A WDR Gene Is a Conserved Member of a Chitin Synthase Gene Cluster and Influences the Cell Wall in Aspergillus nidulans
Abstract
:1. Introduction
2. Results
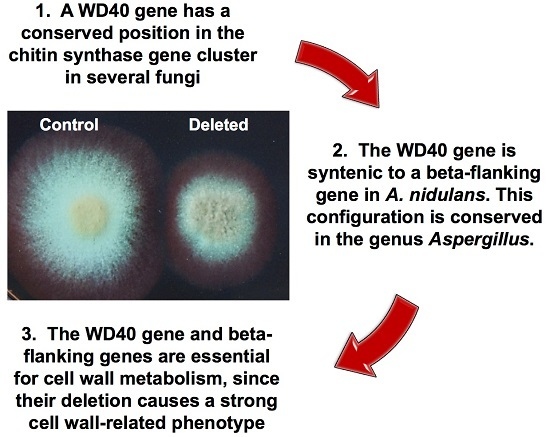
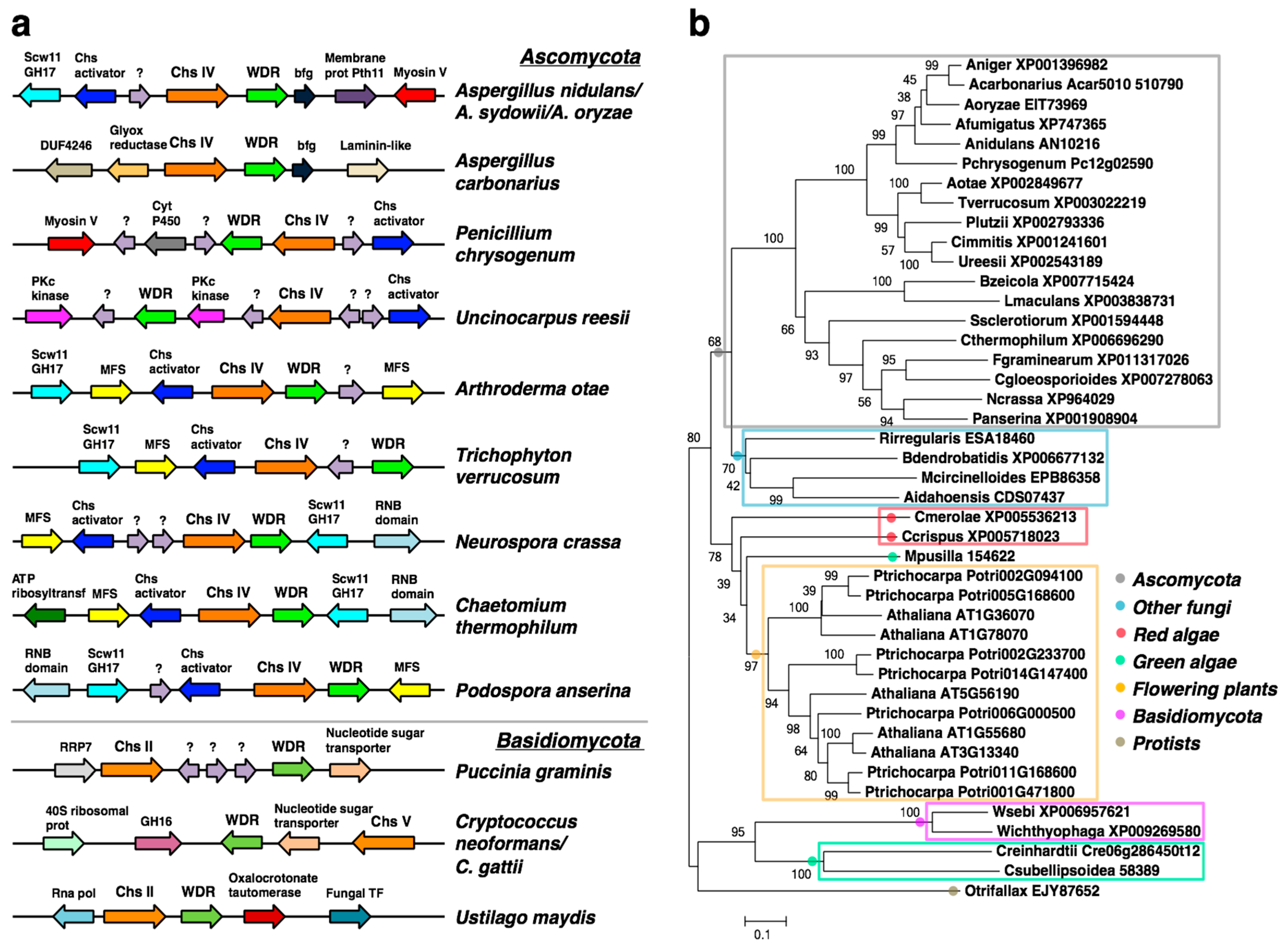
2.1. A WDR Gene Is a Conserved Neighbor of chs Genes in Fungi
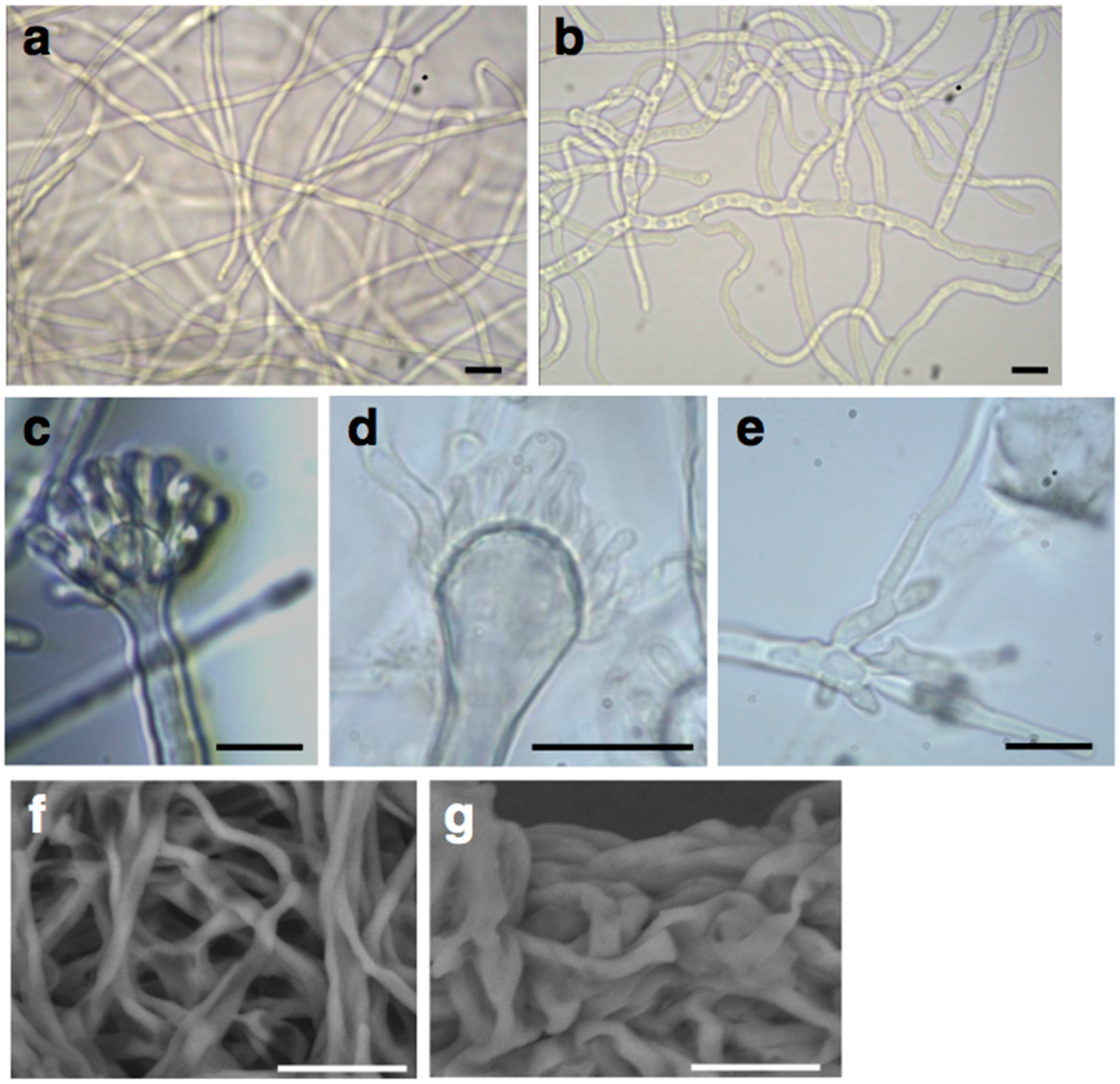
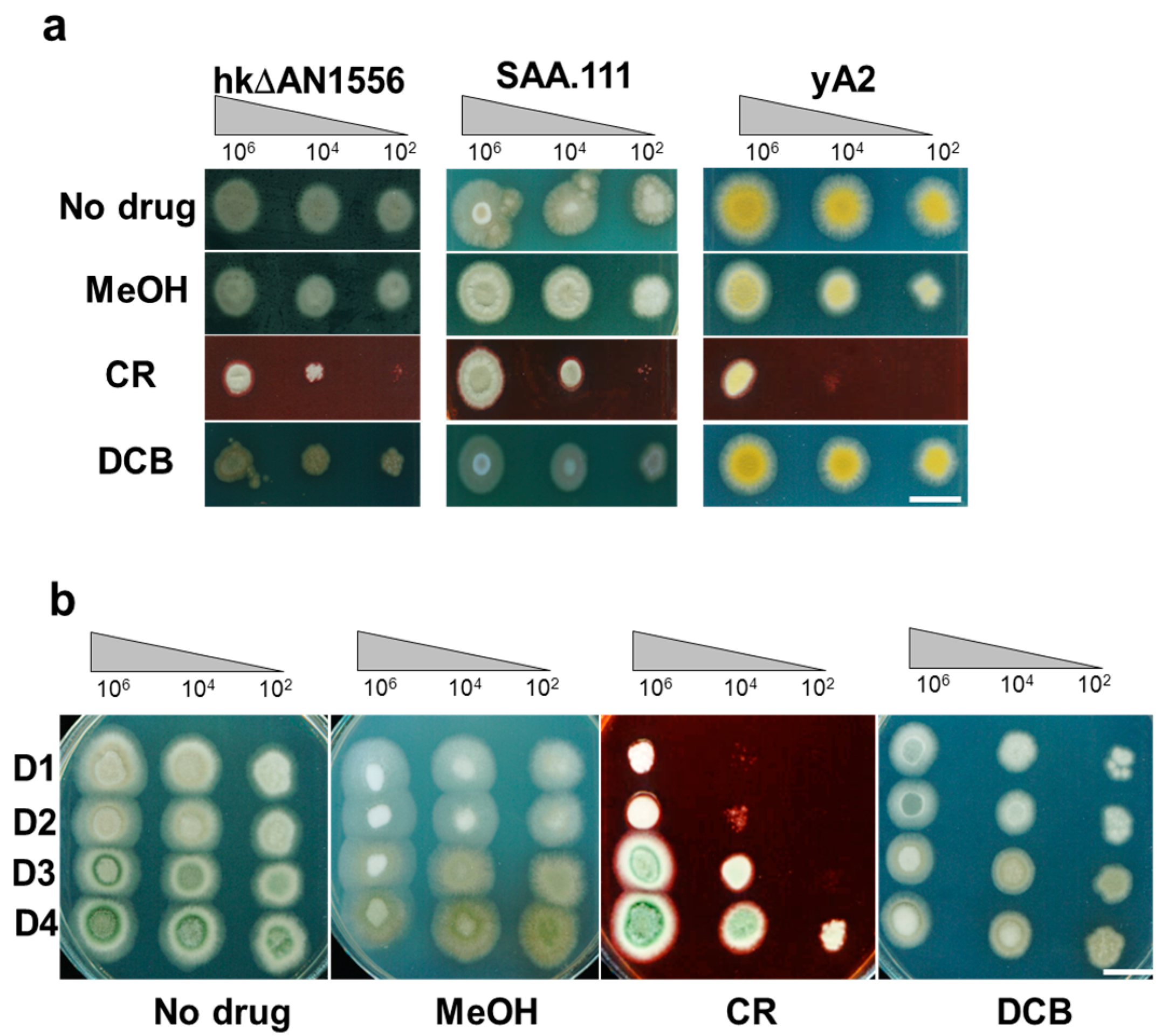
2.2. Deletion of the AN1556.2 Locus Alters Growth and Cell Wall Properties
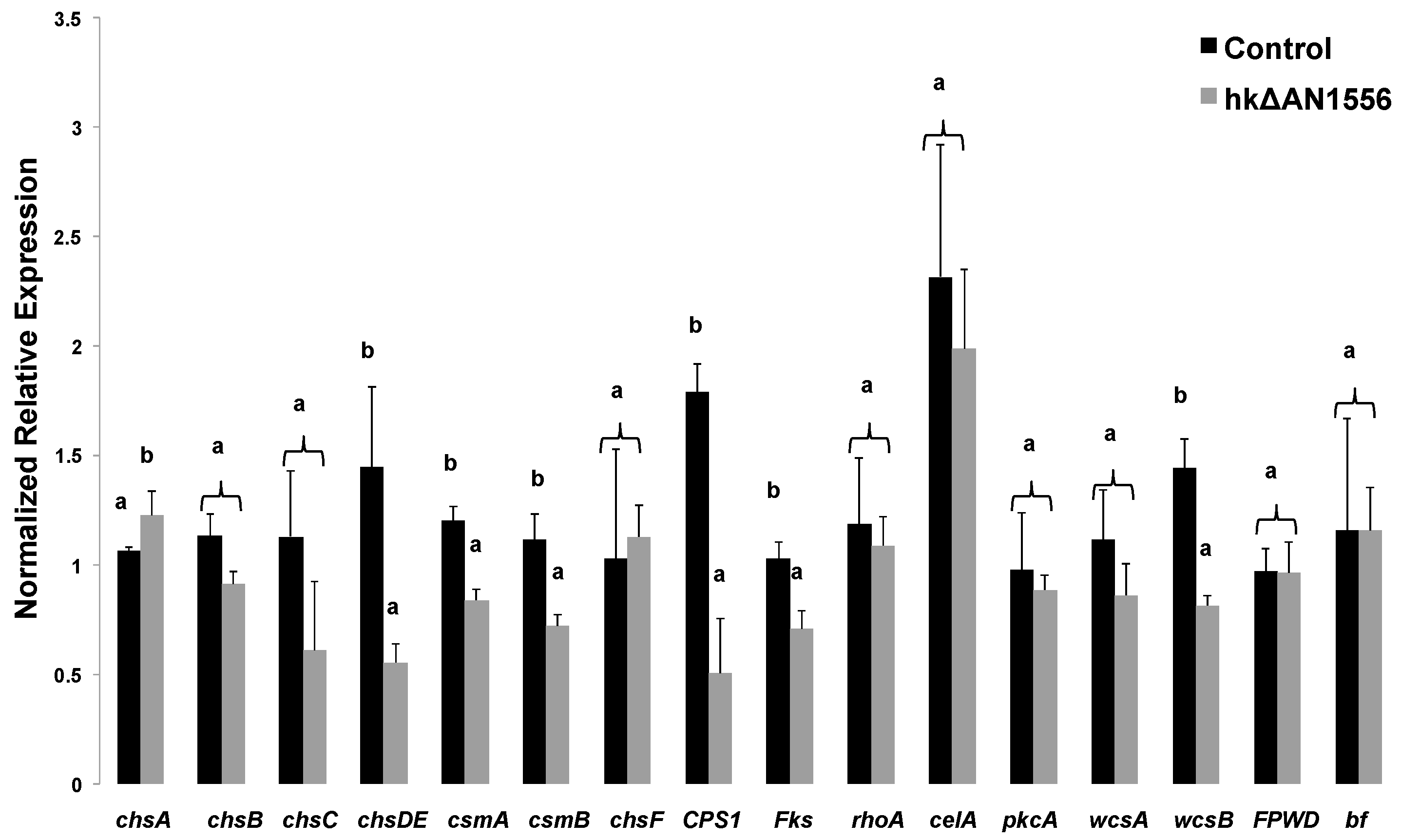
2.3. Expression of Cell Wall-Related Genes
2.4. Genetic Analyses of hkΔAN1556
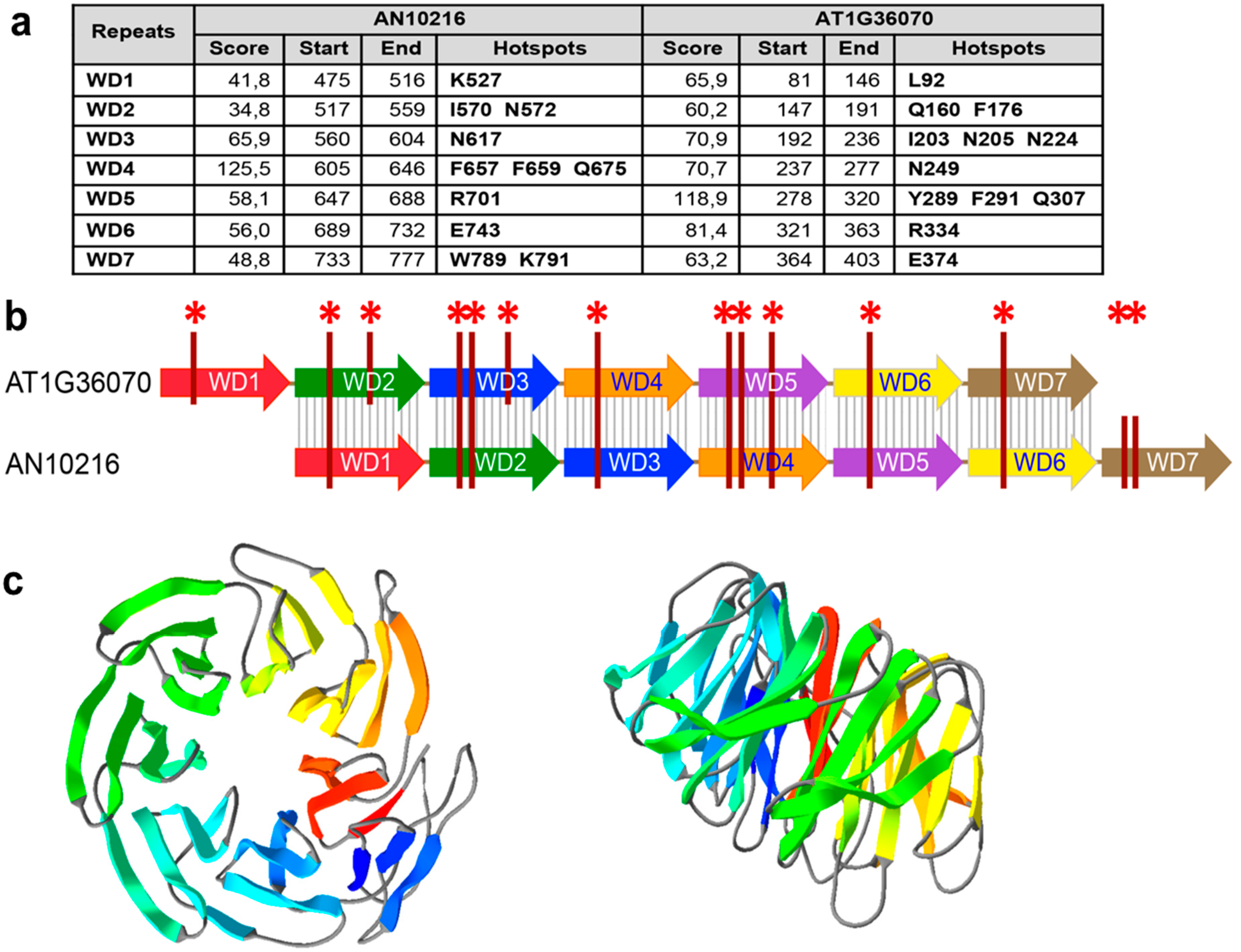
2.5. Bioinformatic Analyses of the bf Gene in the AN1556.2 Locus
3. Discussion
4. Experimental Section
4.1. Bioinformatic Analyses of AN10216
4.2. Fungal Cultivation, Preparation of Gene Replacement Cassette and Transformation
4.3. Southern Blotting and PCR of Genomic DNA
4.4. Growth Tests
4.5. Optical, Confocal, Scanning Electron and Atomic Force Microscopy of A. nidulans Mycelia
4.6. RNA Extraction, cDNA Synthesis and qPCR
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stirnimann, C.U.; Petsalaki, E.; Russell, R.B.; Müller, C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010, 35, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Pöggeler, S.; Kück, U. A WD40 repeat protein regulates fungal cell differentiation and can be replaced functionally by the mammalian homologue striatin. Eukaryot. Cell 2004, 3, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Espagne, E.; Balhadère, P.; Penin, M.L.; Barreau, C.; Turcq, B. HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina. Genetics 2002, 16, 71–81. [Google Scholar]
- Chiu, Y.-H.; Xian, X.; Dawe, A.L.; Morris, N.R. Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol. Biol. Cell 1997, 8, 1735–1749. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Berndt, P.; Xia, X.; Kahnt, J.; Kahmann, R. A seven-WD40 protein related to human RACK1 regulates mating and virulence in Ustilago maydis. Mol. Microbiol. 2011, 81, 1484–1498. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Hausman, J.F.; Ezcurra, I. WD40-repeat proteins in plant cell wall formation: Current evidence and research prospects. Front. Plant Sci. 2015, 6, 1112. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Spadiut, O.; Kerschbamer, C.; Giorno, F.; Baric, S.; Ezcurra, I. Analysis of cellulose synthase genes from domesticated apple identifies collinear genes WDR53 and CesA8A: Partial co-expression, bicistronic mRNA, and alternative splicing of CESA8A. J. Exp. Bot. 2012, 63, 6045–6056. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Arjona, J.R.; Ramirez-Prado, J.H. Large-scale phylogenetic classification of fungal chitin synthases and identification of a putative cell-wall metabolism gene cluster in Aspergillus genomes. PLoS ONE 2014, 9, e104920. [Google Scholar]
- Guerriero, G.; Silvestrini, L.; Obersriebnig, M.; Salerno, M.; Pum, D.; Strauss, J. Sensitivity of Aspergillus nidulans to the cellulose synthase inhibitor dichlobenil: Insights from wall-related genes’ expression and ultrastructural hyphal morphologies. PLoS ONE 2013, 8, e80038. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T.; Denness, L. Cell wall integrity maintenance in plants: Lessons to be learned from yeast? Plant Signal. Behav. 2011, 6, 1706–1709. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. 2011 Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- De Groot, P.W.; Brandt, B.W.; Horiuchi, H.; Ram, A.F.; de Koster, C.G.; Klis, F.M. Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 2009, 46, S72–S81. [Google Scholar] [CrossRef] [PubMed]
- Teepe, A.G.; Loprete, D.M.; He, Z.; Hoggard, T.A.; Hill, T.W. The protein kinase C orthologue PkcA plays a role in cell wall integrity and polarized growth in Aspergillus nidulans. Fungal Genet. Biol. 2007, 44, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Ichinomiya, M.; Uchida, H.; Koshi, Y.; Ohta, A.; Horiuchi, H. A protein kinase C-encoding gene, pkcA, is essential to the viability of the filamentous fungus Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007, 71, 2787–2799. [Google Scholar] [CrossRef] [PubMed]
- Futagami, T.; Nakao, S.; Kido, Y.; Oka, T.; Kajiwara, Y.; Takashita, H.; Omori, T.; Furukawa, K.; Goto, M. Putative stress sensors WscA and WscB are involved in hypo-osmotic and acidic pH stress tolerance in Aspergillus nidulans. Eukaryot. Cell 2011, 10, 1504–1515. [Google Scholar] [CrossRef] [PubMed]
- Futagami, T.; Goto, M. Putative cell wall integrity sensor proteins in Aspergillus nidulans. Commun. Integr. Biol. 2012, 5, 206–208. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, E.B.; Timberlake, W.E. Molecular characterization of the Aspergillus nidulans yA locus. Genetics 1989, 121, 249–254. [Google Scholar] [PubMed]
- Cerqueira, G.C.; Arnaud, M.B.; Inglis, D.O.; Skrzypek, M.S.; Binkley, G.; Simison, M.; Miyasato, S.R.; Binkley, J.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database: Multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014, 42, D705–D710. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.B.; Cerqueira, G.C.; Inglis, D.O.; Skrzypek, M.S.; Binkley, J.; Chibucos, M.C.; Crabtree, J.; Howarth, C.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database (AspGD): Recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012, 40, D653–D659. [Google Scholar] [CrossRef] [PubMed]
- Kües, U.; James, T.Y.; Vilgalys, R.; Challen, M.P. The chromosomal region containing pab-1, mip, and the A mating type locus of the secondarily homothallic basidiomycete Coprinus bilanatus. Curr. Genet. 2001, 39, 16–24. [Google Scholar] [PubMed]
- Walsh, A.J.; Martin, T.; di Domenico, T.; Vullo, A.; Pollastri, G.; Tosatto, S.C. CSpritz: Accurate prediction of protein disorder segments with annotation for homology, secondary structure and linear motifs. Nucleic Acids Res. 2011, 39, W190–W196. [Google Scholar] [CrossRef] [PubMed]
- Dinkel, H.; Michael, S.; Weatheritt, R.J.; Davey, N.E.; van Roey, K.; Altenberg, B.; Toedt, G.; Uyar, B.; Seiler, M.; Budd, A.; et al. ELM—The database of eukaryotic linear motifs. Nucleic Acids Res. 2012, 40, D242–D251. [Google Scholar] [CrossRef] [PubMed]
- Gerstenberger, J.P.; Occhipinti, P.; Gladfelter, A.S. Heterogeneity in mitochondrial morphology and membrane potential is independent of the nuclear division cycle in multinucleate fungal cells. Eukaryot. Cell 2012, 11, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Nützmann, H.-W.; Osbourn, A. Regulation of metabolic gene clusters in Arabidopsis thaliana. New Phytol. 2015, 205, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Bashline, L.; Li, S.; Zhu, X.; Gu, Y. The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 12870–12875. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, F.; Zhuo, Z.; Wu, X.H.; Wu, Y.D. A method for WD40 repeat detection and secondary structure prediction. PLoS ONE 2013, 8, e65705. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.H.; Wang, Y.; Zhuo, Z.; Jiang, F.; Wu, Y.D. Identifying the hotspots on the top faces of WD40-repeat proteins from their primary sequences by β-bulges and DHSW tetrads. PLoS ONE 2012, 7, e43005. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Bernreiter, A.; Ramon, A.; Fernández-Martínez, J.; Berger, H.; Araújo-Bazan, L.; Espeso, E.A.; Pachlinger, R.; Gallmetzer, A.; Anderl, I.; Scazzocchio, C.; et al. Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol. Cell. Biol. 2007, 27, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Nayak, T.; Szewczyk, E.; Oakley, C.E.; Osmani, A.; Ukil, L.; Murray, S.L.; Hynes, M.J.; Osmani, S.A.; Oakley, B.R. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 2006, 172, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Davis, M.A.; Hynes, M.J. Genetic manipulation of Aspergillus nidulans: Heterokaryons and diploids for dominance, complementation and haploidization analyses. Nat. Protoc. 2007, 2, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Church, G.M.; Gilbert, W. Genomic sequencing. Proc. Natl. Acad. Sci. USA 1984, 81, 1991–1995. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Legay, S.; Hausman, J.F.; Guerriero, G. Analysis of cell wall-related genes in organs of Medicago sativa L. under different abiotic stresses. Int. J. Mol. Sci. 2015, 16, 16104–16124. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerriero, G.; Silvestrini, L.; Obersriebnig, M.; Hausman, J.-F.; Strauss, J.; Ezcurra, I. A WDR Gene Is a Conserved Member of a Chitin Synthase Gene Cluster and Influences the Cell Wall in Aspergillus nidulans. Int. J. Mol. Sci. 2016, 17, 1031. https://doi.org/10.3390/ijms17071031
Guerriero G, Silvestrini L, Obersriebnig M, Hausman J-F, Strauss J, Ezcurra I. A WDR Gene Is a Conserved Member of a Chitin Synthase Gene Cluster and Influences the Cell Wall in Aspergillus nidulans. International Journal of Molecular Sciences. 2016; 17(7):1031. https://doi.org/10.3390/ijms17071031
Chicago/Turabian StyleGuerriero, Gea, Lucia Silvestrini, Michael Obersriebnig, Jean-Francois Hausman, Joseph Strauss, and Inés Ezcurra. 2016. "A WDR Gene Is a Conserved Member of a Chitin Synthase Gene Cluster and Influences the Cell Wall in Aspergillus nidulans" International Journal of Molecular Sciences 17, no. 7: 1031. https://doi.org/10.3390/ijms17071031
APA StyleGuerriero, G., Silvestrini, L., Obersriebnig, M., Hausman, J.-F., Strauss, J., & Ezcurra, I. (2016). A WDR Gene Is a Conserved Member of a Chitin Synthase Gene Cluster and Influences the Cell Wall in Aspergillus nidulans. International Journal of Molecular Sciences, 17(7), 1031. https://doi.org/10.3390/ijms17071031