Long Non-Coding RNA Profiling in a Non-Alcoholic Fatty Liver Disease Rodent Model: New Insight into Pathogenesis
Abstract
:1. Introduction
2. Results
2.1. Animal Model of Non-Alcoholic Fatty Liver Disease
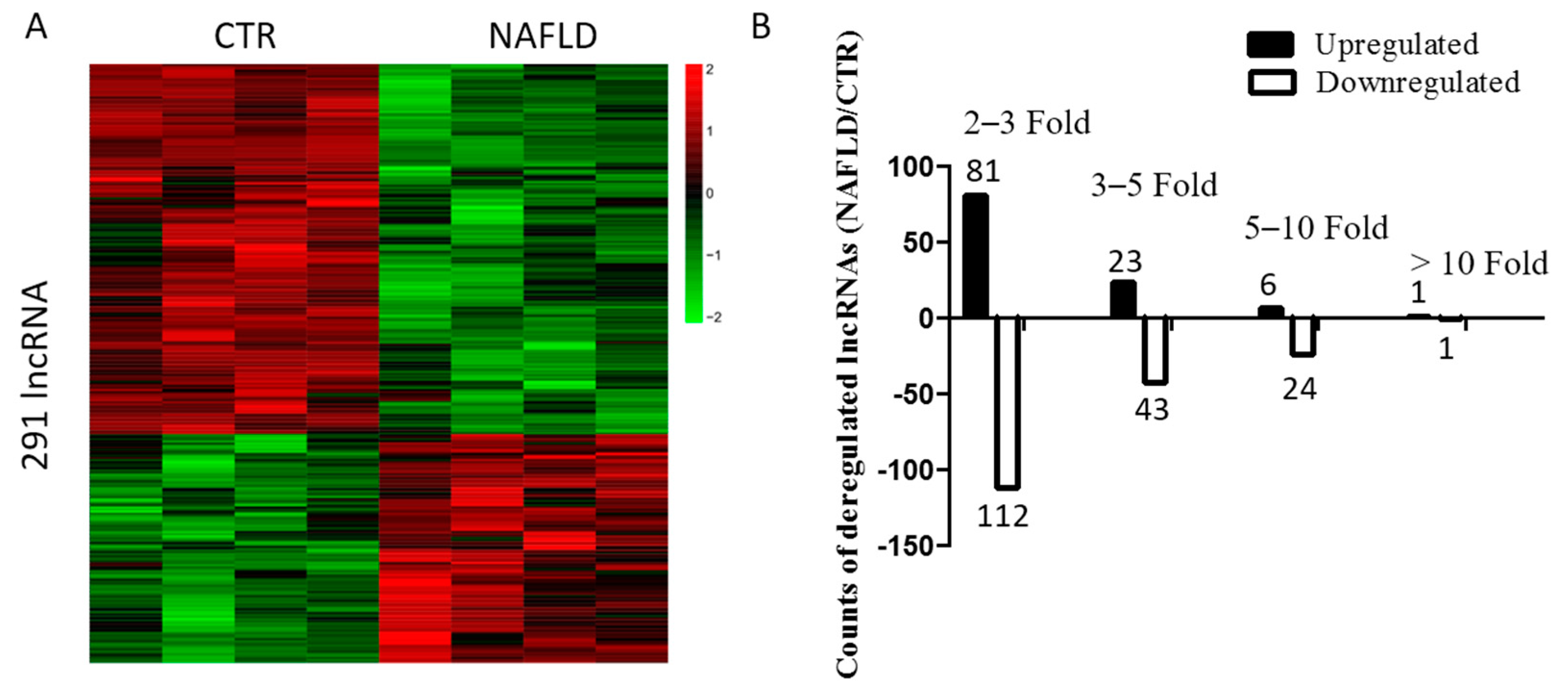
2.2. Long Non-Coding RNA Profile in Non-Alcoholic Fatty Liver Disease
2.3. Bioinformatic Analysis and Characterization of Differentially Expressed lncRNAs
2.4. Global Changes in Gene Expression Profiles of Non-Alcoholic Fatty Liver Disease and Bioinformatic Analysis
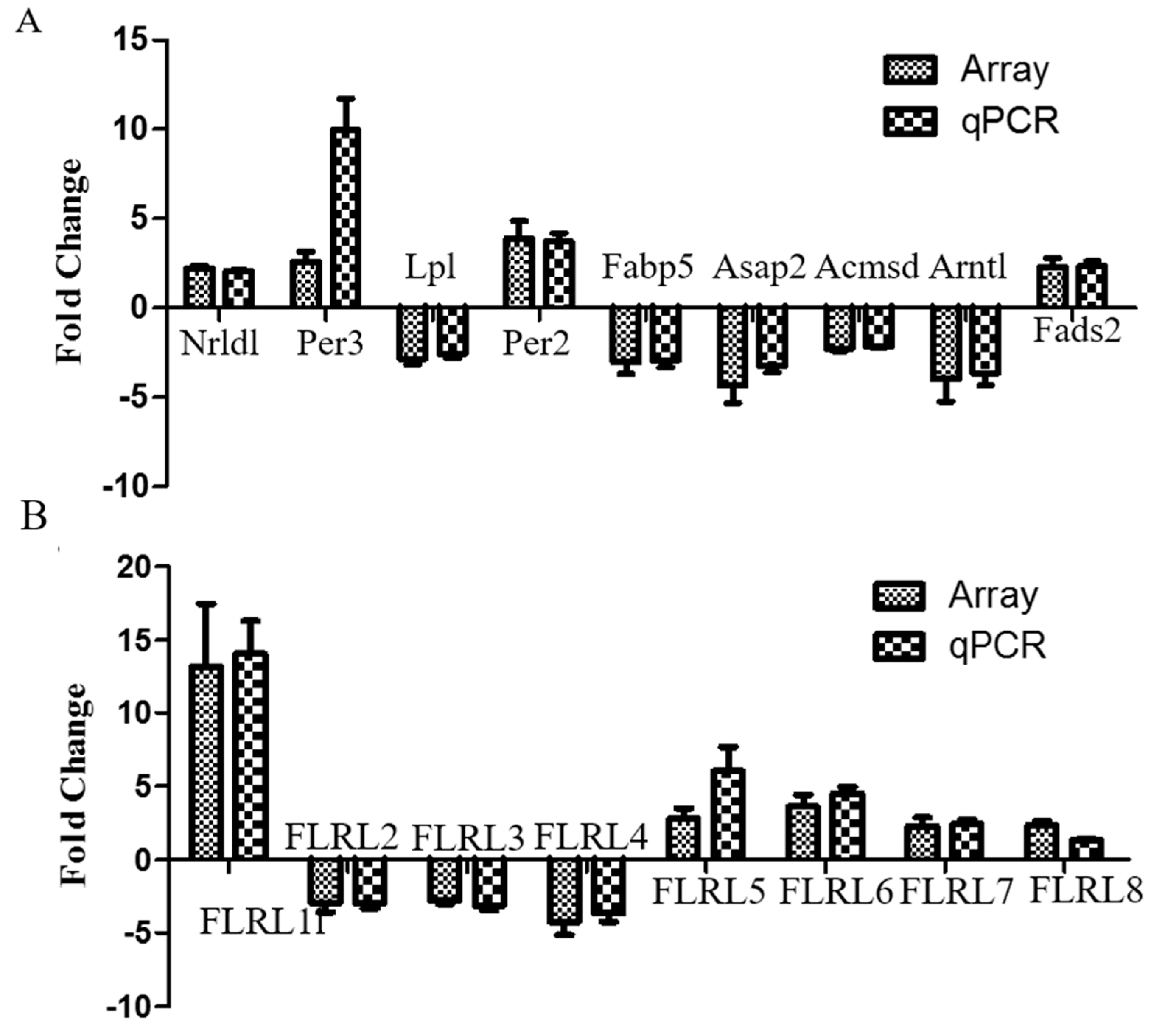
2.5. Quantitative Polymerase Chain Reaction Validation of Microarray Data
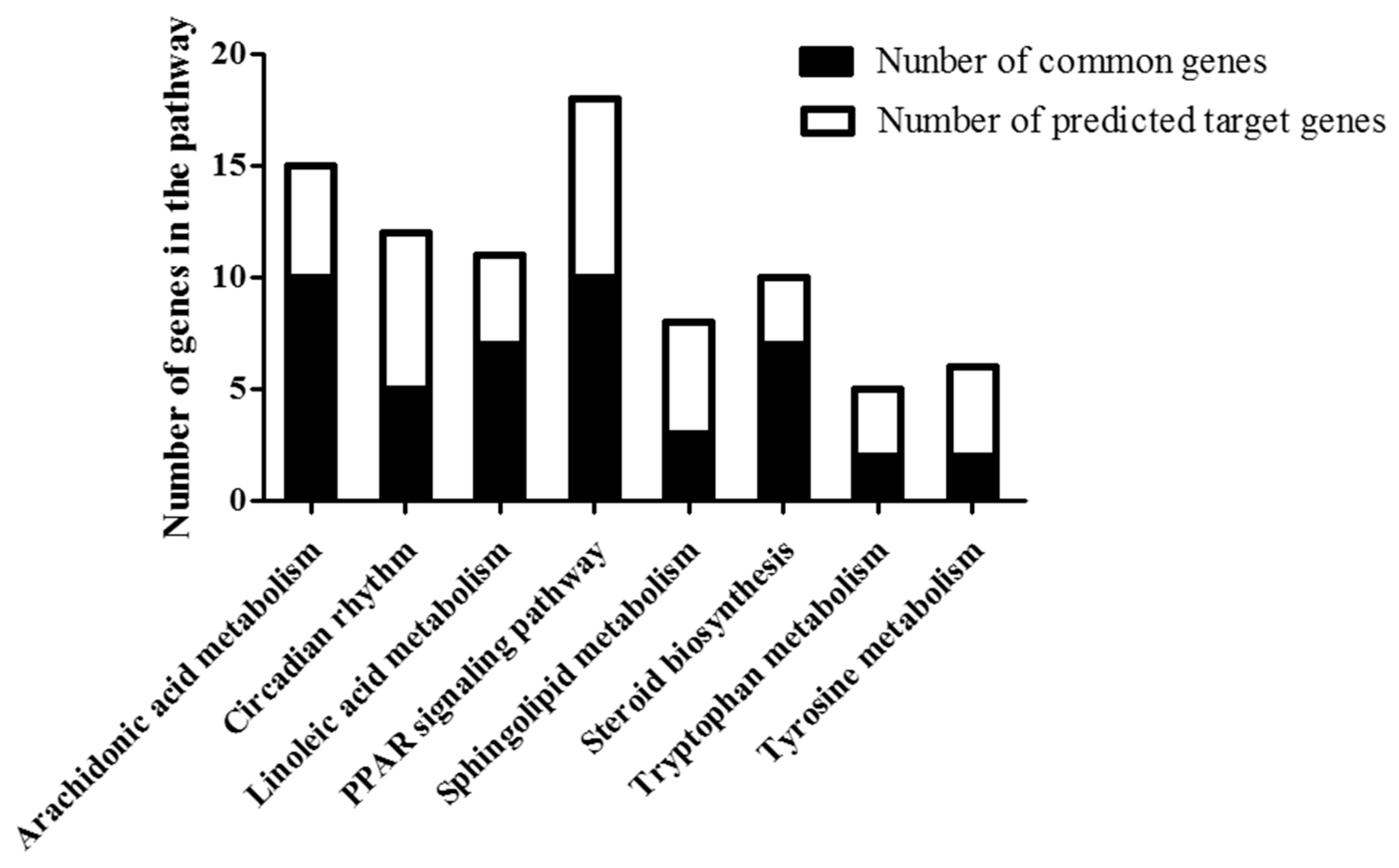
2.6. Common Pathway of Different Expressed lncRNA and Corresponding mRNA
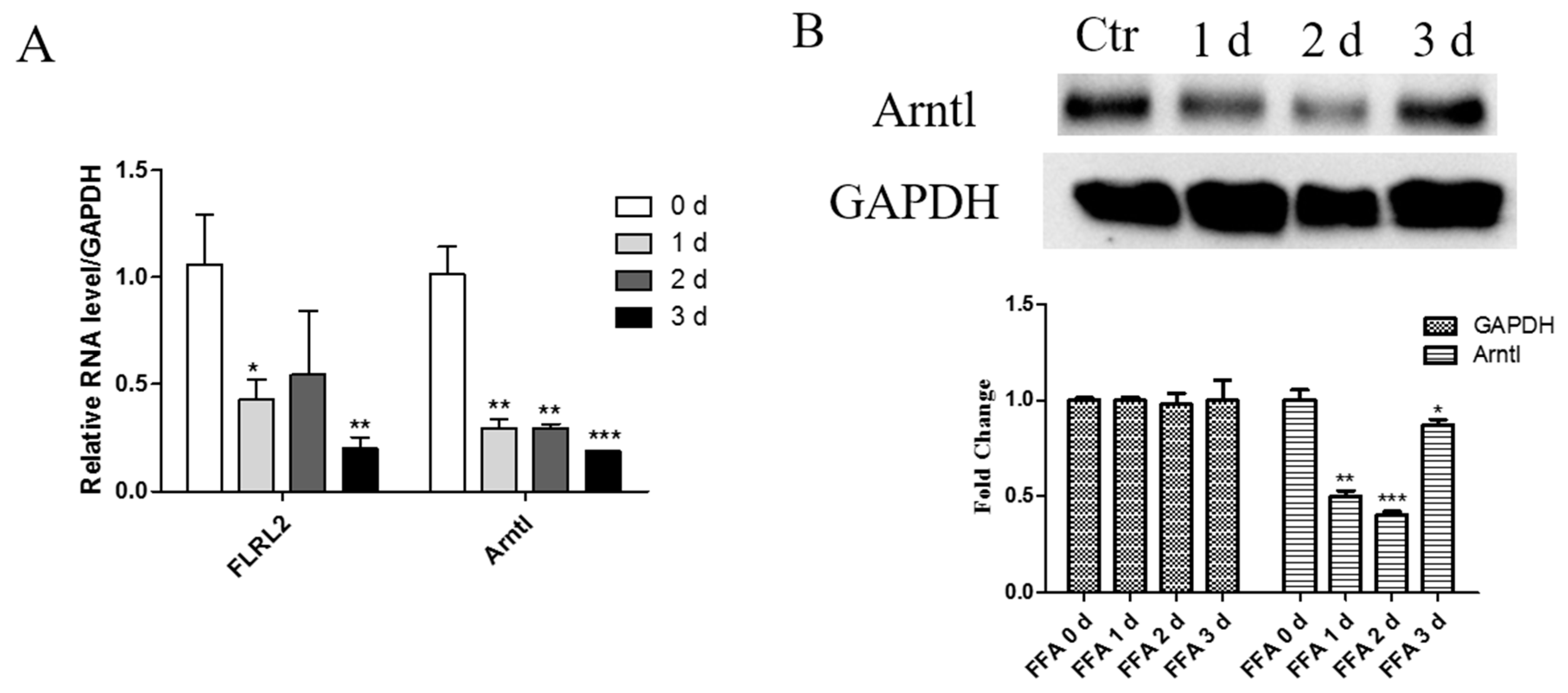
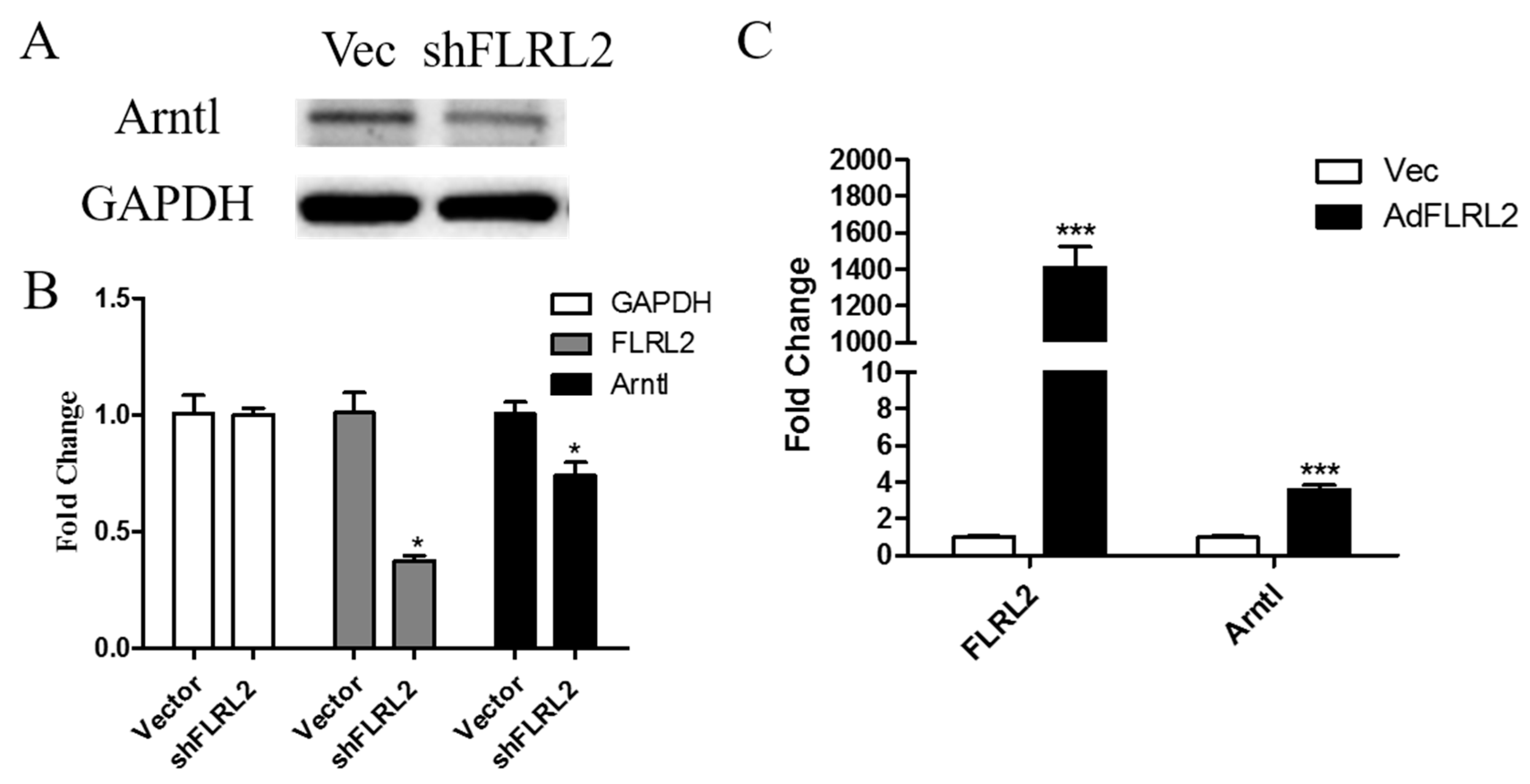
2.7. Knockdown of lncRNA FLRL2 Increases Predicted-Target Arntl Expression and Overexpression Vice Versa
3. Discussion
4. Materials and Methods
4.1. Animal Model of Non-Alcoholic Fatty Liver Disease and Liver Samples
4.2. RNA Extraction and Purification
4.3. LncRNA and mRNA Microarray
4.4. Quantitative Polymerase Chain Reaction Validation
4.5. Cell Culture, Non-Alcoholic Fatty Liver Disease Cellular Model and Plasmid Transfection
4.6. Protein Extraction and Western Blot
4.7. Lipid Staining and Intracellular Triglyceride Quantification
4.8. Bio-Informatic Analysis of LncRNA Targets and Associated Pathways
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology 2012, 142, 1592–1609. [Google Scholar] [CrossRef] [PubMed]
- Sattar, N.; Forrest, E.; Preiss, D. Non-alcoholic fatty liver disease. BMJ 2014, 349, g4596. [Google Scholar] [CrossRef] [PubMed]
- Starley, B.Q.; Calcagno, C.J.; Harrison, S.A. Nonalcoholic fatty liver disease and hepatocellular carcinoma: A weighty connection. Hepatology 2010, 51, 1820–1832. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P.; James, O.F. Steatohepatitis: A tale of two “hits”? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef]
- Del Ben, M.; Polimeni, L.; Baratta, F.; Pastori, D.; Loffredo, L.; Angelico, F. Modern approach to the clinical management of non-alcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 8341–8350. [Google Scholar] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.; Duerksen, J.D. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol. Cell. Biochem. 1975, 9, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.; Haga, H.; Patel, T. Long noncoding RNA in liver diseases. Hepatology 2014, 60, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, F.; Yuan, J.H.; Yuan, S.X.; Zhou, W.P.; Huo, X.S.; Xu, D.; Bi, H.S.; Wang, F.; Sun, S.H. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Kong, G.; You, X.; Zhang, S.; Zhang, T.; Gao, Y.; Ye, L.; Zhang, X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J. Biol. Chem. 2012, 287, 26302–26311. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012, 29, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, L.; Wu, L.M.; Lai, M.C.; Xie, H.Y.; Zhang, F.; Zheng, S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011, 18, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.F.; Guo, Y.J.; Zhao, C.X.; Yuan, S.X.; Wang, Y.; Tang, G.N.; Zhou, W.P.; Sun, S.H. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 2013, 57, 1882–1892. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, M.; Thulluru, H.K.; Mulders, J.; Michel, O.J.; Poutsma, A.; Windhorst, S.; Kleiverda, G.; Sie, D.; Lachmeijer, A.M.; Oudejans, C.B. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J. Clin. Investig. 2012, 122, 4003–4011. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, X.; Yi, Z.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L.; Huang, F. Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease. IUBMB Life 2015, 67, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Arellanes-Licea, E.; Caldelas, I.; de Ita-Perez, D.; Diaz-Munoz, M. The circadian timing system: A recent addition in the physiological mechanisms underlying pathological and aging processes. Aging Dis. 2014, 5, 406–418. [Google Scholar] [PubMed]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.H. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform. 2016. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci. Rep. 2015, 5, 13186. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.A.; Wang, X.S.; You, Z.H.; Chan, K.C. FMLNCSIM: Fuzzy measure-based lncRNA functional similarity calculation model. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; You, Z.H.; Yan, G.Y.; Gong, D.W. IRWRLDA: Improved random walk with restart for lncRNA-disease association prediction. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.A.; Chen, X.; You, Z.H.; Huang, D.S.; Chan, K.C. ILNCSIM: Improved lncRNA functional similarity calculation model. Oncotarget 2016, 7, 25902–25914. [Google Scholar] [CrossRef] [PubMed]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, B.; Bellet, M.M.; Katada, S.; Astarita, G.; Hirayama, J.; Amin, R.H.; Granneman, J.G.; Piomelli, D.; Leff, T.; Sassone-Corsi, P. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010, 12, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Paschos, G.K.; Ibrahim, S.; Song, W.L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, C.A.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 2012, 18, 1768–1777. [Google Scholar] [CrossRef] [PubMed]
- Shimba, S.; Ogawa, T.; Hitosugi, S.; Ichihashi, Y.; Nakadaira, Y.; Kobayashi, M.; Tezuka, M.; Kosuge, Y.; Ishige, K.; Ito, Y.; et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS ONE 2011, 6, e25231. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X. Long Noncoding RNA HULC Modulates Abnormal Lipid Metabolism in Hepatoma Cells through an miR-9-Mediated RXRA Signaling Pathway. Cancer Res. 2015, 75, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Belury, M.A. Dietary conjugated linoleic acid in health: Physiological effects and mechanisms of action. Annu. Rev. Nutr. 2002, 22, 505–531. [Google Scholar] [CrossRef] [PubMed]
- Zuo, R.; Ai, Q.; Mai, K.; Xu, W. Effects of conjugated linoleic acid on growth, non-specific immunity, antioxidant capacity, lipid deposition and related gene expression in juvenile large yellow croaker (Larmichthys crocea) fed soyabean oil-based diets. Br. J. Nutr. 2013, 110, 1220–1232. [Google Scholar] [CrossRef] [PubMed]
- Van Heek, M.; Compton, D.S.; France, C.F.; Tedesco, R.P.; Fawzi, A.B.; Graziano, M.P.; Sybertz, E.J.; Strader, C.D.; Davis, H.R., Jr. Diet-induced obese mice develop peripheral, but not central, resistance to leptin. J. Clin. Investig. 1997, 99, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Ferdowsian, H. Human and animal research guidelines: Aligning ethical constructs with new scientific developments. Bioethics 2011, 25, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, M.J.; Schriever, S.C.; Roscher, A.A.; Ensenauer, R. Digital image analysis approach for lipid droplet size quantitation of Oil Red O-stained cultured cells. Anal. Biochem. 2014, 445, 87–89. [Google Scholar] [CrossRef] [PubMed]
- ImageJ. Available online: https://imagej.nih.gov/ij/ (accessed on 16 February 2016).
- Xu, C.; Wan, X.; Xu, L.; Weng, H.; Yan, M.; Miao, M.; Sun, Y.; Xu, G.; Dooley, S.; Li, Y.; et al. Xanthine oxidase in non-alcoholic fatty liver disease and hyperuricemia: One stone hits two birds. J. Hepatol. 2015, 62, 1412–1419. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Yang, J.; Hou, Y.; Zhang, H.; Zeng, Z.; Zhao, L.; Yu, T.; Tang, X.; Tu, G.; Cui, X.; et al. LncRNA expression signatures of twist-induced epithelial-to-mesenchymal transition in MCF10A cells. Cell Signal. 2014, 26, 83–93. [Google Scholar] [CrossRef] [PubMed]
- UCSC Genome Browser. Available online: http://genome.ucsc.edu/ (accessed on 16 February 2016).
- Matkovich, S.J.; Edwards, J.R.; Grossenheider, T.C.; de Guzman Strong, C.; Dorn, G.W., 2nd. Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc. Natl. Acad. Sci. USA 2014, 111, 12264–12269. [Google Scholar] [CrossRef] [PubMed]
- Tafer, H.; Hofacker, I.L. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Hu, Z.; Chen, F.; Zhu, R.; Deng, Y.; Shao, X.; Li, Y.; Zhao, J.; Li, H.; Zhang, B.; et al. Transcriptome analysis of long non-coding RNAs of the nucleus accumbens in cocaine-conditioned mice. J. Neurochem. 2012, 123, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhu, J.; Liu, Y.; Chen, Y.; Li, Y.; Huang, L.; Chen, S.; Li, T.; Dang, Y.; Chen, T. Methamphetamine induces alterations in the long non-coding RNAs expression profile in the nucleus accumbens of the mouse. BMC Neurosci. 2015, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Genecodis. Available online: http://genecodis.cnb.csic.es/ (accessed on 16 February 2016).
- Nogales-Cadenas, R.; Carmona-Saez, P.; Vazquez, M.; Vicente, C.; Yang, X.; Tirado, F.; Carazo, J.M.; Pascual-Montano, A. GeneCodis: Interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009, 37, W317–W322. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Huang, H.; Xu, C.; Yu, C.; Li, Y. Long Non-Coding RNA Profiling in a Non-Alcoholic Fatty Liver Disease Rodent Model: New Insight into Pathogenesis. Int. J. Mol. Sci. 2017, 18, 21. https://doi.org/10.3390/ijms18010021
Chen Y, Huang H, Xu C, Yu C, Li Y. Long Non-Coding RNA Profiling in a Non-Alcoholic Fatty Liver Disease Rodent Model: New Insight into Pathogenesis. International Journal of Molecular Sciences. 2017; 18(1):21. https://doi.org/10.3390/ijms18010021
Chicago/Turabian StyleChen, Yi, Haixiu Huang, Chengfu Xu, Chaohui Yu, and Youming Li. 2017. "Long Non-Coding RNA Profiling in a Non-Alcoholic Fatty Liver Disease Rodent Model: New Insight into Pathogenesis" International Journal of Molecular Sciences 18, no. 1: 21. https://doi.org/10.3390/ijms18010021
APA StyleChen, Y., Huang, H., Xu, C., Yu, C., & Li, Y. (2017). Long Non-Coding RNA Profiling in a Non-Alcoholic Fatty Liver Disease Rodent Model: New Insight into Pathogenesis. International Journal of Molecular Sciences, 18(1), 21. https://doi.org/10.3390/ijms18010021




