Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers
Abstract
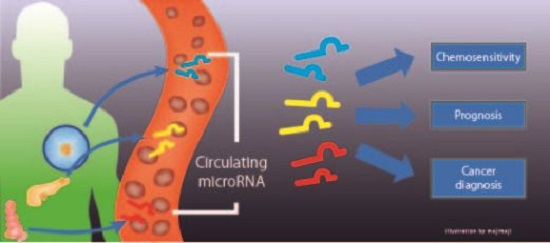
:1. Introduction
2. The Biology and Detection of Circulating miRNAs
3. Circulating miRNAs as Cancer Diagnostic Tools
3.1. Esophageal Cancer
3.2. Gastric Cancer
3.3. Colorectal Cancer
3.4. Hepatocellular Cancer
3.5. Pancreatic Cancer
4. Malignant Potential, Tumor Recurrence, and Prognostic Biomarkers
4.1. Esophageal Cancer (ECa)
4.2. Gastric Cancer (GC)
4.3. Colorectal Cancer (CRC)
4.4. Hepatocellular Carcinoma (HCC)
4.5. Pancreatic Cancer (PCa)
5. Predicting Tool for Tumor Chemosensitivity
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Chim, S.S.; Shing, T.K.; Hung, E.C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Pigati, L.; Yaddanapudi, S.C.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, D.; Komatsu, S.; Konishi, H.; Otsuji, E. Circulating microRNA in digestive tract cancers. Gastroenterology 2012, 142, 1074–1078. [Google Scholar] [CrossRef] [PubMed]
- Etheridge, A.; Lee, I.; Hood, L.; Galas, D.; Wang, K. Extracellular microRNA: A new source of biomarkers. Mutat. Res. 2011, 717, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Tsujiura, M.; Ichikawa, D.; Konishi, H.; Komatsu, S.; Shiozaki, A.; Otsuji, E. Liquid biopsy of gastric cancer patients: Circulating tumor cells and cell-free nucleic acids. World J. Gastroenterol. 2014, 20, 3265–3286. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Das, S. Profiling cell-free and circulating miRNA: A clinical diagnostic tool for different cancers. Tumour Biol. 2016, 37, 5705–5714. [Google Scholar] [CrossRef] [PubMed]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sultmann, H. Serum microRNAs as non-invasive biomarkers for cancer. Mol. Cancer 2010, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, C.; Chen, X.; Yang, C.; Li, K.; Wang, J.; Dai, J.; Hu, Z.; Zhou, X.; Chen, L.; et al. Expression profile of microRNAs in serum: A fingerprint for esophageal squamous cell carcinoma. Clin. Chem. 2010, 56, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Morimura, R.; Nagata, H.; Kosuga, T.; Iitaka, D.; Konishi, H.; Shiozaki, A.; et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer 2011, 105, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liao, J.; Yang, M.; Shi, Y.; Peng, Y.; Wang, Y.; Pan, E.; Guo, W.; Pu, Y.; Yin, L. Circulating miR-155 expression in plasma: A potential biomarker for early diagnosis of esophageal cancer in humans. J. Toxicol. Environ. Health 2012, 75, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Hirajima, S.; Kawaguchi, T.; Miyamae, M.; Okajima, W.; Ohashi, T.; Arita, T.; Konishi, H.; Shiozaki, A.; et al. Plasma microRNA profiles: Identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br. J. Cancer 2014, 111, 1614–1624. [Google Scholar] [CrossRef] [PubMed]
- Hirajima, S.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Konishi, H.; Shiozaki, A.; Morimura, R.; Tsujiura, M.; Nagata, H.; Kawaguchi, T.; et al. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 108, 1822–1829. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, N.; Hoshino, I.; Mori, M.; Akutsu, Y.; Hanari, N.; Yoneyama, Y.; Ikeda, N.; Isozaki, Y.; Maruyama, T.; Akanuma, N.; et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 108, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Dong, S.; Dong, C.; Sun, K.; Meng, W.; Lv, P.; Yin, H.; Ming, L.; He, F. Predictive value of plasma miRNA-718 for esophageal squamous cell carcinoma. Cancer Biomark. 2016, 16, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Chiam, K.; Wang, T.; Watson, D.I.; Mayne, G.C.; Irvine, T.S.; Bright, T.; Smith, L.; White, I.A.; Bowen, J.M.; Keefe, D.; et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J. Gastrointest. Surg. 2015, 19, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Tsujiura, M.; Ichikawa, D.; Komatsu, S.; Shiozaki, A.; Takeshita, H.; Kosuga, T.; Konishi, H.; Morimura, R.; Deguchi, K.; Fujiwara, H.; et al. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer 2010, 102, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Ichikawa, D.; Komatsu, S.; Shiozaki, A.; Tsujiura, M.; Takeshita, H.; Morimura, R.; Nagata, H.; Arita, T.; Kawaguchi, T.; et al. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br. J. Cancer 2012, 106, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Tsujiura, M.; Komatsu, S.; Ichikawa, D.; Shiozaki, A.; Konishi, H.; Takeshita, H.; Moriumura, R.; Nagata, H.; Kawaguchi, T.; Hirajima, S.; et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer 2015, 18, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, L.; Liu, B.; Yang, L.; Meng, X.; Zhang, W.; Ma, Y.; Xiao, H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012, 316, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Cui, L.; Sun, W.; Zhou, H.; Yuan, X.; Huo, M.; Chen, J.; Lou, Y.; Guo, J. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer Biomark. 2011, 10, 71–77. [Google Scholar] [PubMed]
- Valladares-Ayerbes, M.; Reboredo, M.; Medina-Villaamil, V.; Iglesias-Diaz, P.; Lorenzo-Patino, M.J.; Haz, M.; Santamarina, I.; Blanco, M.; Fernandez-Tajes, J.; Quindos, M.; et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J. Transl. Med. 2012, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xiao, B.; Zhou, F.; Deng, H.; Zhang, X.; Lou, Y.; Gong, Z.; Du, C.; Guo, J. MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers 2012, 17, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, J.F.; Cai, Q.; Qiu, Q.Q.; Yan, M.; Liu, B.Y.; Zhu, Z.G. MiRNA-199a-3p: A potential circulating diagnostic biomarker for early gastric cancer. J. Surg. Oncol. 2013, 108, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ge, X.; Zhang, Y.; Xia, H.; Yuan, D.; Tang, Q.; Chen, L.; Pang, X.; Leng, W.; Bi, F. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol. Rep. 2014, 31, 1863–1870. [Google Scholar] [PubMed]
- Fu, Z.; Qian, F.; Yang, X.; Jiang, H.; Chen, Y.; Liu, S. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med. Oncol. 2014, 31, 164. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.X.; Zhao, J.; Rong, Z.H.; Wu, Y.G.; Geng, W.M.; Qin, C.K. Diagnostic and prognostic value of circulating miR-18a in the plasma of patients with gastric cancer. Tumour Biol. 2014, 35, 12119–12125. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ren, C.; Han, J.; Ding, Y.; Du, J.; Dai, N.; Dai, J.; Ma, H.; Hu, Z.; Shen, H.; et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br. J. Cancer 2014, 110, 2291–2299. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kwong, A.; Sihoe, A.; Chu, K.M. Plasma miR-940 may serve as a novel biomarker for gastric cancer. Tumour Biol. 2016, 37, 3589–3597. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, G.; Wang, Z.; Yao, Y.; Chen, R.; Pu, X.; Wang, J. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis. Mark. 2015, 2015, 435656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, Y.; Zhang, C.; Zhi, X.; Fu, H.; Ma, Y.; Chen, Y.; Pan, F.; Wang, K.; Ni, J.; et al. Circulating miR-16–5p and miR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics 2015, 5, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Z.; Wang, C.J.; Niu, L.; Xu, J.; Wang, M.; Cao, H.; Hu, B. Analysis of plasma microRNAs to identifying early diagnostic molecule for gastric cancer. Int. J. Clin. Exp. Med. 2015, 8, 3700–3706. [Google Scholar] [PubMed]
- Ng, E.K.; Chong, W.W.; Jin, H.; Lam, E.K.; Shin, V.Y.; Yu, J.; Poon, T.C.; Ng, S.S.; Sung, J.J. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009, 58, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zhang, L.; Cogdell, D.E.; Zheng, H.; Schetter, A.J.; Nykter, M.; Harris, C.C.; Chen, K.; Hamilton, S.R.; Zhang, W. Circulating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE 2011, 6, e17745. [Google Scholar] [CrossRef] [PubMed]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef] [PubMed]
- Zanutto, S.; Pizzamiglio, S.; Ghilotti, M.; Bertan, C.; Ravagnani, F.; Perrone, F.; Leo, E.; Pilotti, S.; Verderio, P.; Gariboldi, M.; et al. Circulating miR-378 in plasma: A reliable, haemolysis-independent biomarker for colorectal cancer. Br. J. Cancer 2014, 110, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kosaka, N.; Tanaka, M.; Koizumi, F.; Kanai, Y.; Mizutani, T.; Murakami, Y.; Kuroda, M.; Miyajima, A.; Kato, T.; et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 2009, 14, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Tian, Y.; Wen, X.; Zhang, W.; Zhang, P.; Gao, J.; Run, W.; Tian, L.; Jia, X.; Gao, Y. Serum microRNA characterization identifies miR-885–5p as a potential marker for detecting liver pathologies. Clin. Sci. 2011, 120, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Cheng, S.Q.; Wang, H.; Li, N.; Chen, Y.F.; Gao, C.F. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS ONE 2011, 6, e28486. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.M.; Yao, T.J.; Wang, W.; Wong, K.F.; Lee, N.P.; Fan, S.T.; Poon, R.T.; Gao, C.; Luk, J.M. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: A retrospective cohort study. BMJ Open 2012, 2, e000825. [Google Scholar] [CrossRef] [PubMed]
- Tomimaru, Y.; Eguchi, H.; Nagano, H.; Wada, H.; Kobayashi, S.; Marubashi, S.; Tanemura, M.; Tomokuni, A.; Takemasa, I.; Umeshita, K.; et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J. Hepatol. 2012, 56, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wei, X.; Tang, C.; Li, J.; Liu, R.; Shen, A.; Wu, Z. Circulating microRNA-101 as a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Oncol. Lett. 2013, 6, 1811–1815. [Google Scholar] [PubMed]
- Luo, J.; Chen, M.; Huang, H.; Yuan, T.; Zhang, M.; Zhang, K.; Deng, S. Circulating microRNA-122a as a diagnostic marker for hepatocellular carcinoma. Onco Targets Ther. 2013, 6, 577–583. [Google Scholar] [PubMed]
- Shen, J.; Wang, A.; Wang, Q.; Gurvich, I.; Siegel, A.B.; Remotti, H.; Santella, R.M. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: MiR-483-5p as a potential biomarker. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2364–2373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ge, S.; Wang, X.; Yuan, Q.; Yan, Q.; Ye, H.; Che, Y.; Lin, Y.; Zhang, J.; Liu, P. Serum miR-483-5p as a potential biomarker to detect hepatocellular carcinoma. Hepatol. Int. 2013, 7, 199–207. [Google Scholar] [CrossRef] [PubMed]
- El-Garem, H.; Ammer, A.; Shehab, H.; Shaker, O.; Anwer, M.; El-Akel, W.; Omar, H. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J. Hepatol. 2014, 6, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Giray, B.G.; Emekdas, G.; Tezcan, S.; Ulger, M.; Serin, M.S.; Sezgin, O.; Altintas, E.; Tiftik, E.N. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol. Biol. Rep. 2014, 41, 4513–4519. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yao, Q.; Butt, A.M.; Guo, J.; Tian, Z.; Bao, X.; Li, H.; Meng, Q.; Lu, J. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol. Ther. 2014, 15, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Meng, H.; Wang, N.; Liang, L.N.; Liu, L.N.; Lu, S.M.; Luan, Y. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn. Pathol. 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Dhayat, S.A.; Husing, A.; Senninger, N.; Schmidt, H.H.; Haier, J.; Wolters, H.; Kabar, I. Circulating microRNA-200 family as diagnostic marker in hepatocellular carcinoma. PLoS ONE 2015, 10, e0140066. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Ferracin, M.; Trere, D.; Milazzo, M.; Marinelli, S.; Galassi, M.; Venerandi, L.; Pollutri, D.; Patrizi, C.; Borghi, A.; et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS ONE 2015, 10, e0141448. [Google Scholar] [CrossRef] [PubMed]
- Motawi, T.K.; Shaker, O.G.; El-Maraghy, S.A.; Senousy, M.A. Serum microRNAs as potential biomarkers for early diagnosis of hepatitis C virus-related hepatocellular carcinoma in Egyptian patients. PLoS ONE 2015, 10, e0137706. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Lin, B.; Ye, Y.; Wen, D.; Chen, L.; Zhou, X. Differential expression of serum microRNAs in cirrhosis that evolve into hepatocellular carcinoma related to hepatitis B virus. Oncol. Rep. 2015, 33, 2863–2870. [Google Scholar] [PubMed]
- Wang, Y.; Gao, Y.; Shi, W.; Zhai, D.; Rao, Q.; Jia, X.; Liu, J.; Jiao, X.; Du, Z. Profiles of differential expression of circulating microRNAs in hepatitis B virus-positive small hepatocellular carcinoma. Cancer Biomark. 2015, 15, 171–180. [Google Scholar] [PubMed]
- Ghosh, A.; Ghosh, A.; Datta, S.; Dasgupta, D.; Das, S.; Ray, S.; Gupta, S.; Datta, S.; Chowdhury, A.; Chatterjee, R.; et al. Hepatic miR-126 is a potential plasma biomarker for detection of hepatitis B virus infected hepatocellular carcinoma. Int. J. Cancer 2016, 138, 2732–2744. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.H.; Hu, T.H.; Lu, S.N.; Kuo, F.Y.; Chen, C.H.; Wang, J.H.; Huang, C.M.; Lee, C.M.; Lin, C.Y.; Yen, Y.H.; et al. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int. J. Cancer 2016, 138, 714–720. [Google Scholar] [CrossRef]
- Kong, X.; Du, Y.; Wang, G.; Gao, J.; Gong, Y.; Li, L.; Zhang, Z.; Zhu, J.; Jing, Q.; Qin, Y.; et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: MiR-196a could be a potential marker for poor prognosis. Dig. Dis. Sci. 2011, 56, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Morimura, R.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Nagata, H.; Konishi, H.; Shiozaki, A.; Ikoma, H.; Okamoto, K.; et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br. J. Cancer 2011, 105, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Komatsu, S.; Ichikawa, D.; Morimura, R.; Tsujiura, M.; Konishi, H.; Takeshita, H.; Nagata, H.; Arita, T.; Hirajima, S.; et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br. J. Cancer 2013, 108, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Miyamae, M.; Kawaguchi, T.; Morimura, R.; Hirajima, S.; Okajima, W.; Ohashi, T.; Imamura, T.; Konishi, H.; et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. Expert Opin. Biol. Ther. 2015, 15, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Miyamae, M.; Komatsu, S.; Ichikawa, D.; Kawaguchi, T.; Hirajima, S.; Okajima, W.; Ohashi, T.; Imamura, T.; Konishi, H.; Shiozaki, A.; et al. Plasma microRNA profiles: Identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br. J. Cancer 2015, 113, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, X.; Du, Y.; Yao, W.; Shen, L.; Wang, C.; Hu, Z.; Zhuang, R.; Ning, G.; Zhang, C.; et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin. Chem. 2012, 58, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Yu, J.; Kim, H.; Wolfgang, C.L.; Canto, M.I.; Hruban, R.H.; Goggins, M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin. Cancer Res. 2013, 19, 3600–3610. [Google Scholar] [CrossRef] [PubMed]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, B.; Yue, S.; Galli, U.; Rana, S.; Gross, W.; Muller, M.; Giese, N.A.; Kalthoff, H.; Becker, T.; Buchler, M.W.; et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer 2015, 136, 2616–2627. [Google Scholar] [CrossRef] [PubMed]
- Abue, M.; Yokoyama, M.; Shibuya, R.; Tamai, K.; Yamaguchi, K.; Sato, I.; Tanaka, N.; Hamada, S.; Shimosegawa, T.; Sugamura, K.; et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int. J. Oncol. 2015, 46, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Ganepola, G.A.; Rutledge, J.R.; Suman, P.; Yiengpruksawan, A.; Chang, D.H. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J. Gastrointest. Oncol. 2014, 6, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.S.; Liu, L.X.; Li, G.P.; Chen, Y.; Li, C.Y.; Jin, D.Y.; Wang, X.L. Combined serum CA19–9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev. Res. 2013, 6, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Takeshita, H.; Konishi, H.; Nagata, H.; Hirajima, S.; Kawaguchi, T.; Arita, T.; Shiozaki, A.; Fujiwara, H.; et al. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin. Biol. Ther. 2012, 12, S53–S59. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Yu, Q.; Shi, Z.L.; Li, P.; Fu, S. Circulating microRNAs in esophageal squamous cell carcinoma: Association with locoregional staging and survival. Int. J. Clin. Exp. Med. 2015, 8, 7241–7250. [Google Scholar] [PubMed]
- Odenthal, M.; Hee, J.; Gockel, I.; Sisic, L.; Schmitz, J.; Stoecklein, N.H.; Driemel, C.; Mohlendick, B.; Schmidt, T.; Knoefel, W.T.; et al. Serum microRNA profiles as prognostic/predictive markers in the multimodality therapy of locally advanced adenocarcinomas of the gastroesophageal junction. Int. J. Cancer 2015, 137, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Wei, Y.; Su, L.; Liu, G.; Kulke, M.H.; Wain, J.C.; Christiani, D.C. Whole-miRNome profiling identifies prognostic serum miRNAs in esophageal adenocarcinoma: The influence of Helicobacter pylori infection status. Carcinogenesis 2015, 36, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Tsujiura, M.; Konishi, H.; Takeshita, H.; Nagata, H.; Kawaguchi, T.; Hirajima, S.; Arita, T.; Shiozaki, A.; et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013, 33, 271–276. [Google Scholar] [PubMed]
- Imaoka, H.; Toiyama, Y.; Okigami, M.; Yasuda, H.; Saigusa, S.; Ohi, M.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M. Circulating microRNA-203 predicts metastases, early recurrence, and poor prognosis in human gastric cancer. Gastric Cancer 2016, 19, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Shivapurkar, N.; Weiner, L.M.; Marshall, J.L.; Madhavan, S.; Deslattes Mays, A.; Juhl, H.; Wellstein, A. Recurrence of early stage colon cancer predicted by expression pattern of circulating microRNAs. PLoS ONE 2014, 9, e84686. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Okugawa, Y.; Ide, S.; Imaoka, H.; Boland, C.R.; Goel, A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2015, 65. [Google Scholar] [CrossRef] [PubMed]
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Koberle, V.; Kronenberger, B.; Pleli, T.; Trojan, J.; Imelmann, E.; Peveling-Oberhag, J.; Welker, M.W.; Elhendawy, M.; Zeuzem, S.; Piiper, A.; et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur. J. Cancer 2013, 49, 3442–3449. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Kim, J.K.; Nam, J.S.; Wang, H.J.; Lee, J.H.; Kim, B.W.; Kim, S.S.; Noh, C.K.; Shin, S.J.; Lee, K.M.; et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis B virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin. Biochem. 2015, 48, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.; Xu, L.; Wang, P.; Meng, Z. Serum miR-128–2 serves as a prognostic marker for patients with hepatocellular carcinoma. PLoS ONE 2015, 10, e0117274. [Google Scholar] [CrossRef] [PubMed]
- Kjersem, J.B.; Ikdahl, T.; Lingjaerde, O.C.; Guren, T.; Tveit, K.M.; Kure, E.H. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 2014, 8, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Miyata, H.; Sugimura, K.; Fukuda, S.; Kanemura, T.; Yamashita, K.; Miyazaki, Y.; Takahashi, T.; Kurokawa, Y.; Yamasaki, M.; et al. miR-27 is associated with chemoresistance in esophageal cancer through transformation of normal fibroblasts to cancer-associated fibroblasts. Carcinogenesis 2015, 36, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Miyata, H.; Yamasaki, M.; Sugimura, K.; Takahashi, T.; Kurokawa, Y.; Nakajima, K.; Takiguchi, S.; Mori, M.; Doki, Y. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann. Surg. Oncol. 2013, 20, S607–S615. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Kawaguchi, T.; Miyamae, M.; Okajima, W.; Ohashi, T.; Imamura, T.; Kiuchi, J.; Konishi, H.; Shiozaki, A.; et al. Circulating miR-21 as an independent predictive biomarker for chemoresistance in esophageal squamous cell carcinoma. Am. J. Cancer Res. 2016, 6, 1511–1523. [Google Scholar] [PubMed]
- Wang, P.; Zhuang, L.; Zhang, J.; Fan, J.; Luo, J.; Chen, H.; Wang, K.; Liu, L.; Chen, Z.; Meng, Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol. Oncol. 2013, 7, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Yu, X.J.; Guo, X.Z.; Sun, M.H.; Wang, Z.; Song, Y.; Ni, Q.X.; Li, H.Y.; Mukaida, N.; Li, Y.Y. MicroRNA-33a-mediated downregulation of PIM-3 kinase expression renders human pancreatic cancer cells sensitivity to gemcitabine. Oncotarget 2015, 6, 14440–14455. [Google Scholar] [CrossRef] [PubMed]
| Cancer Type | Expression Level 1 | Cancer Diagnostic Tools | Malignant Potential, Tumor Recurrence, And Prognostic Biomarker | Predicting Tool For Chemosensitivity | References |
|---|---|---|---|---|---|
| Esophegeal cancer | High | miR-10a, miR-22, miR-100, miR-127, miR-133a, miR-148b, miR-223, miR-21, miR-155, miR-18a, miR-1246, miR-16, miR-25, miR-320a, let-7e, miR-15b | miR-21, miR-16, miR-1246, miR-192, miR-222, miR-3935, miR-4286 | miR-200c, miR-27a/b, miR-21 | [13,14,15,16,17,18,19,20,74,75,76,77,87,88,89] |
| Low | miR-375, miR-718, miR-30a/miR-324, miR-17/miR-194 | miR-375, miR-302c | - | ||
| Gastric cancer | High | miR-17-5p, miR-21, miR-106a/b, miR-18a, miR-378, miR-451, miR-486, miR-21, miR-200c, miR-421, miR-199a, miR-122, miR-192, miR-222, miR-16, miR-25, miR-92a, miR-940, miR-223, miR-19b, miR-194 | miR-200c, miR-21, miR-222, miR-18a | - | [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,78,79] |
| Low | miR-141, miR-1233 | miR-203 | - | ||
| Colorectal cancer | High | miR-92a, miR-141, let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, miR-23a, miR-378 | miR-141, miR-320, miR-596, miR-203 | miR-106a, miR-484, miR-130b | [37,38,39,40,80,81,86] |
| Low | - | miR-15a, miR-103, miR-148a, miR451 | - | ||
| Hepatocellular Cancer | High | miR-500, miR-885, miR-122, miR-21, miR-223, miR-15b, miR130b, miR-101, miR-483, miR-125, miR-143, miR-215, miR-939, miR-595 | miR-718, miR-128-2 | - | [41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,82,83,84,85] |
| Low | miR-200, miR-19a, miR-195 | miR-1, miR-122 | - | ||
| Pancreatic cancer | High | miR-21, miR-196a, miR-18a, miR-221, miR-223, miR-20a, miR-24, miR-25, miR-99a, miR-185, miR-191, miR-1290, miR-27a, miR-642b, miR-885-5p, miR-22, miR-21, miR-483, miR-1246, miR-4644, miR-3976, and miR-4306 | miR-196a, miR-21, miR-221, miR-744 | miR-744 | [62,63,64,65,66,67,68,69,70,71,72,73,89,90] |
| Low | miR-375 | - | - |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, T.; Komatsu, S.; Ichikawa, D.; Tsujiura, M.; Takeshita, H.; Hirajima, S.; Miyamae, M.; Okajima, W.; Ohashi, T.; Imamura, T.; et al. Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers. Int. J. Mol. Sci. 2016, 17, 1459. https://doi.org/10.3390/ijms17091459
Kawaguchi T, Komatsu S, Ichikawa D, Tsujiura M, Takeshita H, Hirajima S, Miyamae M, Okajima W, Ohashi T, Imamura T, et al. Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers. International Journal of Molecular Sciences. 2016; 17(9):1459. https://doi.org/10.3390/ijms17091459
Chicago/Turabian StyleKawaguchi, Tsutomu, Shuhei Komatsu, Daisuke Ichikawa, Masahiro Tsujiura, Hiroki Takeshita, Shoji Hirajima, Mahito Miyamae, Wataru Okajima, Takuma Ohashi, Taisuke Imamura, and et al. 2016. "Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers" International Journal of Molecular Sciences 17, no. 9: 1459. https://doi.org/10.3390/ijms17091459
APA StyleKawaguchi, T., Komatsu, S., Ichikawa, D., Tsujiura, M., Takeshita, H., Hirajima, S., Miyamae, M., Okajima, W., Ohashi, T., Imamura, T., Kiuchi, J., Konishi, H., Shiozaki, A., Okamoto, K., & Otsuji, E. (2016). Circulating MicroRNAs: A Next-Generation Clinical Biomarker for Digestive System Cancers. International Journal of Molecular Sciences, 17(9), 1459. https://doi.org/10.3390/ijms17091459