Screening of α-Tocopherol Transfer Protein Sensitive Genes in Human Hepatoma Cells (HepG2)
Abstract
:1. Introduction
2. Results
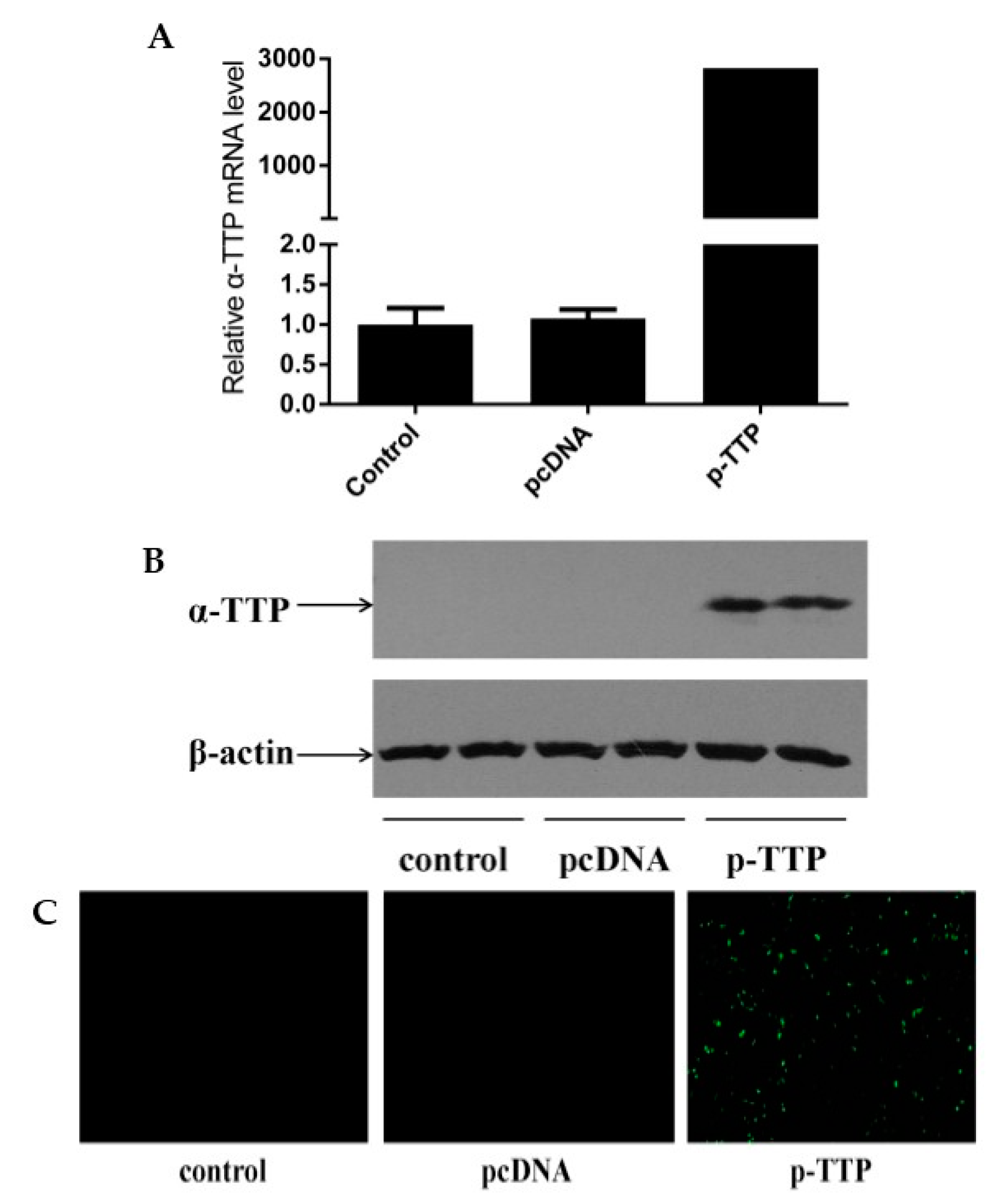
2.1. Expression of Human α-Tocopherol Transfer Protein (α-TTP) Eukaryotic Expression Vector in Human Hepatoma Cells (HepG2)
2.2. Screening Genes Related to α-TTP
2.3. Verification of the Transport-Related Genes
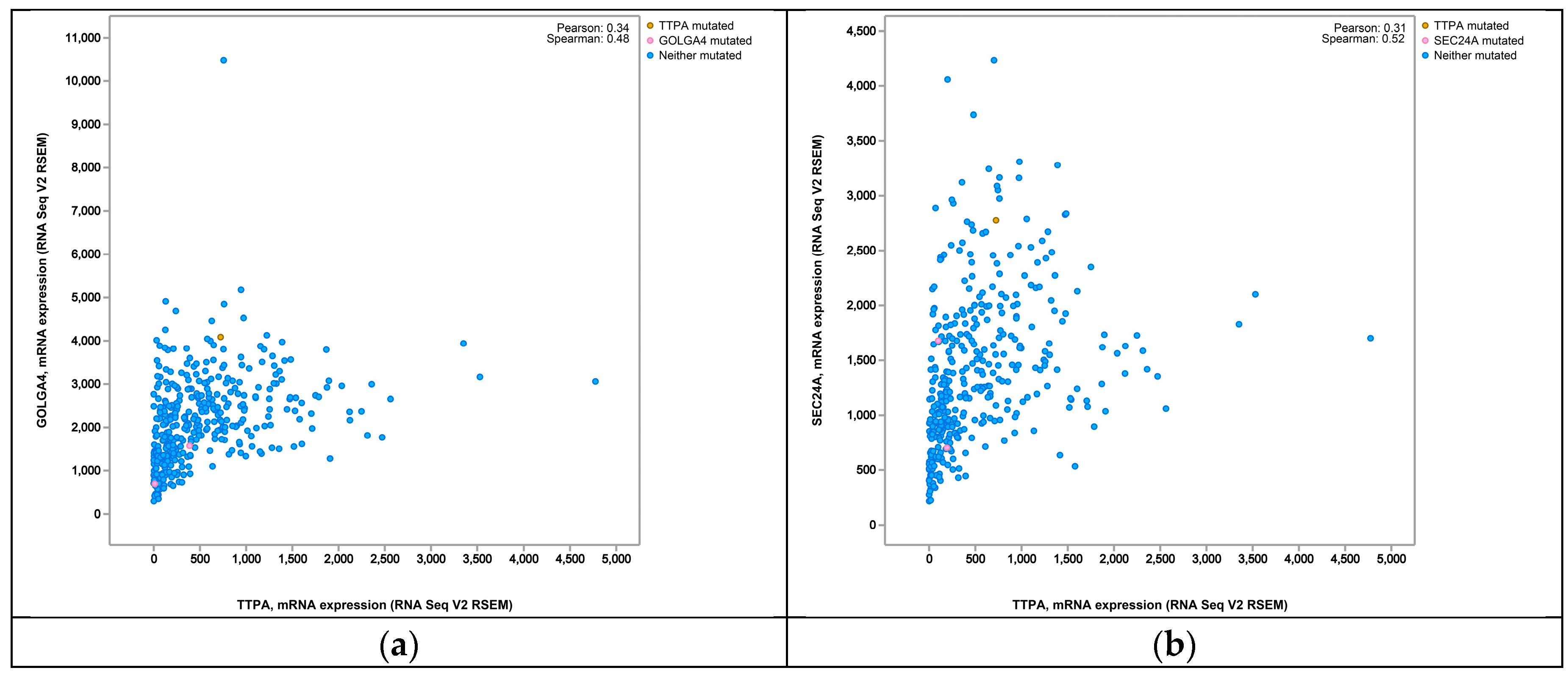
3. Discussion
4. Materials and Methods
4.1. Construction of a Human Eukaryotic Expression Vector
4.2. Expression of Human α-TTP Eukaryotic Expression Vector in HepG2 Cells
4.3. RNA Extraction, Microarray Analysis and Statistics
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| α-TTP | α-tocopherol transfer protein |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| SEC23A | sec23 homolog A |
| RTP4 | receptor transporter protein 4 |
| CLIC3 | chloride intracellular channel 3 |
| CENPE | centromere protein E |
| KCNQ1 | potassium voltage-gated channel, KQT-like subfamily |
| GOLGA4 | golgi autoantigen, golgin subfamily a, 4 |
| BNIP3 | bcl2/adenovirus E1B 19 kDa interacting protein 3 |
| CENPF | centormere protein F, 350/400 ka (mitosin) |
References
- Evans, H.M.; Bishop, K.S. On the existence of a hitherto unrecognized dietary factor essential for reproduction. Science 1922, 56, 650–651. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.W.; Joyce, A.; Ingold, K.U. Is vitamin E the only lipid-soluble, chain-breaking antioxidant in human blood plasma and erythrocyte membranes? Arch. Biochem. Biophys. 1983, 221, 281–290. [Google Scholar] [CrossRef]
- Schneider, C. Chemistry and biology of vitamin E. Mol. Nutr. Food Res. 2005, 49, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Zingg, J.M. Modulation of signal transduction by vitamin E. Mol. Asp. Med. 2007, 28, 481–506. [Google Scholar] [CrossRef] [PubMed]
- Azzi, A.; Gysin, R.; Kempna, P.; Munteanu, A.; Negis, Y.; Villacorta, L.; Visarius, T.; Zingg, J.M. Vitamin E mediates cell signaling and regulation of gene expression. Ann. N. Y. Acad. Sci. 2004, 1031, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Azzi, A. Molecular mechanism of α-tocopherol action. Free Radic. Biol. Med. 2007, 43, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Kono, N.; Arai, H. Intracellular transport of fat-soluble vitamins A and E. Traffic 2015, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Catignani, G.L. An α-tocopherol binding protein in rat liver cytoplasm. Biochem. Biophys. Res. Commun. 1975, 67, 66–72. [Google Scholar] [CrossRef]
- Qian, J.; Morley, S.; Wilson, K.; Nava, P.; Atkinson, J.; Manor, D. Intracellular trafficking of vitamin E in hepatocytes: The role of tocopherol transfer protein. J. Lipid Res. 2005, 46, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Hagiwara, K.; Arai, H.; Inoue, K. Purification and characterization of the α-tocopherol transfer protein from rat liver. FEBS Lett. 1991, 288, 41–45. [Google Scholar] [CrossRef]
- Arita, M.; Sato, Y.; Miyata, A.; Tanabe, T.; Takahashi, E.; Kayden, H.J.; Arai, H.; Inoue, K. Human α-tocopherol transfer protein: cDNA cloning, expression and chromosomal localization. Biochem. J. 1995, 306, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Arai, H.; Miyata, A.; Tokita, S.; Yamamoto, K.; Tanabe, T.; Inoue, K. Primary structure of α-tocopherol transfer protein from rat liver. Homology with cellular retinaldehyde-binding protein. J. Biol. Chem. 1993, 268, 17705–17710. [Google Scholar] [PubMed]
- Ouahchi, K.; Arita, M.; Kayden, H.; Hentati, F.; Ben, H.M.; Sokol, R.; Arai, H.; Inoue, K.; Mandel, J.L.; Koenig, M. Ataxia with isolated vitamin E deficiency is caused by mutations in the α-tocopherol transfer protein. Nat. Genet. 1995, 9, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Jishage, K.; Arita, M.; Igarashi, K.; Iwata, T.; Watanabe, M.; Ogawa, M.; Ueda, O.; Kamada, N.; Inoue, K.; Arai, H.; et al. Α-tocopherol transfer protein is important for the normal development of placental labyrinthine trophoblasts in mice. J. Biol. Chem. 2001, 276, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Arai, H.; Arita, M.; Sato, Y.; Ogihara, T.; Inoue, K.; Mino, M.; Tamai, H. Effect of α-tocopherol status on α-tocopherol transfer protein expression and its messenger RNA level in rat liver. Free Radic. Res. 1998, 28, 87–92. [Google Scholar] [PubMed]
- Shaw, H.M.; Huang, C. Liver α-tocopherol transfer protein and its mRNA are differentially altered by dietary vitamin E deficiency and protein insufficiency in rats. J. Nutr. 1998, 128, 2348–2354. [Google Scholar] [PubMed]
- Fechner, H.; Schlame, M.; Guthmann, F.; Stevens, P.A.; Rustow, B. α- and δ-tocopherol induce expression of hepatic α-tocopherol-transfer-protein mRNA. Biochem. J. 1998, 331 Pt 2, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Barella, L.; Muller, P.Y.; Schlachter, M.; Hunziker, W.; Stocklin, E.; Spitzer, V.; Meier, N.; de Pascual-Teresa, S.; Minihane, A.M.; Rimbach, G. Identification of hepatic molecular mechanisms of action of α-tocopherol using global gene expression profile analysis in rats. Biochim. Biophys. Acta 2004, 1689, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Li, Y.J.; Wang, M.S.; Kang, Z.C.; Huang, H.L.; Shaw, H.M. Elevation of tissue α-tocopherol levels by conjugated linoleic acid in C57BL/6J mice is not associated with changes in vitamin E absorption or α-carboxyethyl hydroxychroman production. Nutrition 2012, 28, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Lauridsen, C.; Theil, P.K.; Jensen, S.R.K. Composition of α-tocopherol and fatty acids in porcine tissues after dietary supplementation with vitamin E and different fat sources. Anim. Feed Sci. Technol. 2013, 179, 93–102. [Google Scholar] [CrossRef]
- Thakur, V.; Morley, S.; Manor, D. Hepatic α-tocopherol transfer protein: Ligand-induced protection from proteasomal degradation. Biochemistry 2010, 49, 9339–9344. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Luo, H.L.; Yue, D.B.; Ge, S.Y.; Yuan, F.; Yan, L.Y.; Jia, H.N. Molecular cloning and characterization of the sheep α-TTP gene and its expression in response to different vitamin E status. Gene 2012, 494, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Luo, H.L.; Zuo, Z.Y.; Jia, H.N.; Zhang, Y.W.; Chang, Y.F.; Jiao, L.J. Regulation of sheep α-TTP by dietary vitamin E and preparation of monoclonal antibody for sheep α-TTP. Gene 2014, 540, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zuo, Z.Y.; Jia, H.N.; Luo, H.L. Screening of Ovine α-TTP interacting proteins by using Co-immunoprecipitation coupled with Mass spectrometry. Chin. J. Anim. Sci. 2014, 50, 80–83. [Google Scholar]
- Zuo, Z.Y.; Luo, H.L.; Liu, K.; Jia, H.N.; Zhang, Y.W.; Jiao, L.J.; Chang, Y.F. Dietary vitamin E affects α-TTP mRNA levels in different tissues of the Tan sheep. Gene 2014, 541, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Manor, D.; Morley, S. The α-tocopherol transfer protein. Vitam. Horm. 2007, 76, 45–65. [Google Scholar] [PubMed]
- Schock, B.C.; van der Vliet, A.; Corbacho, A.M.; Leonard, S.W.; Finkelstein, E.; Valacchi, G.; Obermueller-Jevic, U.; Cross, C.E.; Traber, M.G. Enhanced inflammatory responses in α-tocopherol transfer protein null mice. Arch. Biochem. Biophys. 2004, 423, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Vasu, V.T.; Hobson, B.; Gohil, K.; Cross, C.E. Genome-wide screening of α-tocopherol sensitive genes in heart tissue from α-tocopherol transfer protein null mice (ATTP−/−). FEBS Lett. 2007, 581, 1572–1578. [Google Scholar] [CrossRef] [PubMed]
- Sedy, J.; Bekiaris, V.; Ware, C.F. Tumor necrosis factor superfamily in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2015, 7, a016279. [Google Scholar] [CrossRef] [PubMed]
- Eddleston, J.; Christiansen, S.C.; Zuraw, B.L. Functional expression of the C–X–C chemokine receptor CXCR4 by human bronchial epithelial cells: Regulation by proinflammatory mediators. J. Immunol. 2002, 169, 6445–6451. [Google Scholar] [CrossRef] [PubMed]
- Fillmore, R.A.; Mitra, A.; Xi, Y.; Ju, J.; Scammell, J.; Shevde, L.A.; Samant, R.S. Nmi (N-mycinteractor) inhibits wnt/β-catenin signaling and retards tumor growth. Int. J. Cancer 2009, 125, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Gadient, R.A.; Patterson, P.H. Leukemia inhibitory factor, Interleukin 6, and other cytokines using the GP130 transducing receptor: Roles in inflammation and injury. Stem Cells 1999, 17, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.R.; Zhang, C. Ran GTPase: A master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 2001, 11, 366–371. [Google Scholar] [CrossRef]
- Stolz, A.; Ertych, N.; Kienitz, A.; Vogel, C.; Schneider, V.; Fritz, B.; Jacob, R.; Dittmar, G.; Weichert, W.; Petersen, I.; et al. The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat. Cell Biol. 2010, 12, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.C.; Lu, T.-Y.; Lin, C.-T.; Kao, C.-F.; Wu, H.-C.; Yeh, N.-H. NOLC1, a transcription factor, synergistically regulates MDM2 expression with TP53 to promote the progression of nasopharyngeal carcinoma. Cancer Res. 2008, 68, LB-170. [Google Scholar]
- Park, M.H. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J. Biochem. 2006, 139, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Hubner, A.; Cavanagh-Kyros, J.; Rincon, M.; Flavell, R.A.; Davis, R.J. Functional cooperation of the proapoptotic Bcl2 family proteins Bmf and Bim in vivo. Mol. Cell. Biol. 2009, 30, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.H.; Hu, J.R.; Wang, L.Y.; Hu, Y.J.; Tan, F.Q.; Zhou, H.; Shao, J.Z.; Yang, W.X. The apoptotic function analysis of p53, Apaf1, Caspase3 and Caspase7 during the spermatogenesis of the Chinese fire-bellied newt Cynops orientalis. PLoS ONE 2012, 7, e39920. [Google Scholar] [CrossRef] [PubMed]
- Gohil, K.; Schock, B.C.; Chakraborty, A.A.; Terasawa, Y.; Raber, J.; Farese, R.J.; Packer, L.; Cross, C.E.; Traber, M.G. Gene expression profile of oxidant stress and neurodegeneration in transgenic mice deficient in α-tocopherol transfer protein. Free Radic. Biol. Med. 2003, 35, 1343–1354. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Yoshida, N.; Manabe, H.; Terasawa, Y.; Takemura, T.; Kondo, M. α-Tocopherol protects against expression of adhesion molecules on neutrophils and endothelial cells. Biofactors 1998, 7, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Gysin, R.; Azzi, A.; Visarius, T. γ-Tocopherol inhibits human cancer cell cycle progression and cell proliferation by down-regulation of cyclins. FASEB J. 2002, 16, 1952–1954. [Google Scholar] [CrossRef] [PubMed]
- Hodul, P.J.; Dong, Y.; Husain, K.; Pimiento, J.M.; Chen, J.; Zhang, A.; Francois, R.; Pledger, W.J.; Coppola, D.; Sebti, S.M.; et al. Vitamin E δ-tocotrienol induces p27KIP1-dependent cell-cycle arrest in pancreatic cancer cells via an E2F-1-dependent mechanism. PLoS ONE 2013, 8, e52526. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1: Mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 2000, 88, 1474–1480. [Google Scholar] [PubMed]
- Ulatowski, L.; Dreussi, C.; Noy, N.; Barnholtz-Sloan, J.; Klein, E.; Manor, D. Expression of the α-tocopherol transfer protein gene is regulated by oxidative stress and common single-nucleotide polymorphisms. Free Radic. Biol. Med. 2012, 53, 2318–2326. [Google Scholar] [CrossRef] [PubMed]
- Ban, R.; Takitani, K.; Kim, H.S.; Murata, T.; Morinobu, T.; Ogihara, T.; Tamai, H. α-Tocopherol transfer protein expression in rat liver exposed to hyperoxia. Free Radic. Res. 2002, 36, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Goubau, D.; Romieu-Mourez, R.; Solis, M.; Hernandez, E.; Mesplede, T.; Lin, R.; Leaman, D.; Hiscott, J. Transcriptional re-programming of primary macrophages reveals distinct apoptotic and anti-tumoral functions of IRF-3 and IRF-7. Eur. J. Immunol. 2009, 39, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Mesmin, B.; Bigay, J.; Moser, V.F.J.; Lacas-Gervais, S.; Drin, G.; Antonny, B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 2013, 155, 830–843. [Google Scholar] [CrossRef] [PubMed]
- Boyadjiev, S.A.; Fromme, J.C.; Ben, J.; Chong, S.S.; Nauta, C.; Hur, D.J.; Zhang, G.; Hamamoto, S.; Schekman, R.; Ravazzola, M.; et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat. Genet. 2006, 38, 1192–1197. [Google Scholar] [CrossRef] [PubMed]
- Erlich, R.; Gleeson, P.A.; Campbell, P.; Dietzsch, E.; Toh, B.H. Molecular characterization of trans-Golgi p230. A human peripheral membrane protein encoded by a gene on chromosome 6p12-22 contains extensive coiled-coil α-helical domains and a granin motif. J. Biol. Chem. 1996, 271, 8328–8337. [Google Scholar] [PubMed]
- cBioPortal for Cancer Genomics. Available online: http://www.cbioportal.org/ (accessed on 21 December 2015).
- Valenzuela, S.M.; Martin, D.K.; Por, S.B.; Robbins, J.M.; Warton, K.; Bootcov, M.R.; Schofield, P.R.; Campbell, T.J.; Breit, S.N. Molecular cloning and expression of a chloride ion channel of cell nuclei. J. Biol. Chem. 1997, 272, 12575–12582. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Guo, J.; Li, N.; Qian, M.; Wang, S.N.; Zhu, X.L. Mitosin/CENP-F is a conserved kinetochore protein subjected to cytoplasmic dynein-mediated poleward transport. Cell Res. 2003, 13, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.N.; Olesen, S.P.; Rasmussen, H.B. Kv7.1 surface expression is regulated by epithelial cell polarization. Am. J. Physiol. Cell Physiol. 2011, 300, C814–C824. [Google Scholar] [CrossRef] [PubMed]
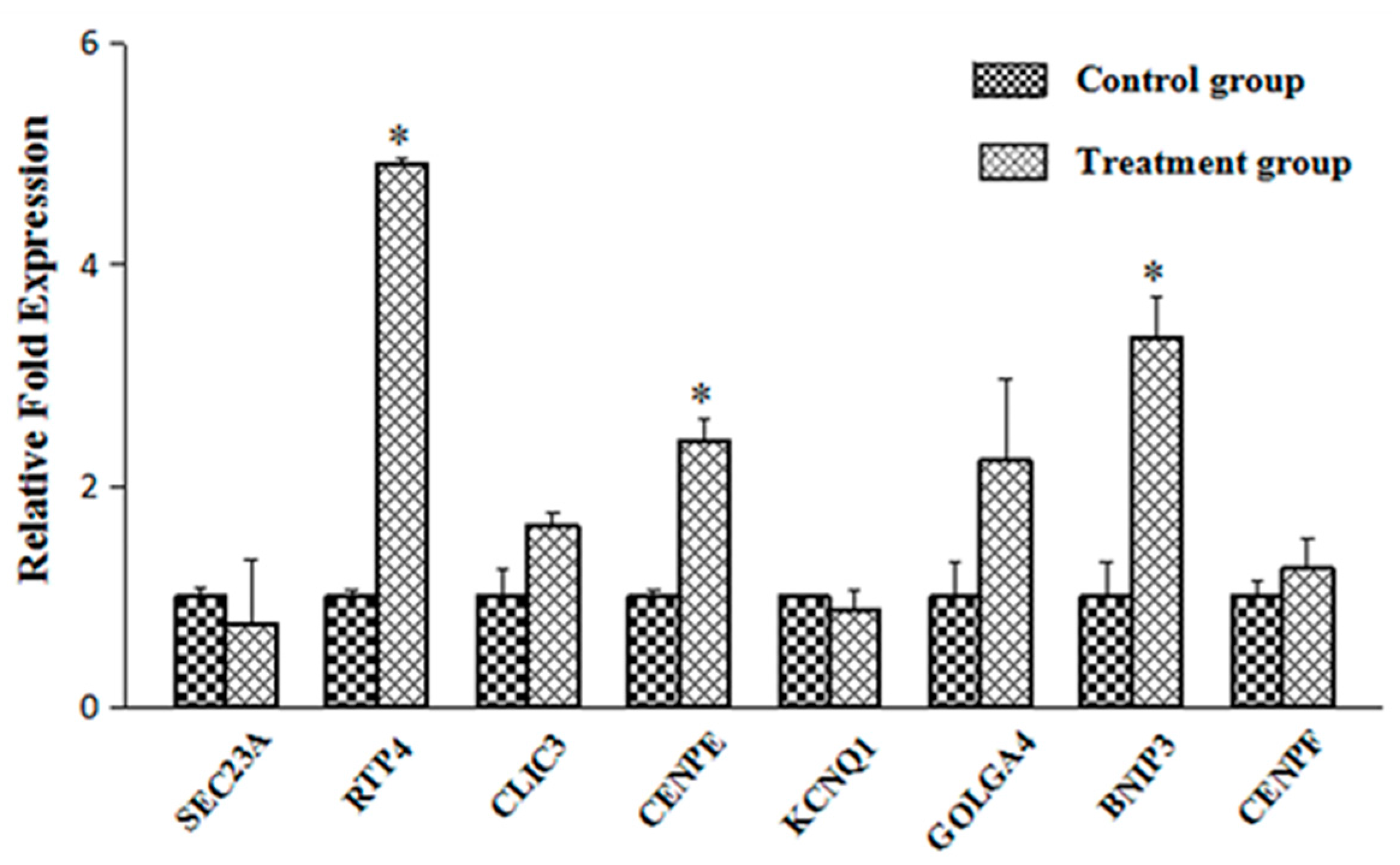
| Classification | Genbank Accession Number | Name of Gene | Fold Change |
|---|---|---|---|
| Genes related to cell inflammation | NM_004688 | N-myc (and STAT) interactor | 1.70 |
| NM_001008540 | Chemokine (C–X–C motif) receptor 4 | 1.67 | |
| NM_139266 | Signal transducer and activator of transcription 1 | 1.65 | |
| NM_002026 | Fibronectin 1 | 1.62 | |
| NM_002310 | Leukemia inhibitory factor receptor α | 1.58 | |
| NM_000877 | Interleukin 1 receptor | 1.51 | |
| NM_001065 | Tumor necrosis factor receptor superfamily, member 1A | 1.58 | |
| NM_001561 | Tumor necrosis factor receptor superfamily, member 9 | −2.00 | |
| NM_001190942 | Tumor necrosis factor receptor superfamily, member 10 | 3.14 | |
| Genes related to cell cycle and cell apoptosis | NM_001030055 | Rho GTPase activating protein 5 | 1.51 |
| NM_022873 | Interferon, α-inducible protein 6 | 2.22 | |
| NM_001206701 | SP100 nuclear antigen | 1.74 | |
| NM_001269 | Regulator of chromosome condensation 1 | −1.56 | |
| NM_181558 | Replication factor C (activator 1) 3 | 1.52 | |
| NM_005531 | Interferon, γ-inducible protein 16 | 1.51 | |
| NM_001003943 | BCL2 modifying factor | 1.55 | |
| NM_001257387 | Checkpoint kinase 2 | 1.60 | |
| NM_004741 | Nucleolar and coiled-body phosphoprotein 1 | −1.54 | |
| NM_001267058 | Caspase 7, apoptosis-related cysteine peptidase | 1.52 | |
| NM_001143762 | Eukaryotic translation initiation factor 5A | −1.73 | |
| NM_004052 | BCL2/adenovirus E1B 19 kDa interacting protein 3 | 1.55 | |
| NM_001813 | Centromere protein E | 1.72 | |
| NM_016343 | Centromere protein F, 350/400 ka (mitosin) | 1.54 | |
| Genes related to cell signaling and gene regulation | NM_004031 | Interferon regulatory factor 7 | 1.84 |
| NM_001243084 | Hypoxia-inducible factor 1 | 1.71 | |
| NM_198318 | Protein arginine methyltransferase | −1.58 | |
| NM_012099 | CD3e molecule, epsilon associated protein | −1.72 | |
| Genes involved in the cellular movement | NM_006364 | Sec23 homolog A | 1.51 |
| NM_022147 | Receptor transporter protein 4 | 3.13 | |
| NM_004669 | Chloride intracellular channel 3 | 1.57 | |
| NM_001813 | Centromere protein E | 1.72 | |
| NM_000218 | Potassium voltage-gated channel subfamily Q member 1 | 1.53 | |
| NM_001172713 | Golgi autoantigen, golgin subfamily a, 4 | 1.70 | |
| NM_004052 | BCL2/adenovirus E1B 19 kDa interacting protein 3 | 1.55 | |
| NM_016343 | Centormere protein F, 350/400 ka (mitosin) | 1.54 |
| Ingredients | Volume |
|---|---|
| DNA | ≤1 µg |
| Kpn I | 1 µL |
| Xba I | 1 µL |
| Buffer | 5 µL |
| ddH2O | X µL |
| Total | 50 µL |
| Ingredients | Volume |
|---|---|
| Vector DNA | 5 µL |
| α-TTP DNA | 3 µL |
| Buffer | 1 µL |
| T4 ligase | 1 µL |
| Total | 10 µL |
| Name of Genes | Primer Sequence | Product Size (bp) |
|---|---|---|
| α-TTP | F: ACAGGAGGTAGAAACTCAGCG | 104 |
| R: TCTTCTTGGCTACGGATGGA | ||
| β-actin | F: CTCACCGAGCGCGGCTACAG | 126 |
| R: GGAGCTGGAAGCAGCCGTGG |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, Y.-H.; Fu, J.-C.; Liu, K.; Zuo, Z.-Y.; Jia, H.-N.; Ma, Y.; Luo, H.-L. Screening of α-Tocopherol Transfer Protein Sensitive Genes in Human Hepatoma Cells (HepG2). Int. J. Mol. Sci. 2016, 17, 1016. https://doi.org/10.3390/ijms17071016
Qu Y-H, Fu J-C, Liu K, Zuo Z-Y, Jia H-N, Ma Y, Luo H-L. Screening of α-Tocopherol Transfer Protein Sensitive Genes in Human Hepatoma Cells (HepG2). International Journal of Molecular Sciences. 2016; 17(7):1016. https://doi.org/10.3390/ijms17071016
Chicago/Turabian StyleQu, Yang-Hua, Jun-Cai Fu, Kun Liu, Zhao-Yun Zuo, Hui-Na Jia, Yong Ma, and Hai-Ling Luo. 2016. "Screening of α-Tocopherol Transfer Protein Sensitive Genes in Human Hepatoma Cells (HepG2)" International Journal of Molecular Sciences 17, no. 7: 1016. https://doi.org/10.3390/ijms17071016
APA StyleQu, Y.-H., Fu, J.-C., Liu, K., Zuo, Z.-Y., Jia, H.-N., Ma, Y., & Luo, H.-L. (2016). Screening of α-Tocopherol Transfer Protein Sensitive Genes in Human Hepatoma Cells (HepG2). International Journal of Molecular Sciences, 17(7), 1016. https://doi.org/10.3390/ijms17071016





