Characterization and Genomic Analysis of a Highly Efficient Dibutyl Phthalate-Degrading Bacterium Gordonia sp. Strain QH-12
Abstract
:1. Introduction
2. Results and Discussion
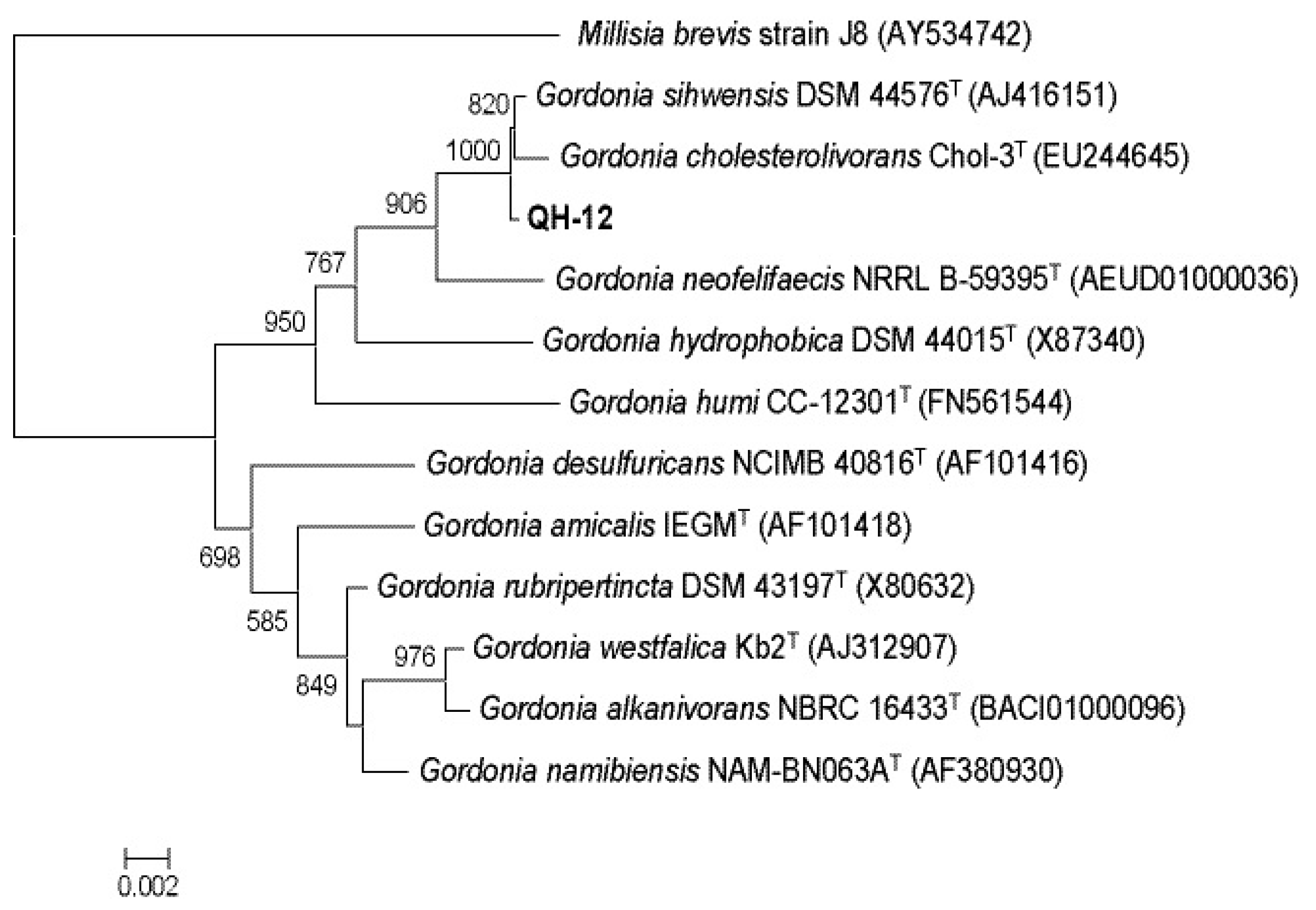
2.1. Isolation and Identification of PAEs-Degrading Strains
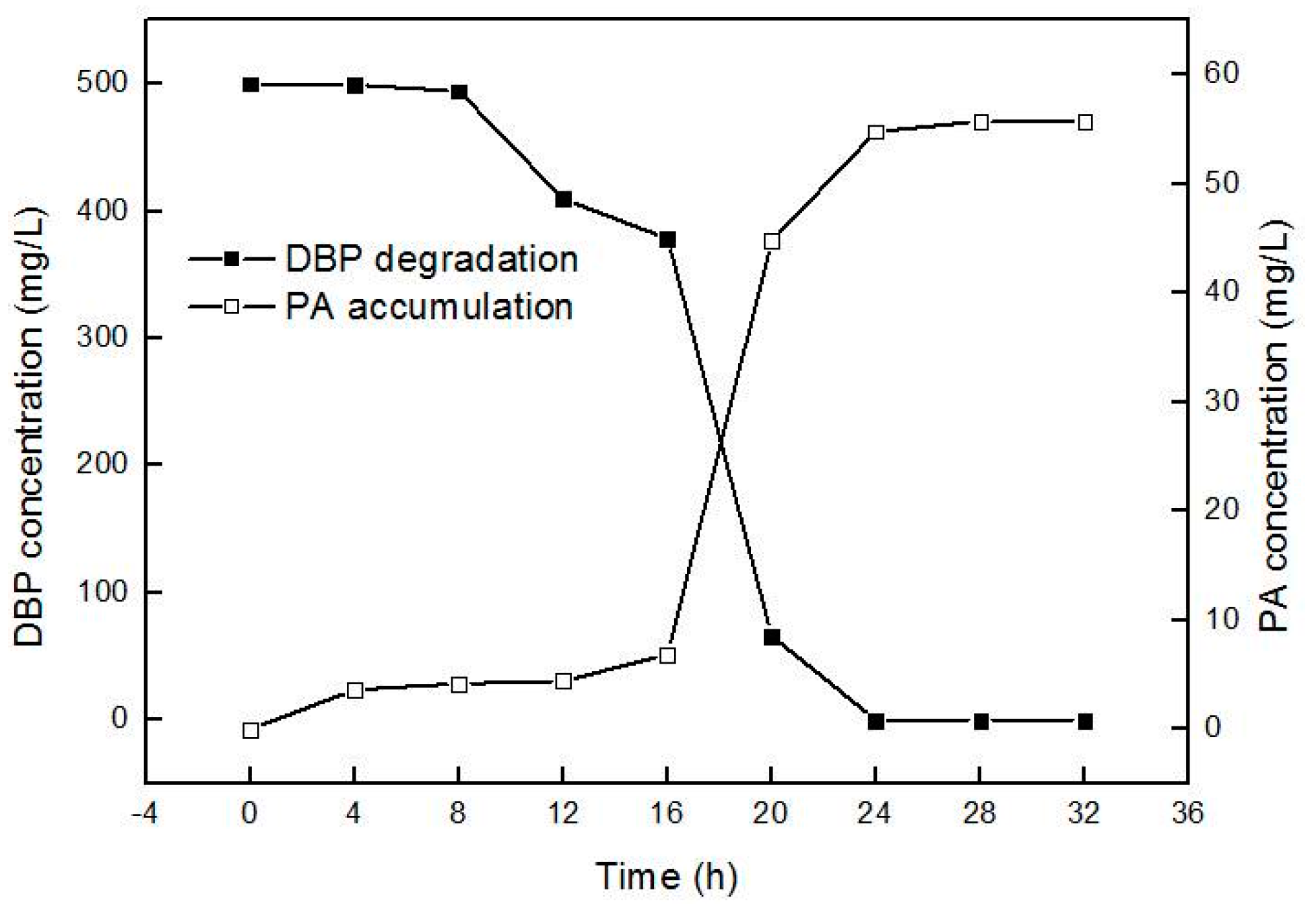
2.2. Biodegradation Kinetics of DBP by Gordonia sp. Strain QH-12
2.3. Substrate Spectrum of Gordonia sp. Strain QH-12
2.4. Overview of the Gordonia sp. Strain QH-12 Genome
2.5. Elucidating the Molecular Mechanism for Inability to Degrade PA in Gordonia Strains
3. Materials and Methods
3.1. Chemicals and Media
3.2. Isolation and Identification of DBP-Degrading Microorganism
3.3. Biodegradation Kinetic of DBP by Gordonia sp. Strain QH-12
3.4. Substrate Utilization Experiments
3.5. Genome Sequencing, Assembly, and Annotation
3.6. Analytical Methods
3.7. Nucleotide Sequence Accession Number
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cartwright, C.D.; Owen, S.A.; Thompson, I.P.; Burns, R.G. Biodegradation of diethyl phthalate in soil by a novel pathway. FEMS Microbiol. Lett. 2000, 186, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Cui, K.; Li, X.; Fu, J.; Sheng, G. Biodegradation kinetics of phthalate esters by Pseudomonas fluoresences FS1. Process. Biochem. 2004, 39, 1125–1129. [Google Scholar] [CrossRef]
- Serrano, S.E.; Braun, J.; Trasande, L.; Dills, R.; Sathyanarayana, S. Phthalates and diet: A review of the food monitoring and epidemiology data. Environ. Health 2014, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Högberg, J.; Hanberg, A.; Berglund, M.; Skerfving, S.; Remberger, M. Phthalate diesters and their metabolites in human breast milk, blood or serum, and urine as biomarkers of exposure in vulnerable populations. Environ. Health Perspect. 2008, 116, 334–339. [Google Scholar] [PubMed]
- Jobling, S.; Reynolds, T.; White, R.; Parker, M.G.; Sumpter, J.P. A variety of environmentally persistent chemicals, including some phthalate plasticizers, are weakly estrogenic. Environ. Health Perspect. 1995, 103, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, C.; Soto, A.M. An updated review of environmental estrogen and androgen mimics and antagonists. J. Steroid Biochem. Mol. Biol. 1998, 65, 143–150. [Google Scholar] [CrossRef]
- Liao, C.; Chen, L.; Chen, B.; Lin, S. Bioremediation of endocrine disruptor di-n-butyl phthalate ester by Deinococcus. radiodurans and Pseudomonas stutzeri. Chemosphere 2010, 78, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Z.; Niu, G.L.; Yang, W.M.; Cao, X.S. Di(2-ethylhexyl) phthalate biodegradation and denitrification by a Pseudoxanthomonas. sp. strain. Bioresour. Technol. 2015, 180, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liao, X.; Yu, F.; Wei, Z.; Yang, L. Cloning of a dibutyl phthalate hydrolase gene from Acinetobacter. Appl. Microbiol. Biotechnol. 2012, 97, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.K.H.; Lam, R.K.W.; Shi, M.Y.; Gu, J.D. Environmental fate of the endocrine disruptors, dimethyl phthalate esters (DMPE), under anoxic sulfate-reducing conditions. Sci. Total Environ. 2007, 381, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.C.; Bai, Z.H.; Chang, D.D.; Hoefel, D.; Jin, B.; Wang, P.; Wei, D.B.; Zhuang, G.Q. Biodegradation of DBP by an isolated Gordonia. sp. strain QH-11 genetic identification and degradation kinetics. J. Hazard. Mater. 2012, 221, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Stewart, G.R.; Mohn, W.W. Involvement of a novel ABC transporter and monoalkyl phthalate ester hydrolase in phthalate ester catabolism by Rhodococcus. jostii RHA1. Appl. Environ. Microbiol. 2010, 76, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.; Suresh, S. Biodegradation of dimethyl phthalate ester using free cells, entrapped cells of Variovorax. sp. BS1 and cell free enzyme extracts: A comparative study. Int. Biodeter. Biodegr. 2015, 97, 179–187. [Google Scholar] [CrossRef]
- Whangsuk, W.; Sungkeeree, P.; Nakasiri, M.; Thiengmag, S.; Mongkolsuk, S.; Loprasert, S. Two endocrine disrupting dibutyl phthalate degrading esterases and their compensatory gene expression in Sphingobium. sp. SM42. Int. Biodeter. Biodegr. 2015, 99, 45–54. [Google Scholar] [CrossRef]
- Chatterjee, S.; Dutta, T.K. Metabolism of butyl benzyl phthalate by Gordonia. sp. strain MTCC 4818. Biochem. Biophys. Res. Commun. 2003, 309, 36–43. [Google Scholar] [CrossRef]
- Wu, X.L.; Liang, R.X.; Dai, Q.Y.; Jin, D.C.; Wang, Y.Y.; Chao, W.L. Complete degradation of di-n-octyl phthalate by biochemical cooperation between Gordonia. sp. strain JDC-2 and Arthrobacter. sp. strain JDC-32 isolated from activated sludge. J. Hazard. Mater. 2010, 176, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.L.; Wang, Y.Y.; Dai, Q.Y.; Liang, R.X.; Jin, D.C. Isolation and characterization of four di-n-butyl phthalate (DBP)-degrading Gordonia. sp. strains and cloning the 3,4-phthalate dioxygenase gene. World J. Microbiol. Biotechnol. 2011, 27, 2611–2617. [Google Scholar] [CrossRef]
- Sarkar, J.; Chowdhury, P.P.; Dutta, T.K. Complete degradation of di-n-octyl phthalate by Gordonia. sp. strain Dop5. Chemosphere 2013, 90, 2571–2577. [Google Scholar] [CrossRef] [PubMed]
- Li, D.D.; Yan, J.L.; Wang, L.; Zhang, Y.Z.; Liu, D.L.; Geng, H.; Xiong, L. Characterization of the phthalate acid catabolic gene cluster in phthalate acid esters transforming bacterium-Gordonia. sp. strain HS-NH1. Int. Biodeter. Biodegr. 2016, 106, 34–40. [Google Scholar] [CrossRef]
- Fang, H.H.P.; Liang, D.W.; Zhang, T. Aerobic degradation of diethyl phthalate by Sphingomonas. sp. Bioresour. Technol. 2007, 98, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Yin, B.; Hong, Y.G.; Gu, J.D. Degradation of dimethyl carboxylic phthalate ester by Burkholderia. cepacia DA2 isolated from marine sediment of South China Sea. Ecotoxicology 2008, 17, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.L.; Wang, Y.Y.; Liang, R.X.; Dai, Q.Y.; Jin, D.C.; Chao, W.L. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Agrobacterium sp. and the biochemical pathway. Process. Biochem. 2011, 46, 1090–1094. [Google Scholar] [CrossRef]
- Fang, C.R.; Yao, J.; Zheng, Y.G.; Jiang, C.J.; Hu, L.F.; Wu, Y.Y.; Shen, D.S. Dibutyl phthalate degradation by Enterobacter. sp. T5 isolated from municipal solid waste in landfill bioreactor. Int. Biodeter. Biodegr. 2010, 64, 442–446. [Google Scholar] [CrossRef]
- Xu, X.R.; Li, H.B.; Gu, J.D. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate ester by Pseudomonas uorescens B-1. Int. Biodeter. Biodegr. 2005, 55, 9–15. [Google Scholar] [CrossRef]
- Nishioka, T.; Iwata, M.; Imaoka, T.; Mutoh, M.; Egashira, Y.; Nishiyama, T.; Shin, T.; Fujii, T. A mono-2-ethylhexyl phthalate hydrolase from a Gordonia. sp. that is able to dissimilate di-2-ethylhexyl phthalate. Appl. Environ. Microbiol. 2006, 72, 2394–2399. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.M.; Wang, C.F.; Xie, Z.R.; Li, J.J.; Yang, Y.J.; Mu, Y.L.; Tang, X.H.; Xu, B.; Zhou, J.P.; Huang, Z.X. Properties of a newly identified esterase from Bacillus sp. K91 and its novel function in diisobutyl phthalate degradation. PLoS ONE 2015, 10, e0119216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Fan, X.; Qiu, Y.J.; Li, C.Y.; Xing, S.; Zheng, Y.T.; Xu, J.H. Properties of a newly identified thermostable esterase from Sulfobacillus acidophilus and its performance in phthalate ester degradation. Appl. Environ. Microbiol. 2014, 80, 6870–6878. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.Y.; Chen, X.; Wang, X.; Liao, X.W.; Xiao, L.; Miao, A.J.; Wu, J.; Yang, L.Y. Identification and characterization of a cold active phthalate esters hydrolase by screening a metagenomic library derived from biofilms of a wastewater treatment plant. PLoS ONE 2013, 8, e75977. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.C.; Liang, R.X.; Dai, Q.Y.; Zhang, R.Y.; Wu, X.L.; Chao, W.L. Biodegradation of di-n-butyl phthalate by Rhodococcus. sp. JDC-11 and molecular detection of 3,4-phthalate dioxygenase gene. J. Microbiol. Biotechnol. 2010, 20, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Manhattan, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Klimke, W.; Maglott, D.R. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009, 37, D32–D36. [Google Scholar] [CrossRef] [PubMed]
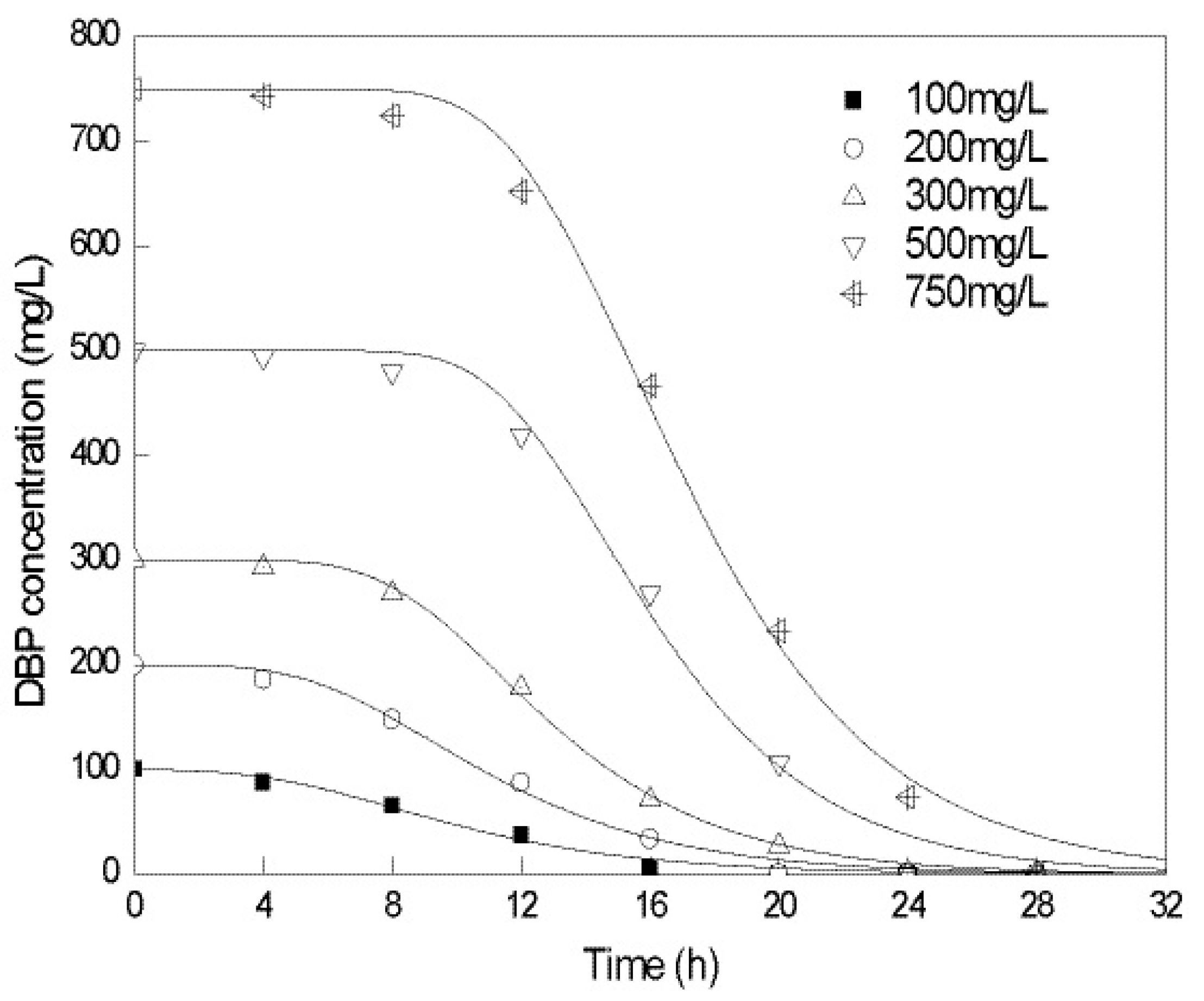

| Initial Concentration/(mg/L) | λ/h | Rm/(mg/L/h) | R2 |
|---|---|---|---|
| 100 | 3.825 | 8.673 | 0.980 |
| 200 | 5.205 | 18.085 | 0.989 |
| 300 | 7.616 | 29.449 | 0.998 |
| 500 | 10.974 | 50.212 | 0.990 |
| 750 | 11.428 | 66.367 | 0.994 |
| Substrate | Utilization | Substrate | Utilization | Substrate | Utilization |
|---|---|---|---|---|---|
| DMP | + | DOP | ++ | MBP | ++ |
| DEP | ++ | DIOP | + | Phthalic acid | − |
| DBP | ++ | DEHP | + | Protocatechuic acid | − |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, D.; Kong, X.; Liu, H.; Wang, X.; Deng, Y.; Jia, M.; Yu, X. Characterization and Genomic Analysis of a Highly Efficient Dibutyl Phthalate-Degrading Bacterium Gordonia sp. Strain QH-12. Int. J. Mol. Sci. 2016, 17, 1012. https://doi.org/10.3390/ijms17071012
Jin D, Kong X, Liu H, Wang X, Deng Y, Jia M, Yu X. Characterization and Genomic Analysis of a Highly Efficient Dibutyl Phthalate-Degrading Bacterium Gordonia sp. Strain QH-12. International Journal of Molecular Sciences. 2016; 17(7):1012. https://doi.org/10.3390/ijms17071012
Chicago/Turabian StyleJin, Decai, Xiao Kong, Huijun Liu, Xinxin Wang, Ye Deng, Minghong Jia, and Xiangyang Yu. 2016. "Characterization and Genomic Analysis of a Highly Efficient Dibutyl Phthalate-Degrading Bacterium Gordonia sp. Strain QH-12" International Journal of Molecular Sciences 17, no. 7: 1012. https://doi.org/10.3390/ijms17071012
APA StyleJin, D., Kong, X., Liu, H., Wang, X., Deng, Y., Jia, M., & Yu, X. (2016). Characterization and Genomic Analysis of a Highly Efficient Dibutyl Phthalate-Degrading Bacterium Gordonia sp. Strain QH-12. International Journal of Molecular Sciences, 17(7), 1012. https://doi.org/10.3390/ijms17071012








