Molecular Characterization and Growth Association of Two Apolipoprotein A-Ib Genes in Common Carp (Cyprinus carpio)
Abstract
:1. Introduction
2. Results
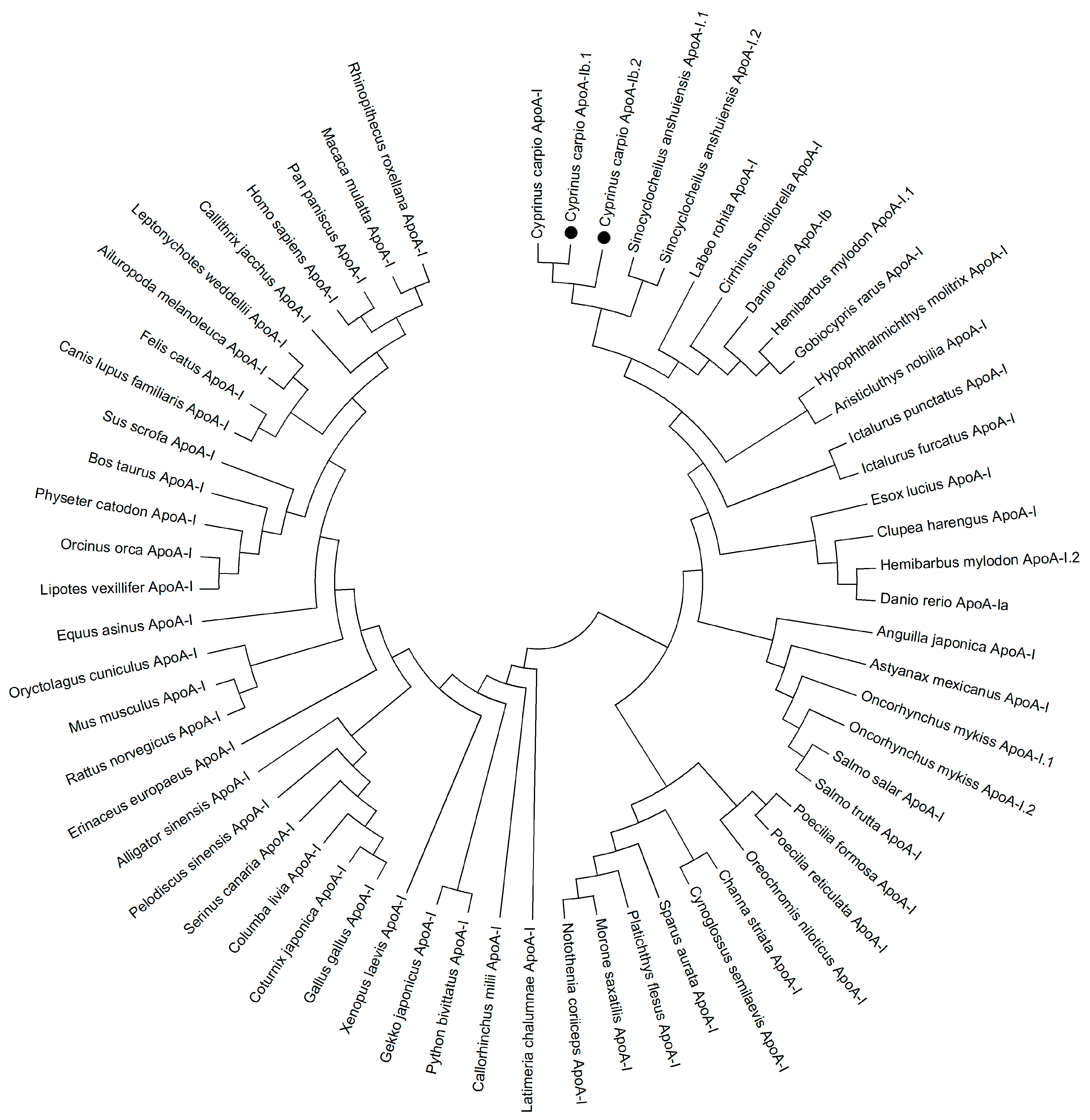
2.1. Characterization of apoA-Ibs
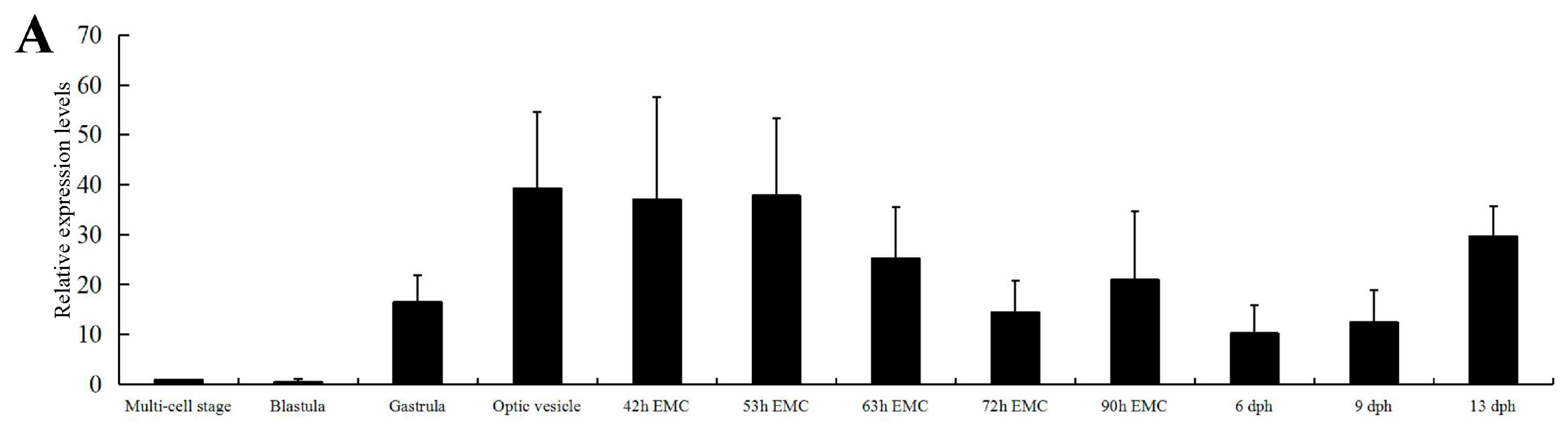
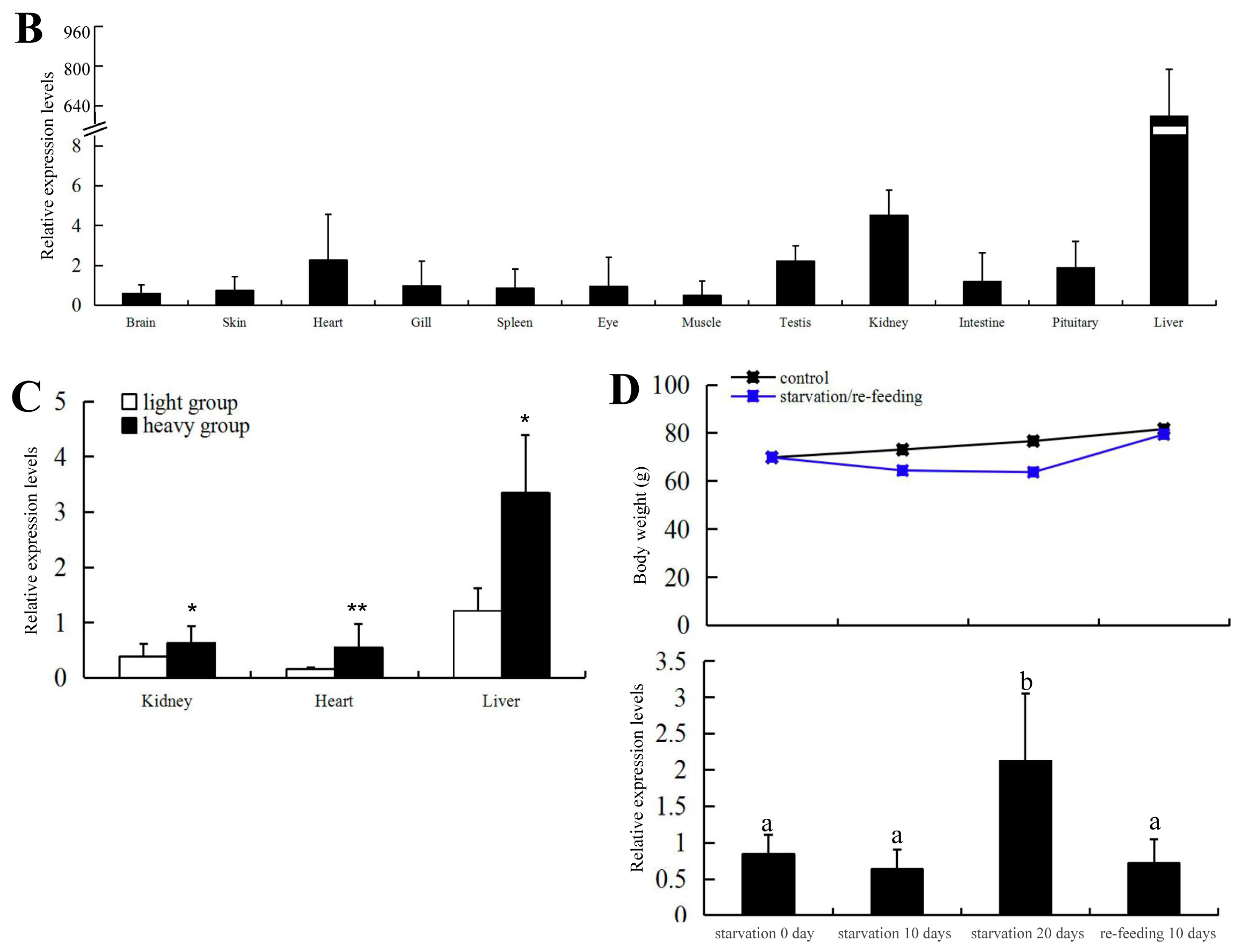
2.2. Expression of C. carpio apoA-Ib
2.3. Polymorphisms of the C. carpio apoA-Ibs
2.4. Genetic Associations between SNPs and Growth Traits
3. Discussion
4. Experimental Section
4.1. Sample Collection and Extraction
4.2. Sequence Analysis of C. carpio apoA-Ib
4.3. Gene Expression Analysis by Quantitative Real-Time PCR
4.4. Compensatory Growth Test
4.5. Identification of Polymorphisms and Association Analysis with Growth Traits
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, W.H.; Tanimura, M.; Luo, C.C.; Datta, S.; Chan, L. The apolipoprotein multigene family—Biosynthesis, structure, structure-function relationships, and evolution. J. Lipid Res. 1988, 29, 245–271. [Google Scholar] [PubMed]
- De Smet, H.; Blust, R.; Moens, L. Absence of albumin in the plasma of the common carp Cyprinus carpio: Binding of fatty acids to high density lipoprotein. Fish Physiol. Biochem. 1998, 19, 71–81. [Google Scholar] [CrossRef]
- Bolanos-Garcia, V.M.; Miguel, R.N. On the structure and function of apolipoproteins: More than a family of lipid-binding proteins. Prog. Biophys. Mol. Biol. 2003, 83, 47–68. [Google Scholar] [CrossRef]
- Srinivas, R.V.; Venkatachalapathi, Y.V.; Rui, Z.; Owens, R.J.; Gupta, K.B.; Srinivas, S.K.; Anantharamaiah, G.M.; Segrest, J.P.; Compans, R.W. Inhibition of virus-induced cell-fusion by apolipoprotein A-I and its amphipathic peptide analogs. J. Cell. Biochem. 1991, 45, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Tada, N.; Sakamoto, T.; Kagami, A.; Mochizuki, K.; Kurosaka, K. Antimicrobial activity of lipoprotein particles containing apolipoprotein-A-l. Mol. Cell. Biochem. 1993, 119, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Burger, D.; Dayer, J. High-density lipoprotein-associated apolipoprotein AI: The missing link between infection and chronic inflammation? Autoimmun. Rev. 2002, 1, 111–117. [Google Scholar] [CrossRef]
- Ma, J.; Liao, X.-L.; Lou, B.; Wu, M.-P. Role of apolipoprotein AI in protecting against endotoxin toxicity. Acta Biochim. Biophys. Sin. 2004, 36, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Brewer, H.; Fairwell, T.; LaRue, A.; Ronan, R.; Houser, A.; Bronzert, T. The amino acid sequence of human ApoA-I, an apolipoprotein isolated from high density lipoproteins. Biochem. Biophys. Res. Commun. 1978, 80, 623–630. [Google Scholar] [CrossRef]
- Ferrari, S.; Drusiani, E.; Calandra, S.; Tarugi, P. Isolation of a cDNA clone for chicken intestinal apolipoprotein AI and its use for detecting apoAI mRNA expression in several chicken tissues. Gene 1986, 42, 209–214. [Google Scholar] [CrossRef]
- Chan, L. The apolipoprotein multigene family—Structure, expression, evolution, and molecular-genetics. Klin. Wochenschr. 1989, 67, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T. Lipid nutrition in fish. Comp. Biochem. Physiol. B Comp. Biochem. 1982, 73, 3–15. [Google Scholar] [CrossRef]
- Kondo, H.; Morinaga, K.; Misaki, R.; Nakaya, M.; Watabe, S. Characterization of the pufferfish Takifugu rubripes apolipoprotein multigene family. Gene 2005, 346, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.; Higgins, D.G.; Wolff, J.; Byrnes, L.; Stack, M.; Sharp, P.M.; Gannon, F. The salmon gene encoding apolipoprotein-A-I—cDNA sequence, sissue sxpression and svolution. Gene 1991, 104, 155–161. [Google Scholar] [CrossRef]
- Llewellyn, L.; Ramsurn, V.P.; Wigham, T.; Sweeney, G.E.; Power, D.M. Cloning, characterisation and expression of the apolipoprotein A-I gene in the sea bream (Sparus aurata). Biochim. Biophys. Acta (BBA)-Gene Struct. Exp. 1998, 1442, 399–404. [Google Scholar] [CrossRef]
- Kondo, H.; Kawazoe, I.; Nakaya, M.; Kikuchi, K.; Aida, K.; Watabe, S. The novel sequences of major plasma apolipoproteins in the eel Anguilla japonica. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2001, 1531, 132–142. [Google Scholar] [CrossRef]
- Concha, M.I.; Lopez, R.; Villanueva, J.; Baez, N.; Amthauer, R. Undetectable apolipoprotein A-I gene expression suggests an unusual mechanism of dietary lipid mobilisation in the intestine of Cyprinus carpio. J. Exp. Biol. 2005, 208, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, F.; Bastias, A.; Casado, A.; Amthauer, R.; Concha, M.I. Apolipoprotein A-I, an antimicrobial protein in Oncorhynchus mykiss: Evaluation of its expression in primary defence barriers and plasma levels in sick and healthy fish. Fish Shellfish Immunol. 2007, 23, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.A.; Adamek, M.; Bilinska, B.; Hejmej, A.; Steinhagen, D.; Ciereszko, A. Characterization, expression and antibacterial properties of apolipoproteins A from carp (Cyprinus carpio L.) seminal plasma. Fish Shellfish Immunol. 2014, 41, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Concha, M.I.; Molina, S.A.; Oyarzún, C.; Villanueva, J.; Amthauer, R. Local expression of apolipoprotein AI gene and a possible role for HDL in primary defence in the carp skin. Fish Shellfish Immunol. 2003, 14, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, J.; Yu, L.; Li, S. SNPs screening of 3′UTR in apoprotein A1-1 gene and its association with growth traits in grass carp. J. Dalian Ocean Univ. 2012, 27, 12–17. [Google Scholar]
- Bostock, J.; McAndrew, B.; Richards, R.; Jauncey, K.; Telfer, T.; Lorenzen, K.; Little, D.; Ross, L.; Handisyde, N.; Gatward, I.; et al. Aquaculture: Global status and trends. Philos. Trans. R. Soc. B 2010, 365, 2897–2912. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Li, J.T.; Zhang, X.F.; Sun, X.W. Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genom. 2012, 13, 96. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, X.F.; Wang, X.M.; Li, J.T.; Liu, G.M.; Kuang, Y.Y.; Xu, J.; Zheng, X.H.; Ren, L.F.; Wang, G.L.; et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Otis, J.P.; Zeituni, E.M.; Thierer, J.H.; Anderson, J.L.; Brown, A.C.; Boehm, E.D.; Cerchione, D.M.; Ceasrine, A.M.; Avraham-Davidi, I.; Tempelhof, H.; et al. Zebrafish as a model for apolipoprotein biology: Comprehensive expression analysis and a role for ApoA-IV in regulating food intake. Dis. Models Mech. 2015, 8, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, M.; Oku, T.; Yamada, S.; Komatsu, M.; Kudoh, K.; Itakura, T.; Ando, S. Isolation and characterization of some novel genes of the apolipoprotein A-I family in Japanese eel, Anguilla japonica. Cent. Eur. J. Biol. 2011, 6, 545–557. [Google Scholar] [CrossRef]
- Delcuve, G.P.; Sun, J.M.; Davie, J.R. Expression of rainbow-trout apolipoprotein-A-I genes in liver and hepatocellular-carcinoma. J. Lipid Res. 1992, 33, 251–262. [Google Scholar] [PubMed]
- Kim, K.Y.; Cho, Y.S.; Bang, I.C.; Nam, Y.K. Isolation and characterization of the apolipoprotein multigene family in Hemibarbus mylodon (Teleostei: Cypriniformes). Comp. Biochem. Phys. B 2009, 152, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Furlaneto, C.J.; Ribeiro, F.P.; Hatanaka, E.; Souza, G.M.; Cassatella, M.A.; Campa, A. Apolipoproteins A-I and A-II downregulate neutrophil functions. Lipids 2002, 37, 925–928. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.F.; Rader, D.J. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ. Res. 2005, 96, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Magnadóttir, B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.H.; Choi, S.H.; Baek, J.S.; Liu, C.; Almazan, F.; Ulrich, F.; Wiesner, P.; Taleb, A.; Deer, E.; Pattison, J.; et al. Control of angiogenesis by AIBP-mediated cholesterol efflux. Nature 2013, 498, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Yun, C.O.; Kwon, O.J.; Choi, E.J.; Song, J.Y.; Choi, I.; Cho, K.H. A proteoliposome containing apolipoprotein A-I mutant (V156K) enhances rapid tumor regression activity of human origin oncolytic adenovirus in tumor-bearing zebrafish and mice. Mol. Cells 2012, 34, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Babin, P.J.; Thisse, C.; Durliat, M.; Andre, M.; Akimenko, M.A.; Thisse, B. Both apolipoprotein E and AI genes are present in a nonmammalian vertebrate and are highly expressed during embryonic development. Proc. Natl. Acad. Sci. USA 1997, 94, 8622–8627. [Google Scholar] [CrossRef] [PubMed]
- Cunha, I.; Galante-Oliveira, S.; Rocha, E.; Urbatzka, R.; Castro, L.F.C. Expression of intercellular lipid transport and cholesterol metabolism genes in eggs and early larvae stages of turbot, Scophthalmus maximus, a marine aquaculture species. Mar. Biol. 2015, 162, 1673–1683. [Google Scholar] [CrossRef]
- Adlouni, A.; Ghalim, N.; Saile, R.; Hda, N.; Parra, H.J.; Benslimane, A. Beneficial effect on serum apo AI, apo B and Lp AI levels of Ramadan fasting. Clin. Chim. Acta 1998, 271, 179–189. [Google Scholar] [CrossRef]
- Akanji, A.O.; Mojiminiyi, O.A.; Abdella, N. Beneficial changes in serum apo A-1 and its ratio to apo B and HDL in stable hyperlipidaemic subjects after Ramadan fasting in Kuwait. Eur. J. Clin. Nutr. 2000, 54, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Afolabi, P.R.; Jahoor, F.; Jackson, A.A.; Stubbs, J.; Johnstone, A.M.; Faber, P.; Lobley, G.; Gibney, E.; Elia, M. The effect of total starvation and very low energy diet in lean men on kinetics of whole body protein and five hepatic secretory proteins. Am. J. Physiol.-Endocrinol. Metab. 2007, 293, E1580–E1589. [Google Scholar] [CrossRef] [PubMed]
- De-Santis, C.; Jerry, D.R. Candidate growth genes in finfish—Where should we be looking? Aquaculture 2007, 272, 22–38. [Google Scholar] [CrossRef]
- Tong, J.G.; Sun, X.W. Genetic and genomic analyses for economically important traits and their applications in molecular breeding of cultured fish. Sci. China Life Sci. 2015, 58, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.H.; Yu, X.M.; Tong, J.G. Polymorphisms in myostatin gene and associations with growth traits in the common carp (Cyprinus carpio L.). Int. J. Mol. Sci. 2012, 13, 14956–14961. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Yu, X.M.; Tong, J.G. Novel single nucleotide polymorphisms of the insulin-like growth factor-I gene and their associations with growth traits in common carp (Cyprinus carpio L.). Int. J. Mol. Sci. 2014, 15, 22471–22482. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Yu, X.M.; Pang, M.X.; Liu, H.Y.; Tong, J.G. Molecular characterization and expression of three preprosomatostatin genes and their association with growth in common carp (Cyprinus carpio). Comp. Biochem. Phys. B 2015, 182, 37–46. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Blum, S.; Feldman, M.W.; Lavi, U.; Hillel, J. Recent duplication of the, common carp (Cyprinus carpio L.) genome as revealed by analyses of microsatellite loci. Mol. Biol. Evol. 2003, 20, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates, Inc.: Sunderland, MA, USA, 1998; Volume 1, p. 980. [Google Scholar]
- Kuhnlein, U.; Ni, L.; Weigend, S.; Gavora, J.S.; Fairfull, W.; Zadworny, D. DNA polymorphisms in the chicken growth hormone gene: Response to selection for disease resistance and association with egg production. Anim. Genet. 1997, 28, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Tokuhiro, S.; Yamada, R.; Chang, X.T.; Suzuki, A.; Kochi, Y.; Sawada, T.; Suzuki, M.; Nagasaki, M.; Ohtsuki, M.; Ono, M.; et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet. 2003, 35, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Pesole, G.; Mignone, F.; Gissi, C.; Grillo, G.; Licciulli, F.; Liuni, S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 2001, 276, 73–81. [Google Scholar] [CrossRef]
- Fackenthal, J.D.; Olopade, O.I. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat. Rev. Cancer 2007, 7, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Ramensky, V.; Bork, P.; Sunyaev, S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002, 30, 3894–3900. [Google Scholar] [CrossRef] [PubMed]
- Nash, R.D.M.; Valencia, A.H.; Geffen, A.J. The origin of Fulton’s condition factor—Setting the record straight. Fisheries 2006, 31, 236–238. [Google Scholar]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; pp. 463–464. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method—A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Gene | Loci | Position | Genotypic Frequency | Allelic Frequency | HO | HE | PHW | ||
|---|---|---|---|---|---|---|---|---|---|
| apoA-Ib.1 | g.501A>T | 5′-UTR | TT | 0.053 | T | 0.321 | 0.537 | 0.438 | 0.034 |
| AA | 0.411 | A | 0.679 | ||||||
| AT | 0.537 | ||||||||
| g.1092A>T | Exon2 | TT | 0.325 | T | 0.575 | 0.525 | 0.492 | 0.648 | |
| AA | 0.163 | A | 0.425 | ||||||
| AT | 0.513 | ||||||||
| g.1114A>T | Exon2 | TT | 0.263 | T | 0.519 | 0.513 | 0.502 | 1.000 | |
| AA | 0.225 | A | 0.481 | ||||||
| AT | 0.513 | ||||||||
| g.1444G>A | Exon3 | AA | 0.673 | A | 0.811 | 0.276 | 0.308 | 0.323 | |
| GG | 0.051 | G | 0.189 | ||||||
| AG | 0.276 | ||||||||
| g.1500C>T | Exon3 | CC | 0.663 | C | 0.832 | 0.337 | 0.281 | 0.066 | |
| CT | 0.337 | T | 0.168 | ||||||
| g.1506G>A | Exon3 | AA | 0.490 | A | 0.719 | 0.459 | 0.241 | 0.219 | |
| GG | 0.051 | G | 0.281 | ||||||
| AG | 0.459 | ||||||||
| g.1518G>A | Exon3 | AA | 0.010 | A | 0.117 | 0.214 | 0.208 | 1.000 | |
| GG | 0.776 | G | 0.883 | ||||||
| AG | 0.214 | ||||||||
| g.1569A>C | Exon3 | AA | 0.898 | A | 0.949 | 0.102 | 0.097 | 1.000 | |
| AC | 0.102 | C | 0.051 | ||||||
| g.1687T>A | Exon3 | TT | 0.060 | T | 0.23 | 0.340 | 0.356 | 0.777 | |
| g.1689T>C | AA | 0.600 | A | 0.77 | |||||
| g.1693A>C | AT | 0.340 | |||||||
| g.1728C>T | Exon3 | CC | 0.663 | C | 0.832 | 0.337 | 0.281 | 0.067 | |
| CT | 0.337 | T | 0.168 | ||||||
| g.1746C>T | Exon3 | CC | 0.571 | C | 0.658 | 0.173 | 0.452 | 0.000 | |
| TT | 0.255 | T | 0.342 | ||||||
| CT | 0.173 | ||||||||
| g.1884C>T | Exon3 | CC | 0.837 | C | 0.908 | 0.163 | 0.168 | 0.577 | |
| TT | 0.010 | T | 0.092 | ||||||
| CT | 0.163 | ||||||||
| g.1961G>C | 3′-UTR | CC | 0.898 | C | 0.949 | 0.102 | 0.097 | 1.000 | |
| g.1966G>C | GC | 0.102 | G | 0.051 | |||||
| g.1985A>G | 3′-UTR | GG | 0.776 | G | 0.888 | 0.224 | 0.200 | 0.602 | |
| GA | 0.224 | A | 0.112 | ||||||
| g.1991C>T | 3′-UTR | CC | 0.133 | C | 0.352 | 0.439 | 0.459 | 0.667 | |
| TT | 0.429 | T | 0.648 | ||||||
| CT | 0.439 | ||||||||
| g.2004A>T | 3′-UTR | AA | 0.735 | A | 0.867 | 0.265 | 0.231 | 0.208 | |
| AT | 0.265 | T | 0.133 | ||||||
| g.2009C>G | 3'-UTR | GG | 0.612 | G | 0.694 | 0.163 | 0.427 | 0.000 | |
| CC | 0.224 | C | 0.306 | ||||||
| GC | 0.163 | ||||||||
| apoA-Ib.2 | g.183A>T | 5′-UTR | TT | 0.135 | T | 0.422 | 0.573 | 0.490 | 0.140 |
| AA | 0.292 | A | 0.578 | ||||||
| AT | 0.573 | ||||||||
| g.1604C>T | Exon3 | CC | 0.530 | C | 0.725 | 0.390 | 0.401 | 0.805 | |
| TT | 0.080 | T | 0.275 | ||||||
| CT | 0.390 | ||||||||
| g.1753C>T | Exon3 | CC | 0.380 | C | 0.64 | 0.520 | 0.463 | 0.279 | |
| TT | 0.100 | T | 0.36 | ||||||
| CT | 0.520 | ||||||||
| g.1767G>C | Exon3 | CC | 0.900 | C | 0.95 | 0.100 | 0.095 | 1.000 | |
| CG | 0.100 | G | 0.05 | ||||||
| g.1850C>A | Exon3 | CC | 0.770 | C | 0.885 | 0.230 | 0.205 | 0.353 | |
| CA | 0.230 | A | 0.115 | ||||||
| g.1862G>A | Exon3 | GG | 0.920 | G | 0.955 | 0.070 | 0.183 | 0.171 | |
| GA | 0.080 | A | 0.045 | ||||||
| g.1949G>C | Exon3 | GG | 0.060 | G | 0.305 | 0.490 | 0.426 | 0.161 | |
| CC | 0.450 | C | 0.695 | ||||||
| GC | 0.490 | ||||||||
| g.2077T>G | 3′-UTR | TT | 0.280 | T | 0.565 | 0.570 | 0.494 | 0.155 | |
| GG | 0.150 | G | 0.435 | ||||||
| TG | 0.570 | ||||||||
| Loci | Populations | Genotypes | n | BL (cm) | BW (g) | K |
|---|---|---|---|---|---|---|
| apoA-Ib.2-g.183 A>T | Yellow River carp | AA | 69 | 17.99 ± 0.15 A | 131.5 ± 2.55 A | 2.26 ± 0.024 |
| TT | 25 | 18.59 ± 0.21 A,B | 141.7 ± 3.61 A,B | 2.20 ± 0.034 | ||
| AT | 102 | 18.45 ± 0.14 B | 140.0 ± 2.36 B | 2.23 ± 0.022 | ||
| Yangtze River carp | AA | 176 | 24.4 ± 2.86 | 378.5 ± 118.7 a | 2.52 ± 0.24 | |
| AT | 21 | 25.6 ± 2.45 | 440.7 ± 122.3 b | 2.55 ± 0.20 | ||
| apoA-Ib.2-g.1753 C>T | Yellow River carp | CC | 82 | 18.42 ± 0.14 a | 139.8 ± 2.44 A | 2.23 ± 0.023 |
| TT | 21 | 18.17 ± 0.23 a,b | 134.6 ± 3.95 A,B | 2.24 ± 0.037 | ||
| CT | 93 | 18.30 ± 0.13 b | 136.4 ± 2.23 B | 2.22 ± 0.021 | ||
| Yangtze River carp | CC | 160 | 24.8 ± 2.68 A | 396.3 ± 115.7 A | 2.53 ± 0.24 | |
| CT | 40 | 23.4 ± 3.14 B | 341.3 ± 127.3 B | 2.56 ± 0.23 |
| Primer Name | Primer Sequence (Forward) | Primer Sequence (Reverse) | Usage |
|---|---|---|---|
| apoA-Ib-qPCR-1 | GAGCCGCCGTCGCAGGTGG | GAAGCCGTTCTGAAAGTAGCC | qRT-PCR |
| β-actin-qPCR | TATCCTATTGAGCACGGTATTG | CCTGTTGGCTTTGGGATTC | qRT-PCR |
| apoA-Ib.1-1 | TCTCTTCCCAGACCAGCTACAGTAAC | ATCTAAACATCCCTTTGACC | SNP identification and genotyping |
| apoA-Ib.1-2 | GCTCTTGCTTGTTTATGCC | TTGCGAGGATGTGTTAGTGT | SNP identification and genotyping |
| apoA-Ib.2-1 | TCTCTTCCCAGACCAGCTACAGTAAC | GCTGATGTTTTGACAGTGTTGAGAG | SNP identification and genotyping |
| apoA-Ib.2-2 | ACATCAGCATTGTTGTCTTC | GAGTGTATTGTGAGCAGGTGTG | SNP identification and genotyping |
| apoA-Ib.1-RFLP-1 | TCTCTTCCCAGACCAGCTACAGTAAC | TTTTTTTTGACGGTGTGGAGAGGATT | genotyping locus apoA-Ib.1-g.501 A>T |
| apoA-Ib.1-RFLP-2 | GCAGTTTTAGTTCTTTTATTGG | TTGCGAGGATGTGTTAGTGT | genotyping locus apoA-Ib.1-g.1693 A>C |
| apoA-Ib.2-RFLP-1 | ACATCAGCATTGTTGTCTTC | GAGCCTTGGTTCTTACTTG | genotyping locus apoA-Ib.2-g.183 A>T |
| apoA-Ib.2-RFLP-2 | ATGGCTACTTTCAGAACGGC | GAGTGTATTGTGAGCAGGTGTG | genotyping locus apoA-Ib.2-g.1753 C>T |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yu, X.; Tong, J. Molecular Characterization and Growth Association of Two Apolipoprotein A-Ib Genes in Common Carp (Cyprinus carpio). Int. J. Mol. Sci. 2016, 17, 1569. https://doi.org/10.3390/ijms17091569
Wang X, Yu X, Tong J. Molecular Characterization and Growth Association of Two Apolipoprotein A-Ib Genes in Common Carp (Cyprinus carpio). International Journal of Molecular Sciences. 2016; 17(9):1569. https://doi.org/10.3390/ijms17091569
Chicago/Turabian StyleWang, Xinhua, Xiaomu Yu, and Jingou Tong. 2016. "Molecular Characterization and Growth Association of Two Apolipoprotein A-Ib Genes in Common Carp (Cyprinus carpio)" International Journal of Molecular Sciences 17, no. 9: 1569. https://doi.org/10.3390/ijms17091569
APA StyleWang, X., Yu, X., & Tong, J. (2016). Molecular Characterization and Growth Association of Two Apolipoprotein A-Ib Genes in Common Carp (Cyprinus carpio). International Journal of Molecular Sciences, 17(9), 1569. https://doi.org/10.3390/ijms17091569