Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses
Abstract
1. Introduction
2. Plant Defense-Related Proteins
2.1. Plant Basal Resistance
2.2. Plant Pathogenesis-Related Proteins and Antimicrobial Peptides
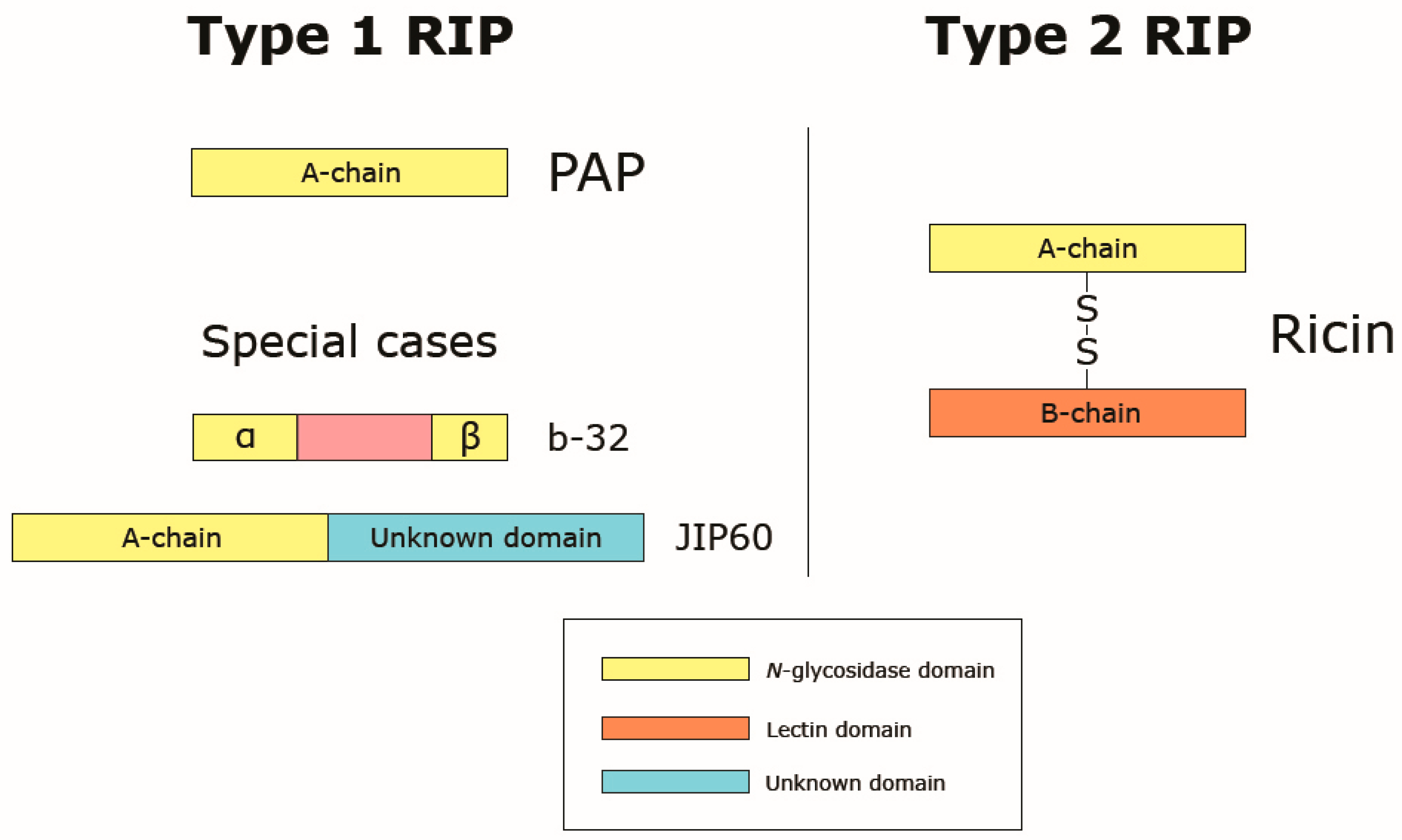
3. Ribosome-Inactivating Proteins
3.1. Classification of RIPs
3.2. Antiviral Activity of RIPs
4. RNA-Binding Proteins
4.1. Structure of RBPs
4.2. Plant RBPs—In Vitro and In Vivo Activity against Viruses
5. Proteins Involved in Innate Antiviral Defense via RNA Silencing Pathway
5.1. Dicer-Like Ribonucleases
5.2. RNA-Dependent RNA Polymerases
5.3. Argonautes
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PR | pathogenesis-related proteins |
| RBP | RNA-binding protein |
| DCL | Dicer-like protein |
| RDR | RNA-dependent RNA polymerase |
| sRNA | small RNA |
| AGO | Argonaute protein |
| RISC | RNA-induced silencing complex |
| RIP | ribosome-inactivating protein |
| AMP | antimicrobial peptide |
| BMV | Brome mosaic virus |
| CaMV | Cauliflower mosaic virus |
| PLRV | Potato leafroll virus |
| RDV | Rice dwarf virus |
| RSV | Rice stripe virus |
| TBSV | Tomato bushy stunt virus |
| TCP | Tomato crinkle virus |
| ToRSV | Tomato ringspot virus |
| TRV | Tobacco rattle virus |
| R | resistance protein |
| Avr | avirulence protein |
| LRR | leucine-rich repeat |
| PRR | pattern recognition receptor |
| PAMP | pathogen-associated molecular pattern |
| HR | hypersensitive response |
| SAR | systemic acquired resistance |
| SA | salicylic acid |
| PAP | pokeweed antiviral protein |
| HSV | Human simplex virus |
| TMV | Tobacco mosaic virus |
| HIV | Human immunodeficiency virus |
| PVY | Potato virus Y |
| PVX | Potato virus X |
| CMV | Cucumber mosaic virus |
| TuMV | Turnip mosaic virus |
| RBD | RNA-binding domain |
| RRM | RNA-recognition motif |
| KH | K homology domain |
| ZnF | zinc finger domain |
| PUF | Pumilio/FBF domain |
| PAZ | Piwi/Argonaute/Zwille domain |
| APUM5 | Arabidopsis Pumilio RNA-binding protein 5 |
| PHD | Pumilio homology domain |
| AtGRP7 | Arabidopsis thaliana glycine-rich protein 7 |
| DRB4 | dsRNA-binding protein 4 |
| TYMV | Turnip yellow mosaic virus |
| TCV | Turnip crinkle virus |
| TLS | tRNA-like structure |
| CaLCuV | Cabbage leaf curl virus |
| BCTV | Beet curly top virus |
| ToMV | Tomato mosaic virus |
| siRNA | small interfering RNA |
| miRNA | microRNA |
| rasiRNA | repeat-associated siRNA |
References
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.A.; Venkatesh, J.; Upadhyaya, C.P.; Nookaraju, A.; Pandey, S.K.; Park, S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 2012, 78, 51–65. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Gen. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Chen, H.; Curtis, C.; Fu, Z.Q. Go in for the kill. Virulence 2014, 5, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Cohn, J.; Sessa, G.; Martin, G.B. Innate immunity in plants. Curr. Opin. Immunol. 2001, 13, 55–62. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Nurnberger, T.; Brunner, F. Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 2002, 5, 318–324. [Google Scholar] [CrossRef]
- Maleck, K.; Dietrich, R.A. Defense on multiple fronts: How do plants cope with diverse enemies? Trends Plant Sci. 1999, 4, 215–219. [Google Scholar] [CrossRef]
- Vlot, A.C.; Klessig, D.F.; Park, S.W. Systemic acquired resistance: The elusive signal(s). Curr. Opin. Plant Biol. 2008, 11, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Lamb, C. Systemic immunity. Curr. Opin. Plant Biol. 2006, 9, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, A.S.; Pooggin, M.M. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses 2012, 4, 2578–2597. [Google Scholar] [CrossRef] [PubMed]
- Zorzatto, C.; Machado, J.P.; Lopes, K.V.; Nascimento, K.J.; Pereira, W.A.; Brustolini, O.J.; Reis, P.A.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.; Lopes, K.V.; Apfata, J.A.; Fontes, E.P. NSP-interacting kinase, NIK: A transducer of plant defence signalling. J. Exp. Bot. 2010, 61, 3839–3845. [Google Scholar] [CrossRef] [PubMed]
- Sels, J.; Mathys, J.; de Coninck, B.M.; Cammue, B.P.; de Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wu, J.; Fan, S.; Li, W.; Dong, L.; Cheng, Q.; Xu, P.; Zhang, S. Isolation and Characterization of a Novel Pathogenesis-Related Protein Gene (GmPRP) with Induced Expression in Soybean (Glycine max) during Infection with Phytophthora sojae. PLoS ONE 2015, 10, e0129932. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Agarwal, P.; Agarwal, P.K. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol. Biol. Rep. 2014, 41, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Edreva, A. Pathogenesis-related proteins: Research progress in the last 15 years. Gen. Appl. Plant Physiol. 2005, 31, 105–124. [Google Scholar]
- Liu, J.-J.; Ekramoddoullah, A.K.M. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 2006, 68, 3–13. [Google Scholar] [CrossRef]
- Shan, Y.; Zheng, Y.; Guan, F.; Zhou, J.; Zhao, H.; Xia, B.; Feng, X. Purification and characterization of a novel anti-HSV-2 protein with antiproliferative and peroxidase activities from Stellaria media. Acta Biochim. Biophys. Sin. 2013, 45, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Kim, K.J.; Shin, R.; Park, J.M.; Shin, Y.C.; Paek, K.H. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. Cell Mol. Biol. 2004, 37, 186–198. [Google Scholar] [CrossRef]
- Sindelarova, M.; Sindelar, L. Isolation of Pathogenesis-Related Proteins from TMV-Infected Tobacco and their Influence on Infectivity of TMV. Plant Prot. Sci. 2005, 41, 52–57. [Google Scholar]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Gozdzicka-Jozefiak, A. Plant antimicrobial peptides. Folia Microbiol. 2014, 59, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.B.; Cheung, R.C.; Wong, J.H.; Ye, X.J. Antimicrobial activity of defensins and defensin-like peptides with special emphasis on those from fungi and invertebrate animals. Curr. Protein Pept. Sci. 2013, 14, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Rivas, L.; Luque-Ortega, J.R.; Fernández-Reyes, M.; Andreu, D. Membrane-active peptides as anti-infectious agents. J. Appl. Biomed. 2010, 8, 159–167. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Peptide antimicrobials: Cell wall as a bacterial target. Ann. N. Y. Acad. Sci. 2013, 1277, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Huang, Y.H.; Rosengren, K.J.; Franquelim, H.G.; Carvalho, F.A.; Johnson, A.; Sonza, S.; Tachedjian, G.; Castanho, M.A.; Daly, N.L.; et al. Decoding the membrane activity of the cyclotide kalata B1: The importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti-HIV activities. J. Biol. Chem. 2011, 286, 24231–24241. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ng, T.B. Limenin, a defensin-like peptide with multiple exploitable activities from shelf beans. J. Pept. Sci. 2006, 12, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Dzianott, A.; Sztuba-Solinska, J.; Bujarski, J.J. Mutations in the antiviral RNAi defense pathway modify Brome mosaic virus RNA recombinant profiles. Mol. Plant Microbe Interact. 2012, 25, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.-B.; Godon, C.; Mourrain, P.; Béclin, C.; Boutet, S.; Feuerbach, F.; Proux, F.; Vaucheret, H. Fertile Hypomorphic ARGONAUTE (ago1) Mutants Impaired in Post-Transcriptional Gene Silencing and Virus Resistance. Plant Cell 2002, 14, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.B.; Jovel, J.; Udomporn, P.; Wang, Y.; Wu, Q.; Li, W.X.; Gasciolli, V.; Vaucheret, H.; Ding, S.W. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 2011, 23, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Ye, X.; Morris, T.J. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 14732–14737. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Correction. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 2015, 27, 944–945. [Google Scholar] [PubMed]
- Ghoshal, B.; Sanfacon, H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology 2014, 456–457, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.J.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstadt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Singh, J.; Li, D.; Qu, F. Temperature-dependent survival of Turnip crinkle virus-infected arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. J. Virol. 2012, 86, 6847–6854. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.S.; Perry, K.L.; Moffett, P. ARGONAUTE2 Mediates RNA-Silencing Antiviral Defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Nicole, M.C.; Meteignier, L.V.; Hong, N.; Wang, G.; Moffett, P. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J. Exp. Bot. 2015, 66, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Gilbert, K.B.; Montgomery, T.A.; Nguyen, T.; Cuperus, J.T.; Carrington, J.C. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 2012, 24, 3613–3629. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Carbonell, A.; Hoyer, J.S.; Fahlgren, N.; Gilbert, K.B.; Takeda, A.; Giampetruzzi, A.; Garcia Ruiz, M.T.; McGinn, M.G.; Lowery, N.; et al. Roles and programming of Arabidopsis Argonaute proteins during Turnip mosaic virus infection. PLoS Pathog. 2015, 11, e1004755. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, H.B.; Alvarado, V.Y.; Vega-Arreguin, J.C.; Ciomperlik, J.; Odokonyero, D.; Brosseau, C.; Jaubert, M.; Zamora, A.; Moffett, P. Identification of an ARGONAUTE for Antiviral RNA Silencing in Nicotiana benthamiana. Plant Physiol. 2011, 156, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef] [PubMed]
- Raja, P.; Jackel, J.N.; Li, S.; Heard, I.M.; Bisaro, D.M. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 2014, 88, 2611–2622. [Google Scholar] [CrossRef] [PubMed]
- Hamera, S.; Song, X.; Su, L.; Chen, X.; Fang, R. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. Cell Mol. Biol. 2012, 69, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Hamera, S.; Yan, Y.; Song, X.; Chaudhary, S.U.; Murtaza, I.; Su, L.; Tariq, M.; Chen, X.; Fang, R. Expression of Cucumber mosaic virus suppressor 2b alters FWA methylation and its siRNA accumulation in Arabidopsis thaliana. Biol. Open 2016, 5, 1727–1734. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Zamora, A.; Azhar, M.T.; Sacco, M.A.; Lambert, L.H.; Moffett, P. Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. Cell Mol. Biol. 2009, 58, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Moffett, P. Functional and Genetic Analysis Identify a Role for Arabidopsis ARGONAUTE5 in Antiviral RNA Silencing. Plant Cell 2015, 27, 1742–1754. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, K.; Ishikawa, M. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology 2008, 380, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Maruyama, K.; Sun, L.; Kondo, H.; Tamada, T.; Suzuki, N. Different Dicer-like protein components required for intracellular and systemic antiviral silencing in Arabidopsis thaliana. Plant Signal. Behav. 2015, 10, e1039214. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, A.; Kanaya, A.; Egami, M.; Nakazawa, Y.; Hiraguri, A.; Moriyama, H.; Fukuhara, T. Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 2011, 17, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Jakubiec, A.; Yang, S.W.; Chua, N.H. Arabidopsis DRB4 protein in antiviral defense against Turnip yellow mosaic virus infection. Plant J. Cell Mol. Biol. 2012, 69, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Jeong, R.-D.; Lim, G.-H.; Yu, K.; Wang, C.; Chandra-Shekara, A.C.; Navarre, D.; Klessig, D.F.; Kachroo, A.; Kachroo, P. Double-Stranded RNA-Binding Protein 4 Is Required for Resistance Signaling against Viral and Bacterial Pathogens. Cell Rep. 2013, 4, 1168–1184. [Google Scholar] [CrossRef] [PubMed]
- Bally, J.; Nakasugi, K.; Jia, F.; Jung, H.; Ho, S.Y.; Wong, M.; Paul, C.M.; Naim, F.; Wood, C.C.; Crowhurst, R.N.; et al. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 2015, 1, 15165. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Garcia, S.; Voinnet, O. Nonsense-Mediated Decay Serves as a General Viral Restriction Mechanism in Plants. Cell Host Microbe 2014, 16, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Iwasaki, S.; Watanabe, T.; Utsumi, M.; Watanabe, Y. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 2008, 49, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Paek, K.H. Plant RNA binding proteins for control of RNA virus infection. Front. Physiol. 2013, 4, 397. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kim, J.S.; Yoo, S.J.; Kang, E.Y.; Han, S.H.; Yang, K.Y.; Kim, Y.C.; McSpadden Gardener, B.; Kang, H. Different roles of glycine-rich RNA-binding protein7 in plant defense against Pectobacterium carotovorum, Botrytis cinerea, and tobacco mosaic viruses. Plant Physiol. Biochem. 2012, 60, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Lodge, J.K.; Kaniewski, W.K.; Tumer, N.E. Broad-spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc. Natl. Acad. Sci. USA 1993, 90, 7089–7093. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zoubenko, O.; Tumer, N.E. Reduced toxicity and broad spectrum resistance to viral and fungal infection in transgenic plants expressing pokeweed antiviral protein II. Plant Mol. Biol. 1998, 38, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Tumer, N.E.; Hwang, D.J.; Bonness, M. C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci. USA 1997, 94, 3866–3871. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.H.; Song, S.K.; Choi, K.W.; Lee, J.S. Expression of a cDNA encoding Phytolacca insularis antiviral protein confers virus resistance on transgenic potato plants. Mol. Cells 1997, 7, 807–815. [Google Scholar] [PubMed]
- Rakhshandehroo, F.; Takeshita, M.; Squires, J.; Palukaitis, P. The influence of RNA-dependent RNA polymerase 1 on potato virus Y infection and on other antiviral response genes. Mol. Plant Microbe Interact. 2009, 22, 1312–1318. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.-H.; Wong, Y.-S.; Wang, B.; Wong, R.N.S.; Yeung, H.-W.; Shaw, P.-C. Use of trichosanthin to reduce infection by turnip mosaic virus. Plant Sci. 1996, 114, 111–117. [Google Scholar] [CrossRef]
- Gustafson, K.R.; Sowder, R.C.; Henderson, L.E.; Parsons, I.C.; Kashman, Y.; Cardellina, J.H.; McMahon, J.B.; Buckheit, R.W.; Pannell, L.K.; Boyd, M.R. Circulins A and B. Novel human immunodeficiency virus (HIV)-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 1994, 116, 9337–9338. [Google Scholar] [CrossRef]
- Daly, N.L.; Clark, R.J.; Plan, M.R.; Craik, D.J. Kalata B8, a novel antiviral circular protein, exhibits conformational flexibility in the cystine knot motif. Biochem. J. 2006, 393, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ng, T.B. Lunatusin, a trypsin-stable antimicrobial peptide from lima beans (Phaseolus lunatus L.). Peptides 2005, 26, 2086–2092. [Google Scholar] [CrossRef] [PubMed]
- Ngai, P.H.K.; Ng, T.B. Phaseococcin, an antifungal protein with antiproliferative and anti-HIV-1 reverse transcriptase activities from small scarlet runner beans. Biochem. Cell Biol. 2005, 83, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ng, T.B. Sesquin, a potent defensin-like antimicrobial peptide from ground beans with inhibitory activities toward tumor cells and HIV-1 reverse transcriptase. Peptides 2005, 26, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Wong, J.H.; Ng, T.B. A defensin with highly potent antipathogenic activities from the seeds of purple pole bean. Biosci. Rep. 2010, 30, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Hao, Q.; Van Damme, E.J. Ribosome-inactivating proteins from plants: More than RNA N-glycosidases? FASEB J. 2001, 15, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, N.L.; Yadav, O.P.; Lodha, M.L. Ribonuclease, deoxyribonuclease, and antiviral activity of Escherichia coli-expressed Bougainvillea xbuttiana antiviral protein 1. Biochem. Biokhimiia 2008, 73, 273–277. [Google Scholar] [CrossRef]
- He, W.J.; Liu, W.Y. Both N- and C-terminal regions are essential for cinnamomin A-chain to deadenylate ribosomal RNA and supercoiled double-stranded DNA. Biochem. J. 2004, 377, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Citores, L.; Di Maro, A.; Ferreras, J.M. Biological activities of the antiviral protein BE27 from sugar beet (Beta vulgaris L.). Planta 2015, 241, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Knight, B. Ricin—A potent homicidal poison. Br. Med. J. 1979, 1, 350–351. [Google Scholar] [PubMed]
- Nielsen, K.; Boston, R.S. Ribosome-Inactivating Proteins: A Plant Perspective. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 785–816. [Google Scholar] [CrossRef] [PubMed]
- Olsnes, S.; Pihl, A. Isolation and properties of abrin: A toxic protein inhibiting protein synthesis. Evidence for different biological functions of its two constituent-peptide chains. Eur. J. Biochem. FEBS 1973, 35, 179–185. [Google Scholar] [CrossRef]
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Polito, L. Ribosome-Inactivating Proteins from Plants: A Historical Overview. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Irvin, J.D. Pokeweed antiviral protein. Pharmacol. Ther. 1983, 21, 371–387. [Google Scholar] [CrossRef]
- Chuethong, J.; Oda, K.; Sakurai, H.; Saiki, I.; Leelamanit, W. Cochinin B, a novel ribosome-inactivating protein from the seeds of Momordica cochinchinensis. Biol. Pharm. Bull. 2007, 30, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Lee-Huang, S.; Huang, P.L.; Nara, P.L.; Chen, H.C.; Kung, H.F.; Huang, P.; Huang, H.I.; Huang, P.L. MAP 30: A new inhibitor of HIV-1 infection and replication. FEBS Lett. 1990, 272, 12–18. [Google Scholar] [CrossRef]
- Lee-Huang, S.; Kung, H.F.; Huang, P.L.; Huang, P.L.; Li, B.Q.; Huang, P.; Huang, H.I.; Chen, H.C. A new class of anti-HIV agents: GAP31, DAPs 30 and 32. FEBS Lett. 1991, 291, 139–144. [Google Scholar] [CrossRef]
- Stirpe, F.; Gasperi-Campani, A.; Barbieri, L.; Falasca, A.; Abbondanza, A.; Stevens, W.A. Ribosome-inactivating proteins from the seeds of Saponaria officinalis L. (soapwort), of Agrostemma githago L. (corn cockle) and of Asparagus officinalis L. (asparagus), and from the latex of Hura crepitans L. (sandbox tree). Biochem. J. 1983, 216, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Girbes, T.; de Torre, C.; Iglesias, R.; Miguel Ferreras, J.; Mendez, E. RIP for viruses. Nature 1996, 379, 777–778. [Google Scholar] [CrossRef]
- Iglesias, R.; Perez, Y.; Citores, L.; Ferreras, J.M.; Mendez, E.; Girbes, T. Elicitor-dependent expression of the ribosome-inactivating protein beetin is developmentally regulated. J. Exp. Bot. 2008, 59, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, S.; Mollenhauer, B.; Reinbothe, C. JIPs and RIPs: The regulation of plant gene expression by jasmonates in response to environmental cues and pathogens. Plant Cell 1994, 6, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Rippmann, J.F.; Michalowski, C.B.; Nelson, D.E.; Bohnert, H.J. Induction of a ribosome-inactivating protein upon environmental stress. Plant Mol. Biol. 1997, 35, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Song, S.K.; Choi, Y.; Moon, Y.H.; Kim, S.G.; Choi, Y.D.; Lee, J.S. Systemic induction of a Phytolacca insularis antiviral protein gene by mechanical wounding, jasmonic acid, and abscisic acid. Plant Mol. Biol. 2000, 43, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Tartarini, A.; Pittaluga, E.; Marcozzi, G.; Testone, G.; Rodrigues-Pousada, R.A.; Giannino, D.; Spano, L. Differential expression of saporin genes upon wounding, ABA treatment and leaf development. Physiol. Plant. 2010, 140, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Polito, L.; Bolognesi, A.; Ciani, M.; Pelosi, E.; Farini, V.; Jha, A.K.; Sharma, N.; Vivanco, J.M.; Chambery, A.; et al. Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta 2006, 1760, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F. Ribosome-inactivating proteins. Toxicon 2004, 44, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Girbes, T.; Ferreras, J.M.; Arias, F.J.; Stirpe, F. Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria. Mini Rev. Med. Chem. 2004, 4, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Puri, M.; Kaur, I.; Perugini, M.A.; Gupta, R.C. Ribosome-inactivating proteins: Current status and biomedical applications. Drug Discov. Today 2012, 17, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F.; Battelli, M.G. Ribosome-inactivating proteins: Progress and problems. Cell. Mol. Life Sci. 2006, 63, 1850–1866. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.M.; Hao, Q.; Chen, Y.; Barre, A.; Vandenbussche, F.; Desmyter, S.; Rougé, P.; Peumans, W.J. Ribosome-Inactivating Proteins: A Family of Plant Proteins That Do More Than Inactivate Ribosomes. Crit. Rev. Plant Sci. 2001, 20, 395–465. [Google Scholar] [CrossRef]
- Walsh, T.A.; Morgan, A.E.; Hey, T.D. Characterization and molecular cloning of a proenzyme form of a ribosome-inactivating protein from maize. Novel mechanism of proenzyme activation by proteolytic removal of a 2.8-kilodalton internal peptide segment. J. Biol. Chem. 1991, 266, 23422–23427. [Google Scholar] [PubMed]
- Chaudhry, B.; Muller-Uri, F.; Cameron-Mills, V.; Gough, S.; Simpson, D.; Skriver, K.; Mundy, J. The barley 60 kDa jasmonate-induced protein (JIP60) is a novel ribosome-inactivating protein. Plant J. Cell Mol. Biol. 1994, 6, 815–824. [Google Scholar] [CrossRef]
- De Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins 2010, 2, 2699–2737. [Google Scholar] [CrossRef] [PubMed]
- Wiley, R.G.; Lappi, D.A. Ribosome-Inactivating Proteins. In Molecular Neurosurgery with Targeted Toxins, 1st ed.; Humana Press: Totowa, NJ, USA, 2005; p. 9. [Google Scholar]
- Stevens, W.A.; Spurdon, C.; Onyon, L.J.; Stirpe, F. Effect of inhibitors of protein synthesis from plants on tobacco mosaic virus infection. Experientia 1981, 37, 257–259. [Google Scholar] [CrossRef]
- Vivanco, J.M.; Tumer, N.E. Translation Inhibition of Capped and Uncapped Viral RNAs Mediated by Ribosome-Inactivating Proteins. Phytopathology 2003, 93, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Foa-Tomasi, L.; Campadelli-Fiume, G.; Barbieri, L.; Stirpe, F. Effect of ribosome-inactivating proteins on virus-infected cells. Inhibition of virus multiplication and of protein synthesis. Arch. Virol. 1982, 71, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Aron, G.M.; Irvin, J.D. Inhibition of herpes simplex virus multiplication by the pokeweed antiviral protein. Antimicrob. Agents Chemother. 1980, 17, 1032–1033. [Google Scholar] [CrossRef] [PubMed]
- Kaur, I.; Puri, M.; Ahmed, Z.; Blanchet, F.P.; Mangeat, B.; Piguet, V. Inhibition of HIV-1 replication by balsamin, a ribosome inactivating protein of Momordica balsamina. PLoS ONE 2013, 8, e73780. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.D.; Bonos, S.; Guo, Z.; Meyer, W.A.; Day, P.R.; Belanger, F.C. Expression of pokeweed antiviral proteins in creeping bentgrass. Plant Cell Rep. 2003, 21, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Valbonesi, P.; Bonora, E.; Gorini, P.; Bolognesi, A.; Stirpe, F. Polynucleotide: Adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucleic Acids Res. 1997, 25, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kumon, K.; Sasaki, J.; Sejima, M.; Takeuchi, Y.; Hayashi, Y. Interactions between tobacco mosaic virus, pokeweed antiviral proteins, and tobacco cell wall. Phytopathology 1990, 80, 636–641. [Google Scholar] [CrossRef]
- Stirpe, F.; Barbieri, L.; Gorini, P.; Valbonesi, P.; Bolognesi, A.; Polito, L. Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 1996, 382, 309–312. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Lorkovic, Z.J.; Barta, A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002, 30, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, W.Y.; Kwak, K.J.; Oh, S.H.; Han, Y.S.; Kang, H. Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 2010, 33, 759–768. [Google Scholar] [PubMed]
- Owttrim, G.W. RNA helicases and abiotic stress. Nucleic Acids Res. 2006, 34, 3220–3230. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.P.; Barrette-Ng, I.H.; Simon, D.M.; Tam, M.W.; Ang, A.L.; Muench, D.G. The Puf family of RNA-binding proteins in plants: Phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Liu, J.; Tolia, N.H.; Schneiderman, J.; Smith, S.K.; Martienssen, R.A.; Hannon, G.J.; Joshua-Tor, L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003, 10, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Albà, M.M.; Pagès, M. Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 1998, 3, 15–21. [Google Scholar] [CrossRef]
- Ambrosone, A.; Costa, A.; Leone, A.; Grillo, S. Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci. 2012, 182, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Olivas, W.M. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip. Rev. RNA 2011, 2, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Kim, M.J.; Paek, K.H. Arabidopsis Pumilio protein APUM5 suppresses Cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Streitner, C.; Hennig, L.; Korneli, C.; Staiger, D. Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7. BMC Plant Biol. 2010, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Brown, R.L.; Damann, K.E.; Cleveland, T.E. PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 2010, 11, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Fristensky, B.; Kav, N.N. Constitutive expression of a PR10 protein enhances the germination of Brassica napus under saline conditions. Plant Cell Physiol. 2004, 45, 1320–1324. [Google Scholar] [CrossRef] [PubMed]
- Barton, D.A.; Roovers, E.F.; Gouil, Q.; da Fonseca, G.C.; Reis, R.S.; Jackson, C.; Overall, R.L.; Fusaro, A.F.; Waterhouse, P.M. Live Cell Imaging Reveals the Relocation of dsRNA Binding Proteins Upon Viral Infection. Mol. Plant Microbe Interact. 2017, 30, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Aliyari, R.; Ding, S.W. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 2009, 227, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Katiyar-Agarwal, S.; Jin, H. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 2010, 48, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, F.; Hohn, T. Biogenesis and Biological Activity of Secondary siRNAs in Plants. Scientifica 2013, 2013, 783253. [Google Scholar] [CrossRef] [PubMed]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of Dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef] [PubMed]
- Krautz-Peterson, G.; Skelly, P.J. Schistosoma mansoni: The dicer gene and its expression. Exp. Parasitol. 2008, 118, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Jin, L.; Huang, Y.; Zhang, F.; Kay, M.A. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr. Biol. 2012, 22, 1536–1542. [Google Scholar] [CrossRef] [PubMed]
- Qu, F. Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. Mol. Plant Microbe Interact. 2010, 23, 1248–1252. [Google Scholar] [CrossRef] [PubMed]
- Willmann, M.R.; Endres, M.W.; Cook, R.T.; Gregory, B.D. The Functions of RNA-Dependent RNA Polymerases in Arabidopsis. Arabidopsis Book Am. Soc. Plant Biol. 2011, 9, e0146. [Google Scholar] [CrossRef] [PubMed]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vazquez, F.; Blevins, T.; Farinelli, L.; et al. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 2012, 8, e1002941. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Qian, D.; Zheng, H.; Meng, L.Y.; Chen, J.; Le, W.J.; Zhou, T.; Zhou, Y.J.; Wei, C.H.; Li, Y. RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Res. 2012, 163, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Pandey, S.P. Evolution of structural and functional diversification among plant Argonautes. Plant Signal. Behav. 2015, 10, e1069455. [Google Scholar] [CrossRef] [PubMed]
- El-Shami, M.; Pontier, D.; Lahmy, S.; Braun, L.; Picart, C.; Vega, D.; Hakimi, M.A.; Jacobsen, S.E.; Cooke, R.; Lagrange, T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007, 21, 2539–2544. [Google Scholar] [CrossRef] [PubMed]
- Till, S.; Ladurner, A.G. RNA Pol IV plays catch with Argonaute 4. Cell 2007, 131, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Rigby, R.E.; Rehwinkel, J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 2015, 36, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Sanfacon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive Resistance to Plant Viruses: Potential Resistance Genes Beyond Translation Initiation Factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef] [PubMed]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- Nicaise, V.; German-Retana, S.; Sanjuan, R.; Dubrana, M.P.; Mazier, M.; Maisonneuve, B.; Candresse, T.; Caranta, C.; LeGall, O. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol. 2003, 132, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Gallois, J.L.; Lesage, M.L.; Caranta, C. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Gen. Genom. 2005, 274, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Gallois, J.L.; Moury, B.; Robaglia, C.; Palloix, A.; Caranta, C. Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 2006, 87, 2089–2098. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-S.; Wei, T.; Laliberté, J.-F.; Wang, A. A Host RNA Helicase-Like Protein, AtRH8, Interacts with the Potyviral Genome-Linked Protein, VPg, Associates with the Virus Accumulation Complex, and Is Essential for Infection. Plant Physiol. 2010, 152, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Desmyter, S.; Vandenbussche, F.; Hao, Q.; Proost, P.; Peumans, W.J.; Van Damme, E.J. Type-1 ribosome-inactivating protein from iris bulbs: A useful agronomic tool to engineer virus resistance? Plant Mol. Biol. 2003, 51, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Delaney, B.; Astwood, J.D.; Cunny, H.; Conn, R.E.; Herouet-Guicheney, C.; Macintosh, S.; Meyer, L.S.; Privalle, L.; Gao, Y.; Mattsson, J.; et al. Evaluation of protein safety in the context of agricultural biotechnology. Food Chem. Toxicol. 2008, 46 (Suppl. S2), S71–S97. [Google Scholar] [CrossRef] [PubMed]
- Hammond, B.; Kough, J.; Herouet-Guicheney, C.; Jez, J.M. Toxicological evaluation of proteins introduced into food crops. Crit. Rev. Toxicol. 2013, 43 (Suppl. S2), 25–42. [Google Scholar] [CrossRef] [PubMed]
- Uckun, F.M.; Bellomy, K.; O’Neill, K.; Messinger, Y.; Johnson, T.; Chen, C.L. Toxicity, biological activity, and pharmacokinetics of TXU (anti-CD7)-pokeweed antiviral protein in chimpanzees and adult patients infected with human immunodeficiency virus. J. Pharmacol. Exp. Ther. 1999, 291, 1301–1307. [Google Scholar] [PubMed]
- Kahn, J.O.; Gorelick, K.J.; Gatti, G.; Arri, C.J.; Lifson, J.D.; Gambertoglio, J.G.; Bostrom, A.; Williams, R. Safety, activity, and pharmacokinetics of GLQ223 in patients with AIDS and AIDS-related complex. Antimicrob. Agents Chemother. 1994, 38, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.C.; Ying, W.B.; Xie, H.; Zhang, Z.C.; Yang, Z.H.; Ling, L.Q. Trichosanthin-monoclonal antibody conjugate specifically cytotoxic to human hepatoma cells in vitro. Cancer Res. 1991, 51, 3353–3355. [Google Scholar] [PubMed]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007, 81, 6690–6699. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Matthew, L.; Smith, N.A.; Curtin, S.J.; Dedic-Hagan, J.; Ellacott, G.A.; Watson, J.M.; Wang, M.-B.; Brosnan, C.; Carroll, B.J.; et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006, 7, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
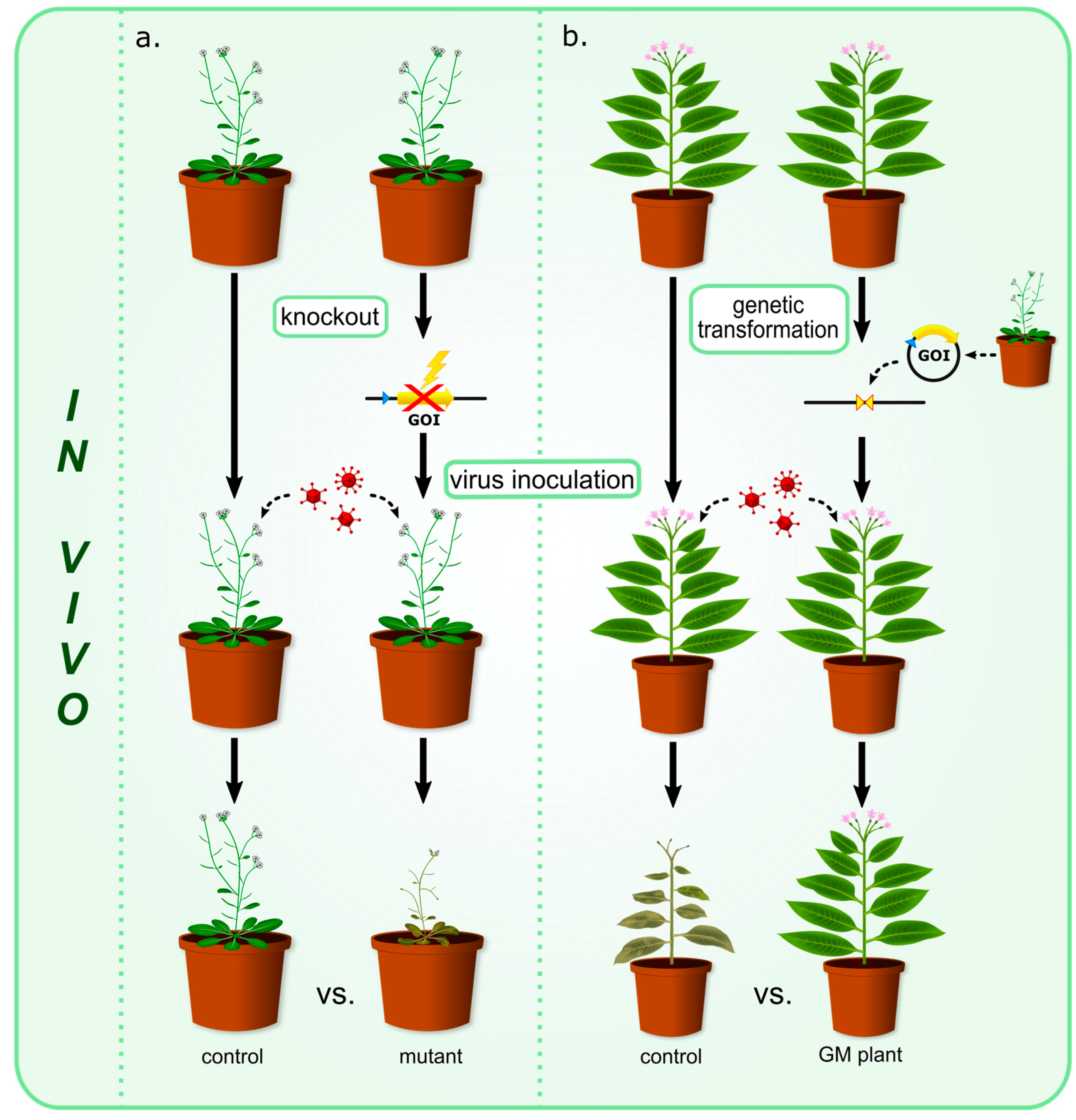

| Protein | Family | Source Plant | Target Virus | Reference |
|---|---|---|---|---|
| a. Proteins with antiviral properties observed in experiments involving mutant plants (Figure 1a) | ||||
| AGO1 | AGO | Arabidopsis thaliana | BMV | [30] |
| CMV | [31,32] | |||
| TCV | [33] | |||
| TuMV | [34] | |||
| Nicotiana benthamiana | ToRSV | [35] | ||
| Oryza sativa | RSV | [36] | ||
| AGO2 | AGO | Arabidopsis thaliana | TCV | [37,38] |
| CMV | [37] | |||
| CMV | [32] | |||
| PVX | [39] | |||
| TRV | [40] | |||
| TuMV | [41,42] | |||
| Nicotiana benthamiana | TBSV | [43] | ||
| AGO4 | AGO | Arabidopsis thaliana | BCTV | [44,45] |
| CMV | [46,47] | |||
| TRV | [40] | |||
| Nicotiana benthamiana | PVX | [48] | ||
| AGO5 | AGO | Arabidopsis thaliana | PVX | [49] |
| TuMV | [42] | |||
| AGO7 | AGO | Arabidopsis thaliana | TCV | [33] |
| TuMV | [42] | |||
| AGO10 | AGO | Arabidopsis thaliana | TuMV | [42] |
| AGO18 | AGO | Oryza sativa | RDV, RSV | [36] |
| BTR1 | RBP | Arabidopsis thaliana | ToMV | [50] |
| DCL2 and DCL4 (together) | DCLs | Arabidopsis thaliana | PVX | [51] |
| TuMV | [34] | |||
| BMV | [30] | |||
| TCV | [33] | |||
| DCL4 | DCL | Arabidopsis thaliana | PVX | [51] |
| TuMV | [34] | |||
| Nicotiana benthamiana | PVX | [51] | ||
| DRB3 | RBP | Arabidopsis thaliana | CaLCuV, BCTV | [45] |
| DRB4 | RBP | Arabidopsis thaliana | TYMV | [52,53] |
| TCV | [54] | |||
| RDR1 | RDR | Arabidopsis thaliana | TuMV | [34] |
| Nicotiana benthamiana | TMV | [55] | ||
| RDR6 | RDR | Arabidopsis thaliana | TCV | [54] |
| TuMV | [34] | |||
| BMV | [30] | |||
| UPF1 | Helicase | Arabidopsis thaliana | PVX | [56] |
| b. Proteins with antiviral properties observed in experiments using GM plants (Figure 1b) | ||||
| AGO2 | AGO | Arabidopsis thaliana | CMV | [57] |
| PVX | [49] | |||
| AGO5 | AGO | Arabidopsis thaliana | CMV | [57] |
| PVX | [49] | |||
| APUM5 | RBP | Arabidopsis thaliana | CMV, TuMV | [58] |
| AtGRP7 | RBP | Arabidopsis thaliana | TMV | [59] |
| BTR1 | RBP | Arabidopsis thaliana | ToMV | [50] |
| CaPR10 | PR-10 | Capsicum annuum | TMV | [22,58] |
| NIK1 | kinase | Arabidopsis thaliana | CaLCuV | [13] |
| PAP | RIP | Phytolacca ameriacana | PVY, PVX, CMV | [60] |
| PAP II | RIP | Phytolacca americana | TMV, PVX | [61] |
| PAP-C | RIP | Phytolacca americana | PVX | [62] |
| PIP | RIP | Phytolacca insularis | PVY, PVX, PLRV | [63] |
| PR2a | PR-2 | Nicotiana tabacum | TMV | [23] |
| PR3 | PR-3 | Nicotiana tabacum | TMV | [23] |
| RDR1 | RDR | Nicotiana tabacum | PVY | [64] |
| Trichosanthin | RIP | Trichosanthes kirilowii | TuMV | [65] |
| c. Proteins exhibiting antiviral activity in vitro (Figure 1c) | ||||
| CirA | AMP | Chassalia parvifolia | HIV | [66] |
| CirB | AMP | Chassalia parvifolia | HIV | [66] |
| Kalata B1 | AMP | Oldenlandia affinis | HIV | [28] |
| Kalata B8 | AMP | Oldenlandia affinis | HIV | [67] |
| Limenin | AMP | Phaseolus limensis | HIV-1 | [29] |
| Lunatusin | AMP | Phaseoluslunatus | HIV-1 | [68] |
| Phaseococcin | AMP | Phaseolus coccineus ‘Minor’ | HIV-1 | [69] |
| Sesquin | AMP | Vigna sesquipedalis cv. | HIV-1 | [70] |
| Stellarmedin A | PR-9 | Stellaria media | HSV-2 | [21] |
| unnamed | AMP | Phaseolus vulgaris cv. | HIV-1 | [71] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musidlak, O.; Nawrot, R.; Goździcka-Józefiak, A. Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses. Int. J. Mol. Sci. 2017, 18, 2300. https://doi.org/10.3390/ijms18112300
Musidlak O, Nawrot R, Goździcka-Józefiak A. Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses. International Journal of Molecular Sciences. 2017; 18(11):2300. https://doi.org/10.3390/ijms18112300
Chicago/Turabian StyleMusidlak, Oskar, Robert Nawrot, and Anna Goździcka-Józefiak. 2017. "Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses" International Journal of Molecular Sciences 18, no. 11: 2300. https://doi.org/10.3390/ijms18112300
APA StyleMusidlak, O., Nawrot, R., & Goździcka-Józefiak, A. (2017). Which Plant Proteins Are Involved in Antiviral Defense? Review on In Vivo and In Vitro Activities of Selected Plant Proteins against Viruses. International Journal of Molecular Sciences, 18(11), 2300. https://doi.org/10.3390/ijms18112300