Transcriptome and Difference Analysis of Fenpropathrin Resistant Predatory Mite, Neoseiulus barkeri (Hughes)
Abstract
:1. Introduction
2. Results
2.1. Resistance Selection
2.2. RNA-Sequencing Analysis
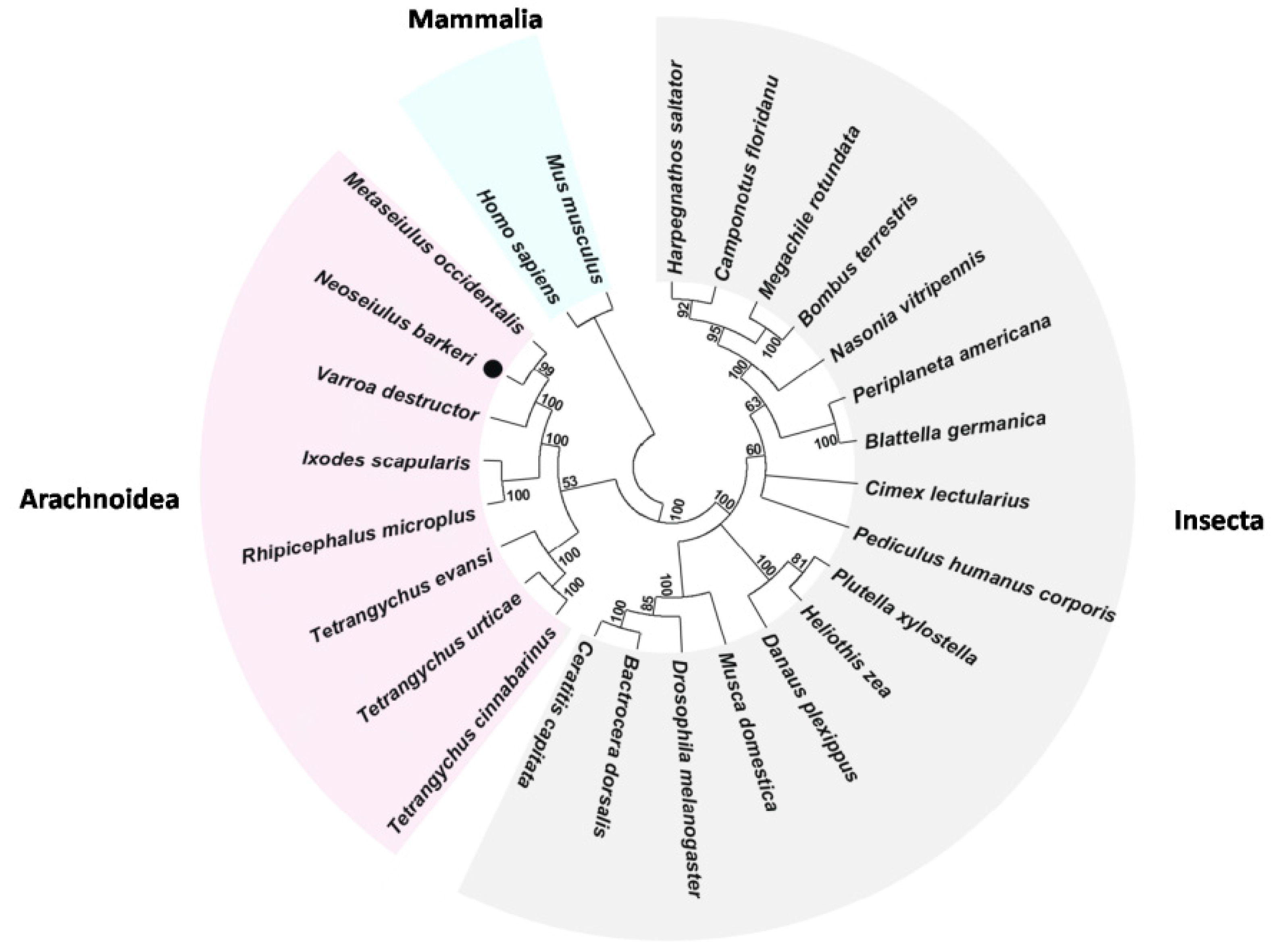
2.3. Annotation of Predicted Proteins
2.4. Functional Annotation
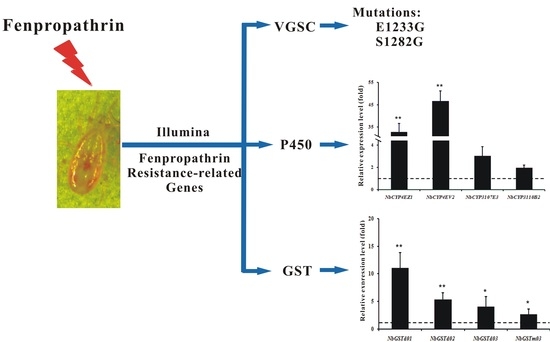
2.5. Cloning of the N. barkeri Sodium Channel Gene and Verification of Point Mutations
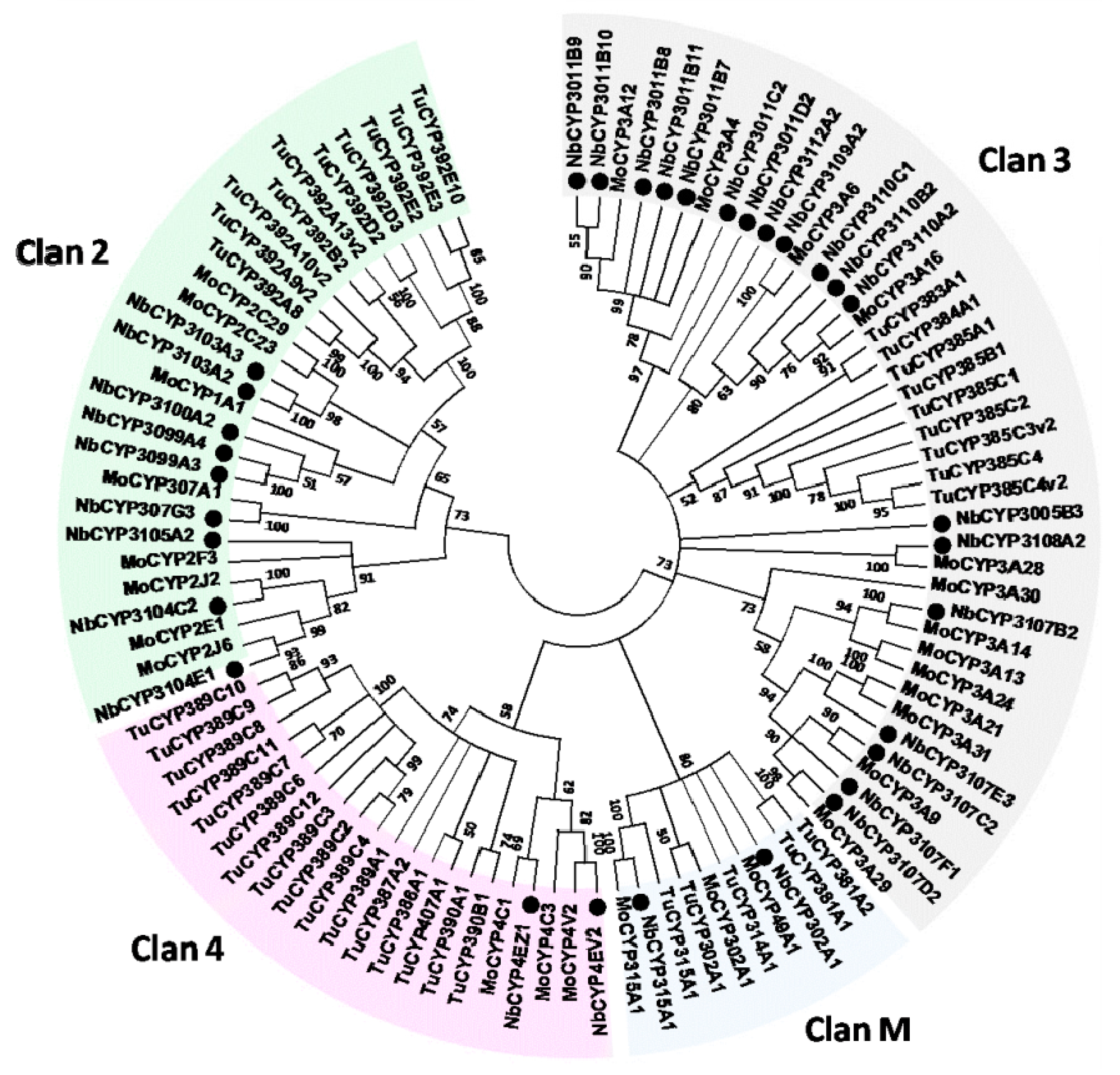
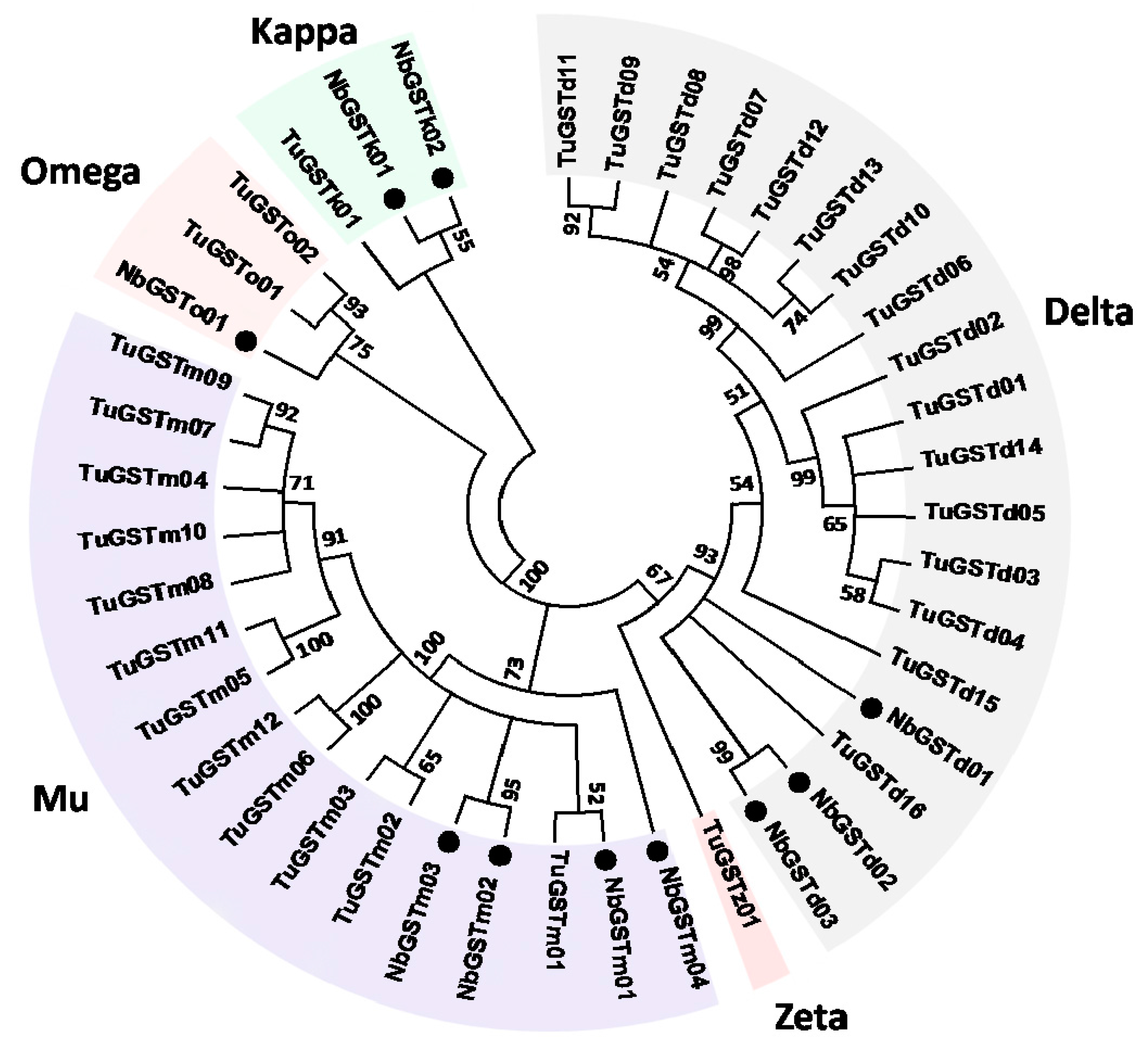
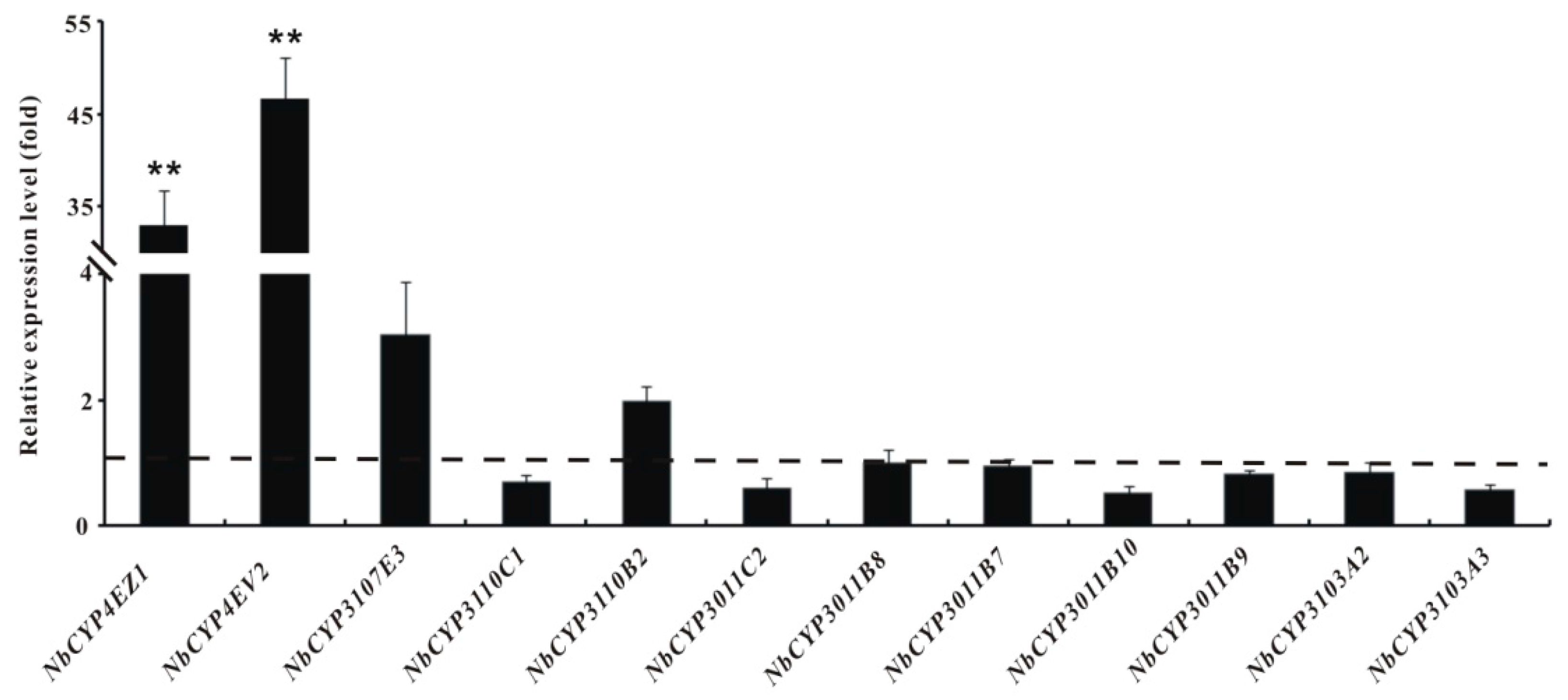
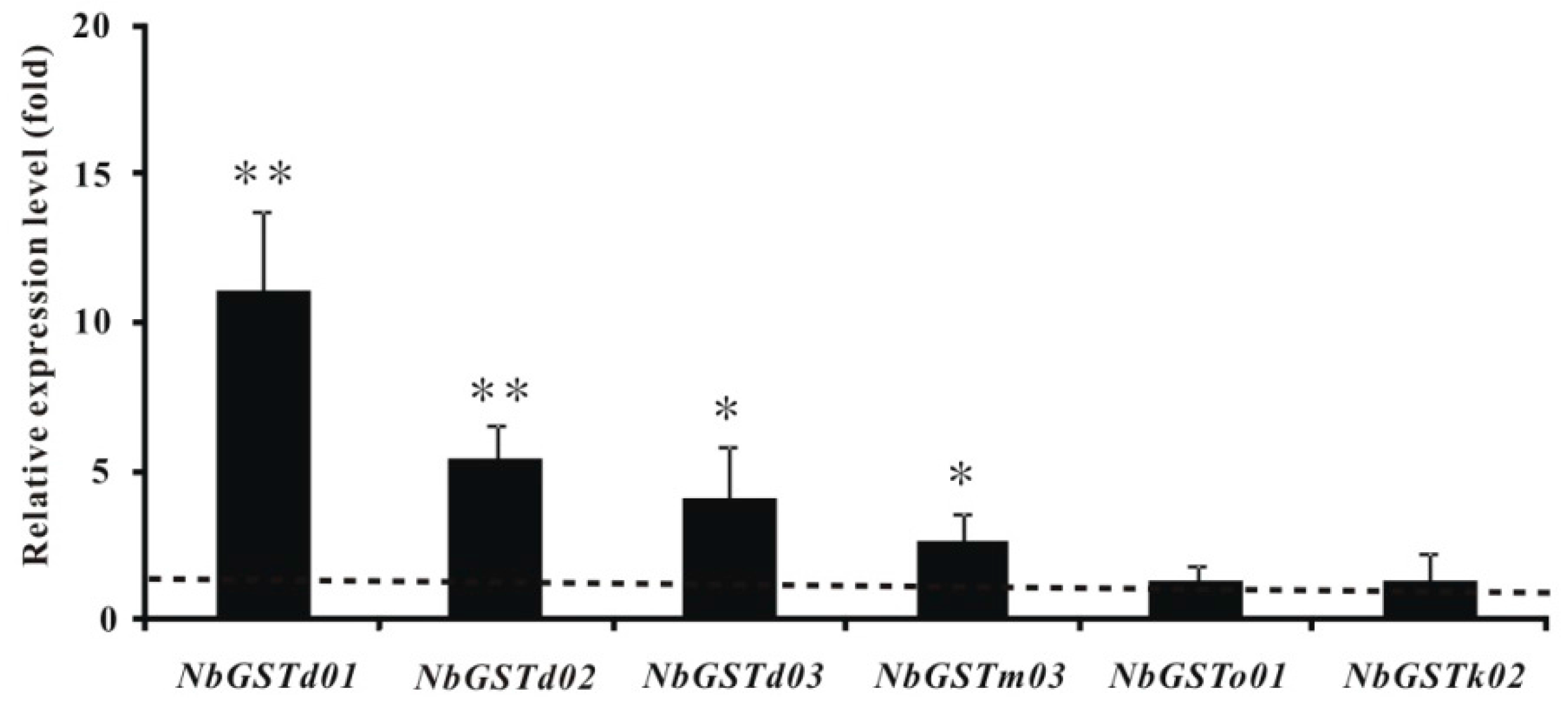
2.6. Identification and Differential Expression Analysis of P450s and GSTs
3. Discussion
4. Materials and Methods
4.1. Colony Rearing and Maintenance
4.2. Resistance Selection
4.3. Toxicity Testing and Determining the LC Value
4.4. RNA Preparation, Library Construction, and Sequencing
4.5. Sequence Analysis, de Novo Assembly, and Identification of Interesting Genes
4.6. Cloning and Mutations Confirmation of Sodium Channel Gene
4.7. Differential Expression Analysis of P450s and GSTs
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| COG | Cluster of Orthologous Groups |
| GO | Gene Ontology |
| GST | Glutathione S-transferase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC50 | Lethal concentration 50 |
| ORF | Open reading frame |
| nr | Nonredundant |
| P450 | Cytochrome P450 |
| qPCR | Quantitative PCR |
| RT-PCR | Reverse transcription PCR |
| VGSC | Voltage-gated sodium channel |
References
- Wu, W.N.; Ou, J.F.; Huang, J.L. Arachnida: Acari: Phytoseiidae. In Fauna Sinica: Invertebrata, 1st ed.; Huo, C.Y., Wu, L.L., Eds.; Science Press: Beijing, China, 2009; Volume 47, pp. 119–120. [Google Scholar]
- Fan, Y.Q.; Petitt, F.L. Functional response of Neoseiulus barkeri Hughes on two-spotted spider mite (Acari: Tetranychidae). Exp. Appl. Acarol. 1994, 18, 613–621. [Google Scholar] [CrossRef]
- Fan, Y.Q.; Petitt, F.L. Biological control of broad mite, Polyphagotarsonemus latus (Banks), by Neoseiulus barkeri Hughes on pepper. Biol. Control. 1994, 4, 390–395. [Google Scholar] [CrossRef]
- Fernando, L.C.P.; Aratchige, N.S.; Kumari, S.L.M.L.; Appuhamy, P.A.L.D.; Hapuarachchi, D.C.L. Development of a method for mass rearing of Neoseiulus baraki, a mite predatory on the coconut mite, Aceria guerreronis. Cocos 2004, 16, 22–36. [Google Scholar] [CrossRef]
- Bakker, F.M.; Sabelis, M.W. How larvae of Thrips tabaci reduce the attack success of phytoseiid predators. Entomol. Exp. Appl. 1989, 50, 47–51. [Google Scholar] [CrossRef]
- Hansen, L.S. Control of Thrips tabaci (Thysanoptera:Thripidae) on glasshouse cucumber using large introductions of predatory mites Amblyseius barkeri (Acarina:Phytoseiidae). Biocontrol 1988, 33, 33–42. [Google Scholar]
- Hessein, N.A.; Parrella, M.P. Predatory mites help control thrips on floriculture crops. Calif. Agric. 1990, 44, 19–21. [Google Scholar]
- BrØdsgaard, H.F.; Hansen, L.S. Effect of Amblyseius cucumeris and Amblyseius barkeri as biological control agents of Thrips tabaci on glasshouse cucumbers. Biocontrol. Sci. Technol. 1992, 2, 215–223. [Google Scholar] [CrossRef]
- Peña, J.E.; Obsorne, L. Biological control of Polyphagotarsonemus latus (Acarina: Tarsonemidae) in greenhouses and field trials using introductions of predacious mites (Acarina: Phytoseiidae). Entomophaga 1996, 41, 279–285. [Google Scholar] [CrossRef]
- Nomikou, M.; Janssen, A.; Schraag, R.; Sabelis, M.W. Phytoseiid predators as potential biological control agents for Bemisia tabacci. Exp. Appl. Acarol. 2001, 25, 271–291. [Google Scholar] [CrossRef] [PubMed]
- Ling, P.; Xia, B.; Li, P.X.; Shu, C.; Zhong, L.; Li, A.H. Functional response of Amblyseius barkeri (Acarina: Phytoseiidae) on Panonychus citri (Acari: Tetranychidae). Acta Arachnol. Sin. 2008, 17, 29–34. [Google Scholar]
- Sato, M.E.; Miyata, T.; Kawai, A.; Nakano, O. Selection for resistance and susceptibility to methidathion and cross resistance in Amblyseius. wormersleyi Schicha (Acari: Phytoseiidae). Appl. Entomol. Zool. 2000, 35, 393–399. [Google Scholar] [CrossRef]
- Auger, P.; Bonafos, R.; Kreiter, S.; Delorme, R. A genetic analysis of mancozeb resistance in Typhlodromus pyri (Acari: Phytoseiidae). Exp. Appl. Acarol. 2005, 37, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Salman, S.Y.; Recep, A.Y. Analysis of hexythiazox resistance mechanisms in a laboratory selected predatory mite Neoseiulus californicus (Acari: Phytoseiidae). Turk. Entomol. Derg. Tu. 2013, 37, 409–422. [Google Scholar]
- Hoy, M.A. Large-scale releases of pesticide-resistant spider mite predators. Calif. Agric. 1982, 36, 8–10. [Google Scholar]
- Hoy, M.A.; Barnett, W.W.; Hendricks, L.C.; Castro, D.; Cahn, D.; Bentley, W.J. Managing spider mites in almonds with pesticide-resistant predators. Calif. Agric. 1984, 38, 18–20. [Google Scholar]
- Hluchý, M.; Zacharda, M.; Gradt, P. Results of large-scale releases of predatory phytoseiid mite, Typhlodromus pyri, an OP-resistance population Mikulov, in apple or chards in South Tyrol and France. Acta Hortic. 1996, 422, 223–225. [Google Scholar] [CrossRef]
- Casida, J.E.; Durkin, K.A. Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annu. Rev. Entomol. 2013, 58, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.D.; Chen, E.H.; Ding, T.B.; Chen, S.C.; Dou, W.; Wang, J.J. De novo assembly, gene annotation, and marker discovery in stored-product pest Liposcelis entomophila (Enderlein) using transcriptome sequences. PLoS ONE 2013, 8, e0080046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Zhang, P.J.; Li, W.D.; Zhang, J.M.; Huang, F.; Yang, J.; Bei, Y.; Lu, Y. De novo transcriptome sequencing in Frankliniella occidentalis to identify genes involved in plant virus transmission and insecticide resistance. Genomics 2013, 101, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.H.; Wei, D.D.; Shen, G.M.; Yuan, G.R.; Bai, P.P.; Wang, J.J. De novo characterization of the Dialeurodes citri transcriptome: Mining genes involved in stress resistance and simple sequence repeats (SSRs) discovery. Insect Mol. Biol. 2014, 23, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C.T.; Maiwald, F.; Schorn, C.; Bass, C.; Ott, M.C.; Nauen, R. A de novo trancriptome of European pollen beetle populations and its analysis, with special reference to insecticide action and resistance. Insect Mol. Biol. 2014, 23, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, H.Y.; Xie, Q.; Bu, W.J. Novel detection of insecticide resistance related P450 genes and transcriptome analysis of the hemimetabolous pest Erthesina fullo (Thunberg) (Hemiptera: Heteroptera). PLoS ONE 2015, 10, e0125970. [Google Scholar] [CrossRef] [PubMed]
- Hoy, M.A.; Yu, F.H.; Meyer, J.M.; Tarazona, O.A.; Jeyaprakash, A.; Wu, K. Transcriptome sequencing and annotation of the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae): A cautionary tale about possible contamination by prey sequences. Exp. Appl. Acarol. 2013, 59, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, A.R.; Donohue, K.V.; Khalil, S.M.S.; Scholl, E.; Opperman, C.; Sonenshine, D.E.; Roe, R.M. New approach for the study of mite reproduction: The first transcriptome analysis of a mite, Phytoseiulus. persimilis (Acari: Phytoseiidae). J. Insect Physiol. 2011, 57, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Grbić, M.; van Leeuween, T.; Clark, R.M.; Rombauts, S.; Rouzé, P.; Grbić, V.; Osborne, E.J.; Dermauw, W.; Thi Ngoc, P.C.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.S.; Denholm, I.; Bell, C.A.; Devonshire, A.L. Knockdown resistance (kdr) to DDT and pyrethroid insecticides maps to a sodium channel gene locus in the housefly (Musca domestica). Mol. Gen. Genet. 1993, 240, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Rinkevich, F.D.; Du, Y.Z.; Dong, K. Diversity and convergence of sodium channel mutations involved in resistance to pyrethroids. Pestic. Biochem. Physiol. 2013, 106, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Du, Y.Z.; Rinkevich, F.; Nomura, Y.; Xu, P.; Wang, L.X.; Kristopher, S.; Boris, S.Z. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Soderlund, D.M.; Knipple, D.C. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem. Mol. Biol. 2003, 33, 563–577. [Google Scholar] [CrossRef]
- Davies, T.E.; O’Reilly, A.O.; Field, L.M.; Wallace, B.A.; Williamson, M.S. Knockdown resistance to DDT and pyrethroids: From target-site mutations to molecular modelling. Pest Manag. Sci. 2008, 64, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, Z.; Dong, K.; Elzen, P.J.; Pettis, J.; Huang, Z.Y. Association of novel mutations in a sodium channel gene with fluvalinate resistance in the mite, Varroa destructor. J. Apic. Res. 2002, 41, 17–25. [Google Scholar]
- Tsagkarakou, A.; Van Leeuwen, T.; Khajehali, J.; Ilian, A.; Grispou, M.; Williamson, M.S.; Tirry, L.; Vontas, J. Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Insect Mol. Biol. 2009, 18, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Vais, H.; Williamson, M.S.; Goodson, S.J.; Devonshire, A.L.; Warmke, J.W.; Usherwood, P.N.; Cohen, C.J. Activation of Drosophila sodium channels promotes modification by deltamethrin-reductions in affinity caused by knock-down resistance mutations. J. Gen. Physiol. 2000, 115, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Smith, T.J.; Knipple, D.C.; Soderlund, D.M. Mutations in the house fly Vssc1 sodium channel gene associated with super-kdr resistance abolish the pyrethroid sensitivity of Vssc1/tipE sodium channels expressed in Xenopus oocytes. Insect Biochem. Mol. Biol. 1999, 29, 185–194. [Google Scholar] [PubMed]
- Kyong, S.Y.; Symington, S.B.; Si, H.L.; Soderlund, D.M.; Clark, J.M. Three mutations identified in the voltage-sensitive sodium channel alpha-subunit gene of permethrin-resistant human head lice reduce the permethrin sensitivity of house fly Vssc1 sodium channels expressed in Xenopus. oocytes. Insect Biochem. Mol. Biol. 2008, 38, 296–306. [Google Scholar]
- Tan, J.; Liu, Z.; Tsai, T.D.; Valles, S.M.; Goldin, A.L.; Dong, K. Novel sodium channel gene mutations in Blattella. germanica reduce the sensitivity of expressed channels to deltamethrin. Insect Biochem. Mol. Biol. 2002, 32, 445–454. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, L.; Li, T.; Zhang, L.; He, L.; Dong, K.; Liu, N.N. Evolutionary adaptation of the amino acid and codon usage of the mosquito sodium channel following insecticide selection in the field mosquitoes. PLoS ONE 2012, 7, e0047609. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.C.; Ponce, G.; Saavedra-Rodriguez, K.; Lopez, B.; Flores, A.E. Frequency of V1016I and F1534C mutations in the voltage-gatedsodiumchannelgene in Aedes aegypti in Venezuela. Pest Manag. Sci. 2015, 71, 863–869. [Google Scholar] [CrossRef] [PubMed]
- González-Cabrera, J.; Davies, T.G.; Field, L.M.; Kennedy, P.J.; Williamson, M.S. An amino acid substitution (L925V) associated with resistance to pyrethroids in Varroa destructor. PLoS ONE 2013, 8, e0082941. [Google Scholar]
- Nyoni, B.N.; Gorman, K.; Mzilahowa, T.; Williamson, M.S.; Navajas, M.; Field, L.M.; Bass, C. Pyrethroid resistance in the tomato red spider mite, Tetranychus evansi, is associated with mutation of the para-type sodium channel. Pest Manag. Sci. 2011, 7, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Clark, J.M.; Lee, S.H. Cloning of a sodium channel gene and identification of mutations putatively associated with fenpropathrin resistance in Tetranychus urtica. Pestic. Biochem. Physiol. 2010, 97, 93–100. [Google Scholar] [CrossRef]
- Feng, Y.N.; Zhao, S.; Sun, W.; Li, M.; Lu, W.C.; He, L. The sodium channel gene in Tetranychus cinnabarinus (Boisduval): Identification and expression analysis of a mutation associated with pyrethroid resistance. Pest Manag. Sci. 2011, 67, 904–912. [Google Scholar] [PubMed]
- Ding, T.B.; Zhong, R.; Jiang, X.Z.; Liao, C.Y.; Xia, W.K.; Liu, B.; Dou, W.; Wang, J.J. Molecular characterisation of asodiumchannelgene and identification of a Phe1538 to Ile mutation in citrus red mite, Panonychus citri. Pest Manag. Sci. 2015, 71, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.W.; Zhu, Y.; Duan, J.; Yu, Q.Y.; Zhang, G.J.; Wan, F.; Xiang, Z.H. Genome-wide analysis of cytochrome P450 monooxygenase genes in the silkworm, Bombyx mori. Gene 2011, 480, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Feyereisen, R. Arthropod CY Pomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta 2011, 1814, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Liu, N.N. Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS ONE 2011, 6, e0029418. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.M.; Dou, W.; Niu, J.Z.; Jiang, H.B.; Yang, W.J.; Jia, F.X.; Hu, F.; Cong, L.; Wang, J.J. Transcriptome analysis of the oriental fruit fly Bactrocera dorsalis. PLoS ONE 2011, 6, e29127. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Jiang, G.F.; Zhang, Y.F.; Li, J.L.; Li, X.J.; Yue, J.S.; Chen, F.; Liu, H.Q.; Li, H.J.; Ran, C. Analysis of transcriptome differences between resistant and susceptible strains of the citrus red mite Panonychus citri (Acari: Tetranychidae). PLoS ONE 2011, 6, e0028516. [Google Scholar] [CrossRef] [PubMed]
- Avicor, S.W.; Wajidi, M.F.F.; El-Garj, F.M.A.; Jaal, Z.; Yahaya, Z.S. Insecticidal activity and expression of cytochrome P450 family 4 genes in Aedes. albopictus after exposure to pyrethroid mosquito coils. Protein J. 2014, 33, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Killiny, N.; Hajeri, S.; Tiwari, S.; Gowda, S.; Stelinski, L.L. Double-stranded RNA uptake through topical application, mediates silencing of five CYP4 genes and suppresses insecticide resistance in Diaphorina citri. PLoS ONE 2014, 9, e0110536. [Google Scholar] [CrossRef] [PubMed]
- Rasool, A.; Joußen, N.; Lorenz, S.; Ellinger, R.; Schneider, B.; Khan, S.A.; Ashfaq, M.; Hechel, D.G. An independent occurrence of the chimeric P450 enzyme CYP337B3 of Helicoverpa armigera confers cypermethrin resistance in Pakistan. Insect Biochem. Mol. Biol. 2014, 53, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Brun-Barale, A.; Hèma, O.; Martin, T.; Suraporn, S.; Audant, P.; Sezutsu, H.; Feyereisen, R. Multiple P450 genes overexpressed in deltamethrin-resistant strains of Helicoverpa armigera. Pest Manag. Sci. 2010, 66, 900–909. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Strode, C.; Vontas, J.; Nikou, D.; Vaughan, A.; Pignatelli, P.M.; Louis, C.; Hemingway, J.; Ranson, H. The Anopheles gambiae detoxification chip: A highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc. Natl. Acad. Sci. USA 2005, 102, 4080–4084. [Google Scholar] [CrossRef] [PubMed]
- Wan, P.J.; Shi, X.Q.; Kong, Y.; Zhou, L.T.; Guo, W.C.; Ahmat, T.; Li, G.Q. Identification of cytochrome P450 monooxygenase genes and their expression profiles in cyhalothrin-treated Colorado potato beetle, Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2013, 107, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Xiao, D.; He, Y.P.; Yao, J.X.; Zhu, G.N.; Zhu, K.Y. Insecticide-mediated up-regulation of cytochrome P450 genes in the red flour beetle (Tribolium castaneum). Int. J. Mol. Sci. 2015, 16, 2078–2098. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, S.; Tsumuki, H. Studies on glutathione S-transferase gene involved in chlorfluazuron resistance of the diamondback moth, Plutella xylostella L. (Lepidoptera: Yponomeutidae). Pestic. Biochem. Physiol. 2005, 82, 94–101. [Google Scholar] [CrossRef]
- Ding, Y.; Ortelli, F.; Rossiter, L.C.; Hemingway, J.; Ranson, H. The Anopheles gambiae glutathione transferase supergene family: Annotation, phylogeny and expression profiles. BMC Genom. 2003, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Samra, A.I.; Kamita, S.G.; Yao, H.W.; Cornel, A.J.; Hammock, B.D. Cloning and characterization of two glutathione S-transferases from pyrethroid resistant Culex pipiens. Pest Manag. Sci. 2012, 68, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.Z.; Sun, J.Y.; Zhou, X.G.; Gao, X.W.; Liang, P. The stability and biochemical basis of fufenozide resistance in a laboratory-selected strain of Plutella xylostella. Pestic. Biochem. Physiol. 2011, 101, 80–85. [Google Scholar] [CrossRef]
- You, Y.C.; Xie, M.; Ren, N.N.; Cheng, X.M.; Li, J.Y.; Ma, X.L.; Zou, M.M.; Liette, V.; Geoff, M.G.; You, M.S. Characterization and expression profiling of glutathione S-transferases in the diamondback moth, Plutella xylostella (L.). BMC Genom. 2015, 16, 152. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.M.; Shi, L.; Xu, Z.F.; He, L. Inducible expression of Mu-class glutathione S-transferases is associated with fenpropathrin resistance in Tetranychus cinnabarinus. Int. J. Mol. Sci. 2014, 15, 22626–22641. [Google Scholar] [CrossRef] [PubMed]
- Nauen, R.; Stumpf, N. Fluorometric microplate assay to measure glutathione S-transferase activity in insects and mites using monochlorobimane. Anal. Biochem. 2002, 30, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Pavlidi, N.; Tseliou, V.; Riga, M.; Nauen, R.; Van Leeuwen, T.; Labrou, N.E.; Vontas, J. Functional characterization ofglutathioneS-transferases associated with insecticide resistance in Tetranychus urticae. Pestic. Boichem. Physiol. 2015, 121, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Rauch, N.; Nauen, R. Spirodiclofen resistance risk assessment in Tetranychus urticae (Acari: Tetranychidae): A biochemical approach. Pestic. Biochem. Physiol. 2002, 74, 91–101. [Google Scholar] [CrossRef]
- He, L.; Xue, C.H.; Wang, J.J.; Li, M.; Lu, W.C.; Zhao, Z.M. Resistance selection and biochemical mechanism of resistance to two Acaricides in Tetranychus cinnabarinus (Boiduval). Pestic. Biochem. Physiol. 2009, 93, 47–52. [Google Scholar]
- Luo, Y.J.; Yang, Z.G.; Xie, D.Y.; Ding, W.; Da, A.S.; Ni, J.; Chai, J.P.; Huang, P.; Jiang, X.J.; Li, S.X. Molecular cloning and expression of glutathione S-transferases involved in propargite resistance of the carmine spider mite, Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Physiol. 2014, 114, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Mounsey, K.E.; Pasay, C.J.; Arlian, L.G.; Morgan, M.S.; Holt, D.C.; Currie, B.J.; Currie, B.J.; Walton, S.F.; McCarthy, J.S. Increased transcription of glutathione S-transferases in acaricide exposed scabies mites. Parasites Vectors 2010, 3, 43. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Dou, W.; Wang, J.J.; Jia, F.X.; Wang, J.J. Multiple glutathione S-transferase genes: Identification and expression in oriental fruit fly, Bactrocera dorsalis. Pest Manag. Sci. 2013, 70, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.H.; Jia, M.; Liu, T.; Xuan, T.; Zhu, K.Y.; Guo, Y.P.; Ma, E.B.; Zhang, J.Z. Identification and characterisation of ten glutathione S-transferase genes from oriental migratory locust, Locusta migratoria manilensis (Meyen). Pest Manag. Sci. 2011, 67, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.E.; Zhang, Y.L. Identification and characterisation of multiple glutathione S-transferase genes from the diamondback moth, Plutella xylostella. Pest Manag. Sci. 2015, 71, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Yang, X.Q.; Zhang, Y.L. Characterization of a lambda-cyhalothrin metabolizing glutathione S-transferase CpGSTd1 from Cydia pomonella (L.). Appl. Microbiol. Biotechnol. 2014, 98, 8947–8962. [Google Scholar] [CrossRef] [PubMed]
- Petrushov, A.Z. Pyrethroid resistance in the predacious mite Amblyseius barkeri. EPPO Bull. 1992, 22, 471–473. [Google Scholar] [CrossRef]
- Duso, C.; Malagnini, V.; Pozzebon, A.; Buzzetti, F.M.; Tirello, P. A method to assess the effects of pesticides on the predatory mite Phytoseiulus persimilis (Acari Phytoseiidae) in the laboratory. Biocontrol. Sci. Technol. 2008, 18, 1027–1040. [Google Scholar] [CrossRef]
- Abbott, W.S. A method of computing the effectiveness of an insecticide. J. Am. Mosq. Control Assoc. 1925, 18, 265–276. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; Chen, Z.H.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 291, 644–652. [Google Scholar] [CrossRef] [PubMed]
- ORF finder. Available online: http://www.ncbi.nlm.nih.gov/gorf/gorf.html (accessed on 25 March 2016).
- ClustalW2. Available online: http://www.ebi.ac.uk/Tools/msa/clustalw2/ (accessed on 25 March 2016).
- Primer 3. Available online: http://primer3.ut.ee/ (accessed on 25 March 2016).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Strains | Regression Equation | X2 | 95% Confidence Interval (mg/L) | R2 | Resistance Ratio |
|---|---|---|---|---|---|
| FS | Y = 3.2619 + 1.1186X | 2.0824 | 35.79 (28.64–44.73) | 0.996 | 1 |
| FR | Y = 0.9489 + 0.9321X | 0.0574 | 22,190.91 (2853.80–172,554.97) | 0.998 | 619.96 |
| Sequencing | |
|---|---|
| Total number of reads | 25,192,607 |
| Total number of nucleotides (nt) | 5,087,856,659 |
| GC percentage (%) | 50.85 |
| Q20 percentage (%) | 100.00 |
| N percentage (%) | 0.01 |
| Number of contigs | 1,385,792 |
| Mean length of contigs (bp) | 74.76 |
| Number of transcripts | 50,462 |
| Mean length of transcripts (bp) | 1494.58 |
| N50 of transcripts (bp) | 3007 |
| Number of unigenes | 34,211 |
| Mean length of unigenes (bp) | 1016.52 |
| N50 of unigenes (bp) | 2126 |
| Total annotated unigenes | 15,987 |
| Unigenes annotations against nr | 15,866 |
| Unigenes annotations against Swiss-Prot | 10,486 |
| Unigenes annotations against KEGG | 5445 |
| Unigenes annotations against COG | 5673 |
| Unigenes annotations against GO | 8707 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, L.; Chen, F.; Yu, S.; Ding, L.; Yang, J.; Luo, R.; Tian, H.; Li, H.; Liu, H.; Ran, C. Transcriptome and Difference Analysis of Fenpropathrin Resistant Predatory Mite, Neoseiulus barkeri (Hughes). Int. J. Mol. Sci. 2016, 17, 704. https://doi.org/10.3390/ijms17060704
Cong L, Chen F, Yu S, Ding L, Yang J, Luo R, Tian H, Li H, Liu H, Ran C. Transcriptome and Difference Analysis of Fenpropathrin Resistant Predatory Mite, Neoseiulus barkeri (Hughes). International Journal of Molecular Sciences. 2016; 17(6):704. https://doi.org/10.3390/ijms17060704
Chicago/Turabian StyleCong, Lin, Fei Chen, Shijiang Yu, Lili Ding, Juan Yang, Ren Luo, Huixia Tian, Hongjun Li, Haoqiang Liu, and Chun Ran. 2016. "Transcriptome and Difference Analysis of Fenpropathrin Resistant Predatory Mite, Neoseiulus barkeri (Hughes)" International Journal of Molecular Sciences 17, no. 6: 704. https://doi.org/10.3390/ijms17060704
APA StyleCong, L., Chen, F., Yu, S., Ding, L., Yang, J., Luo, R., Tian, H., Li, H., Liu, H., & Ran, C. (2016). Transcriptome and Difference Analysis of Fenpropathrin Resistant Predatory Mite, Neoseiulus barkeri (Hughes). International Journal of Molecular Sciences, 17(6), 704. https://doi.org/10.3390/ijms17060704