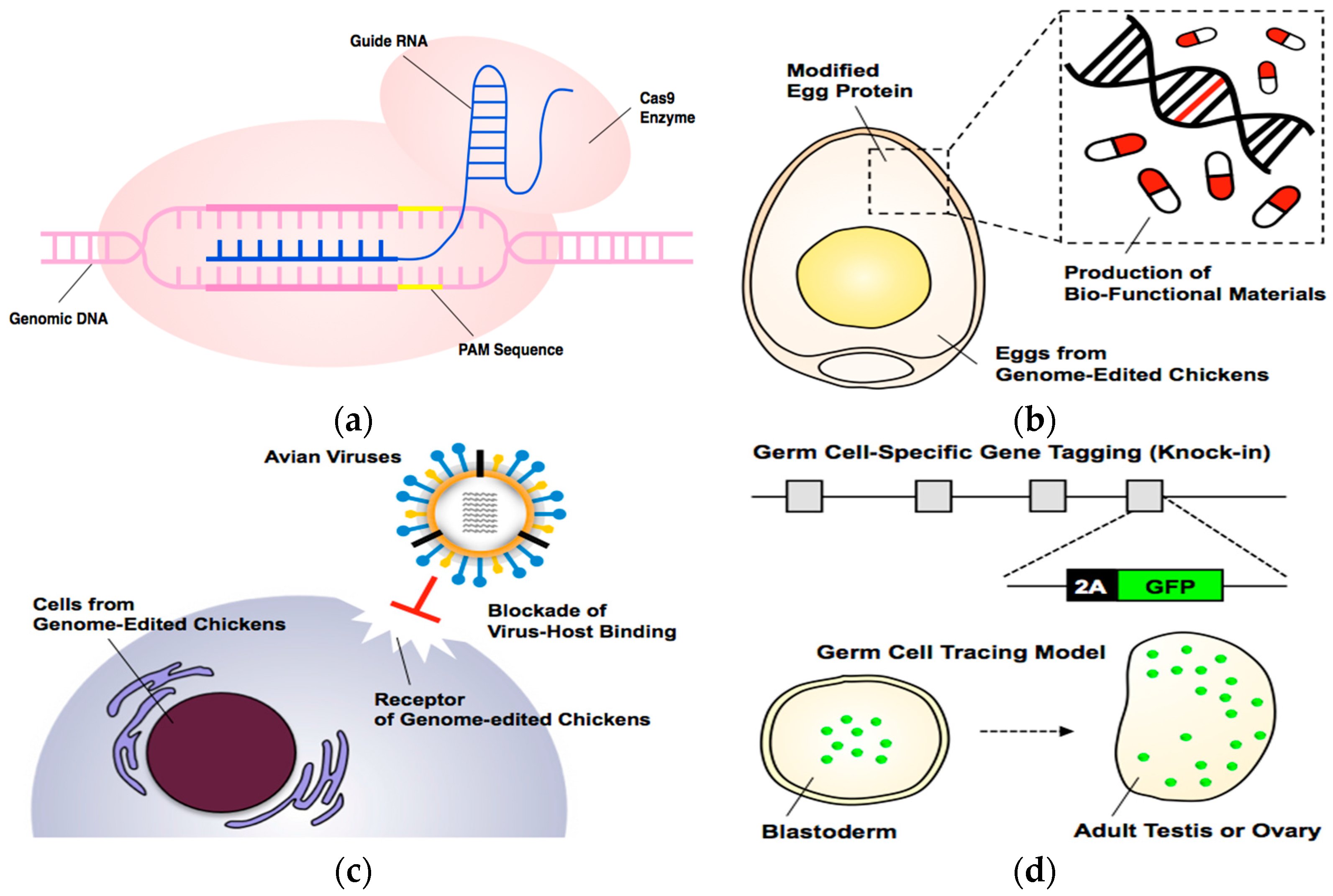
Genome Modification Technologies and Their Applications in Avian Species
Abstract
1. Introduction
2. Development of Transgenic Technologies and Application for Avian Genome Modification
3. Homologous Recombination Technology for Gene Targeting
4. Site-Specific Recombination
5. Programmable Genome Editing Using Endonucleases
6. Future Directions in Avian Biotechnology
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| TALEN | Transcription activator like effector nuclease |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 |
| ADV | Adenoviruses |
| HSV | Herpes simplex viruses |
| AAV | Adeno-associated viruses |
| SV40 | Simian virus 40 |
| kb | Kilobase pairs |
| EG&K | Eyal giladi and Kochav |
| PGC | Primordial germ cell |
| ZFN | Zinc finger nuclease |
| ESC | Embryonic stem cell |
| NHEJ | Non-homologous end joining |
| RMCE | Recombinase mediated gene cassette exchange |
| Flp/FRT | Flipase/Flipase recognition target |
| RVD | Repeat Variable Di-residue |
| crRNA | CRISPR RNA |
| tracrRNA | Trans-activating CRISPR RNA |
| PAM | Protospacer adjacent motif |
| AI | Avian influenza |
| ALV | Avian leukosis virus |
| Cpf1 | CRISPR from Prevotella and Francisella 1 |
| GUIDE-seq | Genome-wide, unbiased identification of double strand breaks enabled by sequencing |
| SSCs | Spermatogonial stem cells |
| HIV | Human immunodeficiency virus |
| STAGE | Sperm transfection-assisted gene editing |
References
- Uchida, N.; Sutton, R.E.; Friera, A.M.; He, D.; Reitsma, M.J.; Chang, W.C.; Veres, G.; Scollay, R.; Weissman, I.L. HIV, but not murine leukemia virus, vectors mediate high efficiency gene transfer into freshly isolated G0/G1 human hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 1998, 95, 11939–11944. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wu, X.; Li, G.; Han, M.; Zhuang, Y.; Xu, T. Efficient transposition of the piggybac (pb) transposon in mammalian cells and mice. Cell 2005, 122, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to fok i cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Tizard, M.; Hallerman, E.; Fahrenkrug, S.; Newell-McGloughlin, M.; Gibson, J.; de Loos, F.; Wagner, S.; Laible, G.; Han, J.Y.; D’Occhio, M.; et al. Strategies to enable the adoption of animal biotechnology to sustainably improve global food safety and security. Transgenic Res. 2016, 25, 575–595. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, H.C.; Han, J.Y. Germline modification and engineering in avian species. Mol. Cells 2015, 38, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Mintz, B. Simian virus 40 DNA sequences in DNA of healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proc. Natl. Acad. Sci. USA 1974, 71, 1250–1254. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.W.; Ruddle, F.H. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science 1981, 214, 1244–1246. [Google Scholar] [CrossRef] [PubMed]
- Bouard, D.; Alazard-Dany, N.; Cosset, F.L. Viral vectors: From virology to transgene expression. Br. J. Pharmacol. 2009, 157, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.P.; Berg, P. Construction of hybrid viruses containing sv40 and lambda phage DNA segments and their propagation in cultured monkey cells. Cell 1976, 9, 695–705. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.O. Integration of retroviral DNA. Curr. Top. Microbiol. Immunol. 1990, 157, 19–48. [Google Scholar] [PubMed]
- Yamashita, M.; Emerman, M. Retroviral infection of non-dividing cells: Old and new perspectives. Virology 2006, 344, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Salter, D.W.; Smith, E.J.; Hughes, S.H.; Wright, S.E.; Fadly, A.M.; Witter, R.L.; Crittenden, L.B. Gene insertion into the chicken germ line by retroviruses. Poult. Sci. 1986, 65, 1445–1458. [Google Scholar] [CrossRef] [PubMed]
- Lillico, S.G.; Sherman, A.; McGrew, M.J.; Robertson, C.D.; Smith, J.; Haslam, C.; Barnard, P.; Radcliffe, P.A.; Mitrophanous, K.A.; Elliot, E.A.; et al. Oviduct-specific expression of two therapeutic proteins in transgenic hens. Proc. Natl. Acad. Sci. USA 2007, 104, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.S.; Kim, T.M.; Kim, S.Y.; Kim, T.W.; Seo, H.W.; Lee, S.K.; Kwon, S.C.; Lee, G.S.; Kim, H.; Lim, J.M.; et al. Generation of transgenic quail through germ cell-mediated germline transmission. FASEB J. 2008, 22, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Kamihira, M.; Kawabe, Y.; Shindo, T.; Ono, K.; Esaka, K.; Yamashita, T.; Nishijima, K.; Iijima, S. Production of chimeric monoclonal antibodies by genetically manipulated chickens. J. Biotechnol. 2009, 141, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Velho, T.A.; Lois, C. Generation of transgenic zebra finches with replication-deficient lentiviruses. Cold Spring Harb. Protoc. 2014, 2014, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Huss, D.; Benazeraf, B.; Wallingford, A.; Filla, M.; Yang, J.; Fraser, S.E.; Lansford, R. A transgenic quail model that enables dynamic imaging of amniote embryogenesis. Development 2015, 142, 2850–2859. [Google Scholar] [CrossRef] [PubMed]
- June Byun, S.; Yuk, S.S.; Jang, Y.J.; Choi, H.; Jeon, M.H.; Erdene-Ochir, T.O.; Kwon, J.H.; Noh, J.Y.; Sun Kim, J.; Gyu Yoo, J.; et al. Transgenic chickens expressing the 3D8 single chain variable fragment protein suppress avian influenza transmission. Sci. Rep. 2017, 7, 5938. [Google Scholar] [CrossRef] [PubMed]
- Kamihira, M.; Ono, K.; Esaka, K.; Nishijima, K.; Kigaku, R.; Komatsu, H.; Yamashita, T.; Kyogoku, K.; Iijima, S. High-level expression of single-chain Fv-Fc fusion protein in serum and egg white of genetically manipulated chickens by using a retroviral vector. J. Virol. 2005, 79, 10864–10874. [Google Scholar] [CrossRef] [PubMed]
- Agate, R.J.; Scott, B.B.; Haripal, B.; Lois, C.; Nottebohm, F. Transgenic songbirds offer an opportunity to develop a genetic model for vocal learning. Proc. Natl. Acad. Sci. USA 2009, 106, 17963–17967. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.C.; Choi, J.W.; Jang, H.J.; Shin, S.S.; Lee, S.K.; Park, T.S.; Choi, I.Y.; Lee, G.S.; Song, G.; Han, J.Y. Production of biofunctional recombinant human interleukin 1 receptor antagonist (rhIL1RN) from transgenic quail egg white. Biol. Reprod. 2010, 82, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Mizuarai, S.; Ono, K.; Yamaguchi, K.; Nishijima, K.; Kamihira, M.; Iijima, S. Production of transgenic quails with high frequency of germ-line transmission using vsv-g pseudotyped retroviral vector. Biochem. Biophys. Res. Commun. 2001, 286, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005, 16, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Bucher, E.; Reinders, J.; Mirouze, M. Epigenetic control of transposon transcription and mobility in arabidopsis. Curr. Opin. Plant Biol. 2012, 15, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Cary, L.C.; Goebel, M.; Corsaro, B.G.; Wang, H.G.; Rosen, E.; Fraser, M.J. Transposon mutagenesis of baculoviruses: Analysis of trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 1989, 172, 156–169. [Google Scholar] [CrossRef]
- Bauser, C.A.; Elick, T.A.; Fraser, M.J. Proteins from nuclear extracts of two lepidopteran cell lines recognize the ends of ttaa-specific transposons piggybac and tagalong. Insect Mol. Biol. 1999, 8, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lin, C.; Lu, D.; Ning, Z.; Cox, T.; Melvin, D.; Wang, X.; Bradley, A.; Liu, P. Chromosomal transposition of piggybac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 9290–9295. [Google Scholar] [CrossRef] [PubMed]
- Koga, A.; Suzuki, M.; Inagaki, H.; Bessho, Y.; Hori, H. Transposable element in fish. Nature 1996, 383, 30. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K. Tol2: A versatile gene transfer vector in vertebrates. Genome Biol. 2007, 8, S7. [Google Scholar] [CrossRef] [PubMed]
- Ivics, Z.; Hackett, P.B.; Plasterk, R.H.; Izsvak, Z. Molecular reconstruction of sleeping beauty, a tc1-like transposon from fish, and its transposition in human cells. Cell 1997, 91, 501–510. [Google Scholar] [CrossRef]
- Glover, J.D.; Taylor, L.; Sherman, A.; Zeiger-Poli, C.; Sang, H.M.; McGrew, M.J. A novel piggybac transposon inducible expression system identifies a role for akt signalling in primordial germ cell migration. PLoS ONE 2013, 8, e77222. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, L.S.; Morris, K.R.; Wise, T.G.; Cummins, D.M.; O’Neil, T.E.; Cao, Y.; Sinclair, A.H.; Doran, T.J.; Smith, C.A. Transgenic chickens overexpressing aromatase have high estrogen levels but maintain a predominantly male phenotype. Endocrinology 2016, 157, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, H.C.; Kim, Y.M.; Hwang, Y.S.; Park, Y.H.; Park, T.S.; Han, J.Y. Site-specific recombination in the chicken genome using flipase recombinase-mediated cassette exchange. FASEB J. 2016, 30, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Taylor, L.; Sherman, A.; Kawakami, K.; Takahashi, Y.; Sang, H.M.; McGrew, M.J. Efficient genetic modification and germ-line transmission of primordial germ cells using piggybac and tol2 transposons. Proc. Natl. Acad. Sci. USA 2012, 109, E1466–E1472. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Han, J.Y. Piggybac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc. Natl. Acad. Sci. USA 2012, 109, 9337–9341. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Lee, H.G.; Moon, J.K.; Lee, H.J.; Yoon, J.W.; Yun, B.N.; Kang, S.C.; Kim, J.; Kim, H.; Han, J.Y.; et al. Deposition of bioactive human epidermal growth factor in the egg white of transgenic hens using an oviduct-specific minisynthetic promoter. FASEB J. 2015, 29, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Tyack, S.G.; Jenkins, K.A.; O’Neil, T.E.; Wise, T.G.; Morris, K.R.; Bruce, M.P.; McLeod, S.; Wade, A.J.; McKay, J.; Moore, R.J.; et al. A new method for producing transgenic birds via direct in vivo transfection of primordial germ cells. Transgenic Res. 2013, 22, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.R.; Capecchi, M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 1987, 51, 503–512. [Google Scholar] [CrossRef]
- Inbar, O.; Liefshitz, B.; Bitan, G.; Kupiec, M. The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 2000, 275, 30833–30838. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, K.M.; Marburger, K.; Intody, Z.; Wilson, J.H. Manipulating the mammalian genome by homologous recombination. Proc. Natl. Acad. Sci. USA 2001, 98, 8403–8410. [Google Scholar] [CrossRef] [PubMed]
- Schusser, B.; Collarini, E.J.; Yi, H.; Izquierdo, S.M.; Fesler, J.; Pedersen, D.; Klasing, K.C.; Kaspers, B.; Harriman, W.D.; van de Lavoir, M.C.; et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci. USA 2013, 110, 20170–20175. [Google Scholar] [CrossRef] [PubMed]
- Skarnes, W.C.; Rosen, B.; West, A.P.; Koutsourakis, M.; Bushell, W.; Iyer, V.; Mujica, A.O.; Thomas, M.; Harrow, J.; Cox, T.; et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011, 474, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Abremski, K.; Hoess, R. Bacteriophage p1 site-specific recombination. Purification and properties of the cre recombinase protein. J. Biol. Chem. 1984, 259, 1509–1514. [Google Scholar] [PubMed]
- Nagy, A. Cre recombinase: The universal reagent for genome tailoring. Genesis 2000, 26, 99–109. [Google Scholar] [CrossRef]
- Schlake, T.; Bode, J. Use of mutated FLPrecognition target (frt) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 1994, 33, 12746–12751. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.; Cole, S.; Laible, G. Site-specific modification of the bovine genome using cre recombinase-mediated gene targeting. Biotechnol. J. 2009, 4, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, M. Development of pronuclear injection-based targeted transgenesis in mice through cre-loxp site-specific recombination. Methods Mol. Biol. 2014, 1194, 3–19. [Google Scholar] [PubMed]
- Oishi, I.; Kim, S.; Yoshii, K.; Esteban, C.R.; Izpisua Belmonte, J.C. Cre-loxp-regulated expression of monoclonal antibodies driven by an ovalbumin promoter in primary oviduct cells. BMC Biotechnol. 2011, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Leighton, P.A.; Pedersen, D.; Ching, K.; Collarini, E.J.; Izquierdo, S.; Jacob, R.; van de Lavoir, M.C. Generation of chickens expressing cre recombinase. Transgenic Res. 2016, 25, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Bateman, J.R.; Lee, A.M.; Wu, C.T. Site-specific transformation of drosophila via phiC31 integrase-mediated cassette exchange. Genetics 2006, 173, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Hartley, J.L.; Temple, G.F.; Brasch, M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000, 10, 1788–1795. [Google Scholar] [CrossRef] [PubMed]
- Schusser, B.; Yi, H.; Collarini, E.J.; Izquierdo, S.M.; Harriman, W.D.; Etches, R.J.; Leighton, P.A. Harnessing gene conversion in chicken b cells to create a human antibody sequence repertoire. PLoS ONE 2013, 8, e80108. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Bibikova, M.; Whitby, F.G.; Reddy, A.R.; Chandrasegaran, S.; Carroll, D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000, 28, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Porteus, M.H.; Carroll, D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005, 23, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A tale nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Mussolino, C.; Cathomen, T. Tale nucleases: Tailored genome engineering made easy. Curr. Opin. Biotechnol. 2012, 23, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Gruetzner, R.; Kandzia, R.; Marillonnet, S. Golden gate shuffling: A one-pot DNA shuffling method based on type iis restriction enzymes. PLoS ONE 2009, 4, e5553. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Cong, L.; Zhou, Y.; Cunniff, M.M.; Feng, G.; Zhang, F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012, 7, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cong, L.; Lodato, S.; Kosuri, S.; Church, G.M.; Arlotta, P. Efficient construction of sequence-specific tal effectors for modulating mammalian transcription. Nat. Biotechnol. 2011, 29, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with tal effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Lee, H.J.; Kim, K.H.; Kim, J.S.; Han, J.Y. Targeted gene knockout in chickens mediated by talens. Proc. Natl. Acad. Sci. USA 2014, 111, 12716–12721. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.; Carlson, D.F.; Nandi, S.; Sherman, A.; Fahrenkrug, S.C.; McGrew, M.J. Efficient talen-mediated gene targeting of chicken primordial germ cells. Development 2017, 144, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. Crispr provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Reardon, S. Welcome to the crispr zoo. Nature 2016, 531, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Abu-Bonsrah, K.D.; Zhang, D.; Newgreen, D.F. Crispr/cas9 targets chicken embryonic somatic cells in vitro and in vivo and generates phenotypic abnormalities. Sci. Rep. 2016, 6, 34524. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Lee, J.; Park, J.Y.; Oh, K.B.; Hwang, S.; Lee, C.W.; Lee, K. Targeted genome editing in a quail cell line using a customized crispr/cas9 system. Poult. Sci. 2017, 96, 1445–1450. [Google Scholar] [CrossRef] [PubMed]
- Veron, N.; Qu, Z.; Kipen, P.A.; Hirst, C.E.; Marcelle, C. Crispr mediated somatic cell genome engineering in the chicken. Dev. Biol. 2015, 407, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Zuo, Q.; Li, D.; Zhang, W.; Wang, F.; Ji, Y.; Jin, J.; Lu, Z.; Wang, M.; et al. Crispr/cas9 mediated chicken stra8 gene knockout and inhibition of male germ cell differentiation. PLoS ONE 2017, 12, e0172207. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Jin, K.; Wang, Y.; Song, J.; Zhang, Y.; Li, B. Crispr/cas9-mediated deletion of c1eis inhibits chicken embryonic stem cell differentiation into male germ cells (gallus gallus). J. Cell. Biochem. 2017, 118, 2380–2386. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, L.; Pedersen, D.; Ching, K.H.; Yi, H.; Collarini, E.J.; Izquierdo, S.; van de Lavoir, M.C.; Leighton, P.A. Germline gene editing in chickens by efficient crispr-mediated homologous recombination in primordial germ cells. PLoS ONE 2016, 11, e0154303. [Google Scholar] [CrossRef] [PubMed]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Kagami, H.; Tagami, T. Targeted mutagenesis in chicken using crispr/cas9 system. Sci. Rep. 2016, 6, 23980. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M.; Mali, P.; Braff, J.L.; Moosburner, M.; Yaung, S.J.; Church, G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 2013, 10, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single rna-guided endonuclease of a class 2 crispr-cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.K.; Kim, K.; Been, K.W.; Baek, G.; Ye, S.; Hur, J.W.; Ryu, S.M.; Lee, Y.S.; Kim, J.S. Targeted mutagenesis in mice by electroporation of cpf1 ribonucleoproteins. Nat. Biotechnol. 2016, 34, 807–808. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H.; Kim, J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, aaf8729. [Google Scholar] [CrossRef] [PubMed]
- Lyall, J.; Irvine, R.M.; Sherman, A.; McKinley, T.J.; Nunez, A.; Purdie, A.; Outtrim, L.; Brown, I.H.; Rolleston-Smith, G.; Sang, H.; et al. Suppression of avian influenza transmission in genetically modified chickens. Science 2011, 331, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Brojatsch, J.; Naughton, J.; Rolls, M.M.; Zingler, K.; Young, J.A. Car1, a tnfr-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 1996, 87, 845–855. [Google Scholar] [CrossRef]
- Klucking, S.; Adkins, H.B.; Young, J.A. Resistance to infection by subgroups b, d, and e avian sarcoma and leukosis viruses is explained by a premature stop codon within a resistance allele of the tvb receptor gene. J. Virol. 2002, 76, 7918–7921. [Google Scholar] [CrossRef] [PubMed]
- Elleder, D.; Stepanets, V.; Melder, D.C.; Senigl, F.; Geryk, J.; Pajer, P.; Plachy, J.; Hejnar, J.; Svoboda, J.; Federspiel, M.J. The receptor for the subgroup c avian sarcoma and leukosis viruses, tvc, is related to mammalian butyrophilins, members of the immunoglobulin superfamily. J. Virol. 2005, 79, 10408–10419. [Google Scholar] [CrossRef] [PubMed]
- Chai, N.; Bates, P. Na+/h+ exchanger type 1 is a receptor for pathogenic subgroup j avian leukosis virus. Proc. Natl. Acad. Sci. USA 2006, 103, 5531–5536. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Gagneux, P. Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 2012, 1253, 16–36. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, K.Y.; Park, Y.H.; Choi, H.J.; Yao, Y.; Nair, V.; Han, J.Y. Acquisition of resistance to avian leukosis virus subgroup B through mutations on tvb cysteine-rich domains in DF-1 chicken fibroblasts. Vet. Res. 2017, 48, 48. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, K.Y.; Jung, K.M.; Park, K.J.; Lee, K.O.; Suh, J.Y.; Yao, Y.; Nair, V.; Han, J.Y. Precise gene editing of chicken Na+/H+ exchange type 1 (chNHE1) confers resistance to avian leukosis virus subgroup J (ALV-J). Dev. Comp. Immunol. 2017, 77, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Paxton, W.A.; Choe, S.; Ceradini, D.; Martin, S.R.; Horuk, R.; MacDonald, M.E.; Stuhlmann, H.; Koup, R.A.; Landau, N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996, 86, 367–377. [Google Scholar] [CrossRef]
- Van de Lavoir, M.C.; Diamond, J.H.; Leighton, P.A.; Mather-Love, C.; Heyer, B.S.; Bradshaw, R.; Kerchner, A.; Hooi, L.T.; Gessaro, T.M.; Swanberg, S.E.; et al. Germline transmission of genetically modified primordial germ cells. Nature 2006, 441, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Kim, S.; Kim, T.M.; Kim, Y.M.; Seo, H.W.; Park, T.S.; Jeong, J.W.; Song, G.; Han, J.Y. Basic fibroblast growth factor activates mek/erk cell signaling pathway and stimulates the proliferation of chicken primordial germ cells. PLoS ONE 2010, 5, e12968. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Glover, J.D.; Taylor, L.; Sang, H.M.; McGrew, M.J. Characterisation and germline transmission of cultured avian primordial germ cells. PLoS ONE 2010, 5, e15518. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Jung, J.G.; Kim, J.N.; Park, T.S.; Kim, T.M.; Shin, S.S.; Kang, D.K.; Lim, J.M.; Han, J.Y. A testis-mediated germline chimera production based on transfer of chicken testicular cells directly into heterologous testes. Biol. Reprod. 2006, 75, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Pramod, R.K.; Lee, B.R.; Kim, Y.M.; Lee, H.J.; Park, Y.H.; Ono, T.; Lim, J.M.; Han, J.Y. Isolation, characterization, and in vitro culturing of spermatogonial stem cells in japanese quail (Coturnix japonica). Stem Cells Dev. 2017, 26, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.A.; Challagulla, A.; Jenkins, K.A.; Wise, T.G.; O’Neil, T.E.; Morris, K.R.; Tizard, M.L.; Doran, T.J. Generation of gene edited birds in one generation using sperm transfection assisted gene editing (STAGE). Transgenic Res. 2017, 26, 331–347. [Google Scholar] [CrossRef] [PubMed]
| Methods | Efficiency of Genome Editing in Chicken PGCs | Efficiency of Germline Transmission (Genome-Edited Chickens) | References |
|---|---|---|---|
| TALEN | 33.3% | 22.3–53.2% (0.0–10.4%) | [66] |
| CRISPR/Cas9 | 0–100% | 67–79% (48–58%) | [76] |
| Homologous recombination | 0.00001% | 0.005–0.2% (NA 1) | [44] |
| TALEN + homologous recombination | 8.1% | NA 1 (0–6%) | [67] |
| CRISPR/Cas9 + homologous recombination | 20–33% | 0–96% (0–48%) | [75] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.J.; Kim, Y.M.; Ono, T.; Han, J.Y. Genome Modification Technologies and Their Applications in Avian Species. Int. J. Mol. Sci. 2017, 18, 2245. https://doi.org/10.3390/ijms18112245
Lee HJ, Kim YM, Ono T, Han JY. Genome Modification Technologies and Their Applications in Avian Species. International Journal of Molecular Sciences. 2017; 18(11):2245. https://doi.org/10.3390/ijms18112245
Chicago/Turabian StyleLee, Hong Jo, Young Min Kim, Tamao Ono, and Jae Yong Han. 2017. "Genome Modification Technologies and Their Applications in Avian Species" International Journal of Molecular Sciences 18, no. 11: 2245. https://doi.org/10.3390/ijms18112245
APA StyleLee, H. J., Kim, Y. M., Ono, T., & Han, J. Y. (2017). Genome Modification Technologies and Their Applications in Avian Species. International Journal of Molecular Sciences, 18(11), 2245. https://doi.org/10.3390/ijms18112245