4 antagonist FP7 inhibits LPS-induced cytokine production and glycolytic reprogramming in dendritic cells, and protects mice from lethal influenza infection. Sci. Rep. 2017, 7, 40791. [Google Scholar] [CrossRef] [PubMed]
- Ciaramelli, C.; Calabrese, V.; Sestito, S.E.; Pérez-Regidor, L.; Klett, J.; Oblak, A.; Jerala, R.; Piazza, M.; Martín-Santamaría, S.; Peri, F. Glycolipid-based TLR4 modulators and fluorescent probes: Rational design, synthesis and biological properties. Chem. Biol. Drug Des. 2016, 88, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Lavado, J.; Sestito, S.E.; Cighetti, R.; Aguilar Moncayo, E.M.; Oblak, A.; Lainšček, D.; Jiménez Blanco, J.L.; García Fernández, J.M.; Ortiz Mellet, C.; Jerala, R.; et al. Trehalose- and Glucose-Derived Glycoamphiphiles: Small-Molecule and Nanoparticle Toll-Like Receptor 4 (TLR4) Modulators. J. Med. Chem. 2014, 57, 9105–9123. [Google Scholar] [CrossRef] [PubMed]
- Fukase, Y.; Fujimoto, Y.; Adachi, Y.; Suda, Y.; Kusumoto, S.; Fukase, K. Synthesis of Rubrivivax gelatinosus lipid A and analogues for investigation of the structural basis for immunostimulating and inhibitory activities. Bull. Chem. Soc. Jpn. 2008, 81, 796–819. [Google Scholar] [CrossRef]
- Artner, D.; Oblak, A.; Ittig, S.; Garate, J.A.; Horvat, S.; Arrieumerlou, C.; Hofinger, A.; Oostenbrink, C.; Jerala, R.; Kosma, P.; et al. Conformationally constrained lipid a mimetics for exploration of structural basis of TLR4/MD-2 activation by lipopolysaccharide. ACS Chem. Biol. 2013, 8, 2423–2432. [Google Scholar] [CrossRef] [PubMed]
- Seydel, U.; Oikawa, M.; Fukase, K.; Kusumoto, S.; Brandenburg, K. Intrinsic conformation of lipid A is responsible for agonistic and antagonistic activity. Eur. J. Biochem. 2000, 267, 3032–3039. [Google Scholar] [CrossRef] [PubMed]
- Fort, M.M.; Mozaffarian, A.; Stover, A.G.; Correia, J.D.S.; Johnson, D.A.; Crane, R.T.; Ulevitch, R.J.; Persing, D.H.; Bielefeldt-Ohmann, H.; Probst, P.; et al. A Synthetic TLR4 Antagonist Has Anti-Inflammatory Effects in Two Murine Models of Inflammatory Bowel Disease. J. Immunol. 2005, 174, 6416–6423. [Google Scholar] [CrossRef] [PubMed]
- Billod, J.-M.; Lacetera, A.; Guzmán-Caldentey, J.; Martín-Santamaría, S. Computational Approaches to Toll-Like Receptor 4 Modulation. Molecules 2016, 21, 994. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, K.; Akashi, S.; Shimazu, R.; Yoshida, T.; Miyake, K.; Nishijima, M. Mouse Toll-like Receptor 4 MD-2 complex Mediates Lipopolysaccharide-mimetic Soìignal Transduction by Taxol. J. Biol. Chem. 2000, 275, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, K.; Gomi, K.; Kawai, Y.; Shiozaki, M.; Nishijima, M. Molecular basis for lipopolysaccharide mimetic action of TaxolTM and flavolipin. J. Endotoxin Res. 2003, 9, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.; Hayashi, T.; Mathewson, R.D.; Nour, A.; Hayashi, Y.; Yao, S.; Tawatao, R.I.; Crain, B.; Tsigelny, I.F.; Kouznetsova, V.L.; et al. Identification of Substituted Pyrimido[5,4-b]indoles as Selective Toll-Like Receptor 4 Ligands. J. Med. Chem. 2013, 56, 4206–4223. [Google Scholar] [CrossRef] [PubMed]
- Nour, A.; Hayashi, T.; Chan, M.; Yao, S.; Tawatao, R.I.; Crain, B.; Tsigelny, I.F.; Kouznetsova, V.L.; Ahmadiiveli, A.; Messer, K.; et al. Bioorganic & Medicinal Chemistry Letters Discovery of substituted 4-aminoquinazolines as selective Toll-like receptor 4 ligands. Bioorg. Med. Chem. Lett. 2014, 24, 4931–4938. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.; Chow, J.C.; Hawkins, L.D.; McGuinness, P.D.; Miyake, K.; Espevik, T.; Gusovsky, F.; Golenbock, D.T. A novel synthetic acyclic lipid A-like agonist activates cells via the lipopolysaccharide/toll-like receptor 4 signaling pathway. J. Biol. Chem. 2001, 276, 1873–1880. [Google Scholar] [CrossRef] [PubMed]
- Morin, M.D.; Wang, Y.; Jones, B.T.; Su, L.; Surakattula, M.M.; Berger, M.; Huang, H.; Beutler, E.K.; Zhang, H.; Beutler, B.; et al. Discovery and Structure—Activity Relationships of the Neoseptins: A New Class of Toll-like Receptor-4(TLR4) Agonists. J. Med. Chem. 2016, 59, 4812–4830. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Su, L.; Morin, M.D.; Jones, B.T.; Whitby, L.R.; Surakattula, M.M.R.P.; Huang, H.; Shi, H.; Choi, J.H.; Wang, K.-W.; et al. TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS. Proc. Natl. Acad. Sci. USA 2016, 113, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Neve, J.E.; Wijesekera, H.P.; Du, S.; Jenkins, I.D.; Ripper, J.A.; Teague, S.J.; Campitelli, M.; Garavelas, A.; Nikolapoulos, G.; Le, P.V.; et al. Euodenine A: A Small-Molecule Agonist of Human TLR4. J. Med. Chem. 2014, 57, 1252–1275. [Google Scholar] [CrossRef] [PubMed]
- Peri, F.; Calabrese, V. Toll-like receptor 4 (TLR4) modulation by synthetic and natural compounds: An update. J. Med. Chem. 2014, 57, 3612–3622. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Gomes, F.P.; Deora, V.; Gregory, K.; Vithanage, T.; Nassar, Z.D.; Cabot, P.J.; Sturgess, D.; Shaw, P.N.; Parat, M.-O. Activation of mu-opioid receptor and Toll-like receptor 4 by plasma from morphine-treated mice. Brain Behav. Immun. 2017, 61, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, A.; Rajoria, S.; George, A.L.; Mittelman, A.; Suriano, R.; Tiwari, R.K. Synthetic Toll Like Receptor-4 (TLR-4) Agonist Peptides as a Novel Class of Adjuvants. PLoS ONE 2012, 7, e30839. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Fukase, K.; Miyake, K.; Shimizu, T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proc. Natl. Acad. Sci. USA 2012, 109, 7421–7426. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Phillips, R.L.; Zhang, D.; Teghanemt, A.; Weiss, J.P.; Gioannini, T.L. NMR studies of hexaacylated endotoxin bound to wild-type and F126A mutant MD-2 and MD-2·TLR4 ectodomain complexes. J. Biol. Chem. 2012, 287, 16346–16355. [Google Scholar] [CrossRef] [PubMed]
- Oblak, A.; Jerala, R. The molecular mechanism of species-specific recognition of lipopolysaccharides by the MD-2/TLR4 receptor complex. Mol. Immunol. 2015, 63, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Hong, M.; Han, G.W.; Wilson, I. A Crystal structure of soluble MD-1 and its interaction with lipid IVa. Proc. Natl. Acad. Sci. USA 2010, 107, 10990–10995. [Google Scholar] [CrossRef] [PubMed]
- Scior, T.; Lozano-Aponte, J.; Figueroa-Vazquez, V.; Yunes-Rojas, J.A.; Zähringer, U.; Alexander, C. Three-Dimensional Mapping of Differential Amino Acids of Human, Murine, Canine and Equine Tlr4/Md-2 Receptor Complexes Conferring Endotoxic Activation By Lipid a, Antagonism By Eritoran and Species-Dependent Activities of Lipid Iva in the Mammalian Lps Se. Comput. Struct. Biotechnol. J. 2013, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Park, B.S.; Kim, J.I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal Structure of the TLR4-MD-2 Complex with Bound Endotoxin Antagonist Eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Holst, O.; Di, L.F.; Callaghan, M.; Nurisso, A.; D’Errico, G.; Zamyatina, A.; Peri, F.; Berisio, R.; Jerala, R.; et al. Chemistry of Lipid A: At the Heart of Innate Immunity. Chemistry 2015, 21, 500–519. [Google Scholar] [CrossRef] [PubMed]
- Peri, F.; Piazza, M. Therapeutic targeting of innate immunity with Toll-like receptor 4 (TLR4) antagonists. Biotechnol. Adv. 2012, 30, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; Parker, A.E.; O’Neill, L.A.J. Targeting Toll-like receptors: Emerging therapeutics? Nat. Rev. Drug Discov. 2010, 9, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Gay, N.J.; Gangloff, M. Structure and Function of Toll Receptors and Their Ligands. Annu. Rev. Biochem. 2007, 76, 141–165. [Google Scholar] [CrossRef] [PubMed]
- Tom, J.K.; Dotsey, E.Y.; Wong, H.Y.; Stutts, L.; Moore, T.; Davies, D.H.; Felgner, P.L.; Esser-Kahn, A.P. Modulation of Innate Immune Responses via Covalently Linked TLR Agonists. ACS Cent. Sci. 2015, 1, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.X.; Azad, M.A.K.; Yuriev, E.; Baker, M.A.; Nation, R.L.; Li, J.; Cooper, M.A.; Velkov, T. Molecular Characterization of Lipopolysaccharide Binding to Human α-1-Acid Glycoprotein. J. Lipids 2012, 2012, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Knirel, Y.A.; Valvano, M.A. Bacterial Lipopolysaccharides; Springer: Berlin, Germany, 2011. [Google Scholar]
- Bennett-guerrero, E.; Mcintosh, T.J.; Robin, G.; Snyder, D.S.; Gibbs, R.J.; Michael, G.; Poxton, I.R.; McIntosh, T.J.; Barclay, G.R.; Mythen, M.G.; et al. Preparation and Preclinical Evaluation of a Novel Liposomal Complete-Core Lipopolysaccharide Vaccine. Infect. Immun. 2000, 68, 6202–6208. [Google Scholar] [CrossRef] [PubMed]
- Di Padova, F.E.; Brade, H.; Barclay, G.R.; Poxton, L.R.; Liehl, E.; Schuetze, E.; Kocher, H.P.; Ramsay, G.; Schreier, M.H.; Mcclelland, D.B.L.; et al. A Broadly Cross-Protective Monoclonal Antibody Binding to Escherichia coli and Salmonella Lipopolysaccharides. Infect. Immun. 1993, 61, 3863–3872. [Google Scholar] [PubMed]
- Luk, J.M.; Lind, S.M.; Tsang, R.S.; Lindberg, A.A. Epitope mapping of four monoclonal antibodies recognizing the hexose core domain of Salmonella lipopolysaccharide. J. Biol. Chem. 1991, 266, 23215–23225. [Google Scholar] [PubMed]
- Galanos, C.; Luderitz, O.; Rietschel, E.T.; Westphal, O.; Brade, H.; Brade, L.; Freudenberg, M.; Schade, U.; Imoto, M.; Yoshimura, H.; et al. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur. J. Biochem. 1985, 148, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Erridge, C.; Bennett-guerrero, E.; Poxton, I.R. Structure and function of lipopolysaccharides. Microbes Infect. 2002, 4, 837–851. [Google Scholar] [CrossRef]
- Muroi, M.; Tanamoto, K. The polysaccharide portion plays an indispensable role in Salmonella LPS-induced activation of NF-kB through human TLR-4. Infect. Immun. 2002, 70, 6043–6047. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.V.; Wolfert, M.A.; Santhanam, B.; Moore, J.N.; Boons, G.J. Synthesis and biological evaluation of Rhizobium sin-1 lipid A derivatives. J. Am. Chem. Soc. 2003, 125, 6103–6112. [Google Scholar] [CrossRef] [PubMed]
- Zughaier, S.M.; Tzeng, Y.; Zimmer, S.M.; Carlson, R.W.; Stephens, D.S.; Datta, A. Neisseria meningitidis Lipooligosaccharide Structure-Dependent Activation of the Pathway Neisseria meningitidis Lipooligosaccharide Structure-Dependent Activation of the Macrophage CD14/Toll-Like Receptor 4 Pathway. Infect. Immun. 2004, 72, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Zughaier, S.; Agrawal, S.; Stephens, D.S.; Pulendran, B. Hexa-acylation and KDO2-glycosylation determine the specific immunostimulatory activity of Neisseria meningitidis lipid a for human monocyte derived dendritic cells. Vaccine 2006, 24, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Vasan, M.; Wolfert, M.A.; Boons, G.-J. Agonistic and antagonistic properties of a Rhizobium sin-1 lipid A modified by an ether-linked lipid. Org. Biomol. Chem. 2007, 5, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wolfert, M.A.; Boons, G.J. The influence of the long chain fatty acid on the antagonistic activities of Rhizobium sin-1 lipid A. Bioorg. Med. Chem. 2007, 15, 4800–4812. [Google Scholar] [CrossRef] [PubMed]
- Zughaier, S.M.; Zimmer, S.M.; Datta, A.; Carlson, R.W.; Stephens, D.S. Differential Induction of the Toll-Like Receptor 4—MyD88-Dependent and -Independent Signaling Pathways by Endotoxins. Infect. Immun. 2005, 73, 2940–2950. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Garrett, T.A.; Reynolds, C.M.; Shaw, W.A.; Moore, J.D.; Smith, D.C.; Ribeiro, A.; Murphy, R.C.; Ulevitch, R.J.; Fearns, C.; et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 2006, 47, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.; Means, T.K.; Heine, H.; Yoshimura, A.; Kusumoto, S.; Fukase, K.; Fenton, M.J.; Oikawa, M.; Qureshi, N.; Monks, B.; et al. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J. Clin. Investig. 2000, 105, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide Endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Brabetz, W.; Müller-Loennies, S.; Holst, O.; Brade, H. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem. 1997, 247, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.C.; Raetz, C.R.H.; Reynolds, C.M.; Barkley, R.M. Mass Spectrometry Advances in Lipidomica: Collision Induced Decomposition of Kdo2-Lipid A. Prostaglandins Other Lipid Mediat. 2005, 77, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Rosner, M.R.; Tang, J.; Barzilay, I.; Khorana, H.G. Structure of the Lipopolysaccharide Mutant from an Escherichia coli Heptose-less Mutant. J. Biol. Chem. 1979, 254, 5906–5917. [Google Scholar] [PubMed]
- Zhou, Z.; Lin, S.; Cotter, R.J.; Raetz, C.R.H. Lipid A Modifications Characteristic of Salmonella Typhimurium Are Induced by NH 4VO3 in Escherichia Coli K1. Biochemistry 1999, 274, 18503–18514. [Google Scholar]
- Zhou, Z.; Ribeiro, A.A.; Lin, S.; Cotter, R.J.; Miller, S.I.; Raetz, C.R.H. Lipid A modifications in polymyxin-resistant Salmonella typhimurium. J. Biol. Chem. 2001, 276, 43111–43121. [Google Scholar] [CrossRef]
- Zhang, Y.; Gaekwad, J.; Wolfert, M.A.; Boons, G.J. Modulation of Innate Immune Responses with Synthetic Lipid A Derivatives. J. Am. Chem. Soc. 2007, 129, 5200–5216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gaekwad, J.; Wolfert, M.A.; Boons, G.J. Innate immune responses of synthetic lipid a derivatives of Neisseria meningitidis. Chemistry 2008, 14, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Weiss, D.S.; Newton, K.; McBride, J.; O’Rourke, K.; Roose-Girma, M.; Lee, W.P.; Weinrauch, Y.; Monack, D.M.; Dixit, V.M. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006, 440, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.I.; Yamasaki, R.; Mandrell, R.E.; Griffiss, J.M. Ability of gonococcal and meningococcal lipooligosaccharides to clot Limulus amebocyte lysate. Infect. Immun. 1992, 60, 762–767. [Google Scholar] [PubMed]
- Schromm, A.B.; Brandenburg, K.; Loppnow, H.; Zähringer, U.; Rietschel, E.T.; Carroll, S.F.; Koch, M.H.; Kusumoto, S.; Seydel, U. The charge of endotoxin molecules influences their conformation and IL-6-inducing capacity. J. Immunol. 1998, 161, 5464–5471. [Google Scholar] [PubMed]
- Vinh, T.; Adler, B.; Faine, S. Ultrastructure and chemical composition of lipopolysaccharide extracted from Leptospira interrogans serovar copenhageni. J. Gen. Microbiol. 1986, 132, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, E.; Perry, M.B.; Conlan, J.W. Structural analysis of Francisella tularensis lipopolysaccharide. Eur. J. Biochem. 2002, 269, 6112–6118. [Google Scholar] [CrossRef] [PubMed]
- Petzold, M.; Thürmer, A.; Menzel, S.; Mouton, J.W.; Heuner, K.; Lück, C. A structural comparison of lipopolysaccharide biosynthesis loci of Legionella pneumophila serogroup 1 strains. BMC Microbiol. 2013, 13, 198. [Google Scholar] [CrossRef] [PubMed]
- Russa, R.; Urbanik-Sypniewska, T.; Choma, A.; Mayer, H. Identification of 3-deoxy-lyxo-2-heptulosaric acid in the core region of lipopolysaccharides from Rhizobiaceae. FEMS Microbiol. Lett. 1991, 68, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Frecer, V.; Ho, B.; Ding, J.L. Interpretation of biological activity data of bacterial endotoxins by simple molecular models of mechanism of action. Eur. J. Biochem. 2000, 267, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Use of Receptor-Based Drug Design and Applications in the Study of Finding Antagonists for MD-2/TLR4, GLP and CXCR4. Ph.D. Thesis, Faculty of the James T. Laney School of Graduate Studies of Emory University, Atlanta, GA, USA, 2012. [Google Scholar]
- Henderson, J.C.; O’Brien, J.P.; Brodbelt, J.S.; Trent, M.S. Isolation and chemical characterization of lipid A from gram-negative bacteria. J. Vis. Exp. 2013, 966, e50623. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cochet, F.; Peri, F. The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling. Int. J. Mol. Sci. 2017, 18, 2318. https://doi.org/10.3390/ijms18112318
Cochet F, Peri F. The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling. International Journal of Molecular Sciences. 2017; 18(11):2318. https://doi.org/10.3390/ijms18112318
Chicago/Turabian StyleCochet, Florent, and Francesco Peri. 2017. "The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling" International Journal of Molecular Sciences 18, no. 11: 2318. https://doi.org/10.3390/ijms18112318
APA StyleCochet, F., & Peri, F. (2017). The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling. International Journal of Molecular Sciences, 18(11), 2318. https://doi.org/10.3390/ijms18112318




