Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers
Abstract
:1. Introduction
2. Brief Description of Biliary Tract Cancers
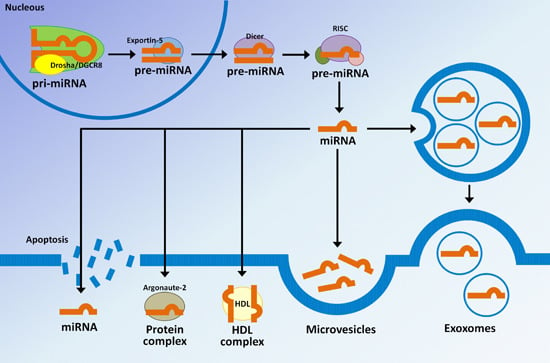
3. MicroRNAs Biogenesis and Their Regulatory Mechanisms
4. MicroRNAs in Extracellular Vesicles
5. miRNAs as Biomarkers in Biliary Tract Cancer: miRNAs in Blood (Plasma and Serum), Bile and Gallstones
5.1. Plasma/Serum miRNA Panels as Biomarkers
5.2. Bile miRNA Panels as Biomarkers
5.3. miRNA Biomarkers in Gallstones for the Identification of High-Risk Patients with Complications in the Biliary Tract
6. Discussion
7. Concluding Remarks
Acknowledgments
Conflicts of Interest
Abbreviations
| 3′UTR | 3′ Untranslated region |
| ABCA1 | ATP-binding cassette transporter A1 |
| AC | Ampulla Vater cancer |
| AGO | Argonaute |
| AQP-5 | Aquaporin-5 |
| AUC | Area under the curve |
| BBD | Benign biliary diseases |
| BBO | Benign biliary obstruction |
| BDL | Bile duct-ligated |
| BTCs | Biliary tract cancers |
| CA19-9 | Carbohydrate 19-9 |
| CCA | Cholangiocarcinoma |
| CEA | Carcinoembryonic antigen |
| CP | Chronic pancreatitis |
| CT | Computed tomography |
| CYP7A1 | Cholesterol 7a-hydroxylase mRNA |
| dCCA | Distal cholangiocarcinoma |
| eCCA | Extrahepatic cholangiocarcinoma |
| ERCP | Endoscopic retrograde cholangiopancreatography |
| EUS | Endoscopic ultrasound |
| EVs | Extracellular vesicles |
| EXOmotif | miRNAs into exosomes attached to specific motifs |
| exRNA | Extracellular RNA |
| FXR | Farnesoid X receptor |
| GBC | Gallbladder cancer |
| HBD | Hilar bile duct cancer |
| HDL | High-density lipoprotein |
| hnRNP | Ribonucleoprotein |
| hnRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 |
| HVs | Healthy volunteers |
| iCCA | Intrahepatic cholangiocarcinoma |
| LncRNAs | Long non-coding RNA |
| LOH | Loss of heterozygosity |
| LXR | Liver X receptors |
| miRNAs | microRNAs |
| miRNA* | Temporary string miRNA |
| MRI | Magnetic resonance imaging |
| NOZ | Gallbladder cancer cell lines |
| PA | Pancreatic adenocarcinoma |
| PBC | Pancreato-biliary cancers |
| pCCA | Perihilarcholangiocarcinoma |
| pre-miRNA | Precursor miRNA |
| pri-miRNA | Primary miRNA |
| PSC | Primary sclerosing cholangitis |
| PTC | Percutaneous transhepatic cholangiography |
| RISC | RNA-induced silencing complex |
| SOD | Sphincter of Oddi dysfunction |
| TNM | Staging system for classification of malignant tumours |
| TUTases | Uridylyltransferases |
| XPO5 | Exportin 5 |
References
- Wu, Q.; Kiguchi, K.; Kawamoto, T.; Ajiki, T.; Traag, J.; Carbajal, S.; Ruffino, L.; Thames, H.; Wistuba, I.; Thomas, M.; et al. Therapeutic effect of rapamycin on gallbladder cancer in a transgenic mouse model. Cancer Res. 2007, 67, 3794–3800. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Jiang, L.; Chen, Y.; She, F.; Han, S.; Zhu, J.; Zhou, L.; Tang, N.; Wang, X.; Li, X. Vascular endothelial growth factor-d promotes growth, lymphangiogenesis and lymphatic metastasis in gallbladder cancer. Cancer Lett. 2012, 314, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Beyazit, Y.; Kekilli, M.; Ibis, M.; Kurt, M.; Sayilir, A.; Onal, I.K.; Purnak, T.; Oztas, E.; Tas, A.; Yesil, Y.; et al. Can red cell distribution width help to discriminate benign from malignant biliary obstruction? A retrospective single center analysis. Hepato-gastroenterology 2012, 59, 1469–1473. [Google Scholar] [PubMed]
- Zhang, L.; Miao, R.; Zhang, X.; Chen, W.; Zhou, Y.; Wang, R.; Zhang, R.; Pang, Q.; Xu, X.; Liu, C. Exploring the diagnosis markers for gallbladder cancer based on clinical data. Front. Med. 2015, 9, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Kau, S.Y.; Shyr, Y.M.; Su, C.H.; Wu, C.W.; Lui, W.Y. Diagnostic and prognostic values of ca 19–9 and cea in periampullary cancers. J. Am. Coll. Surg. 1999, 188, 415–420. [Google Scholar] [CrossRef]
- Strom, B.L.; Maislin, G.; West, S.L.; Atkinson, B.; Herlyn, M.; Saul, S.; Rodriguez-Martinez, H.A.; Rios-Dalenz, J.; Iliopoulos, D.; Soloway, R.D. Serum cea and ca 19–9: Potential future diagnostic or screening tests for gallbladder cancer? Int. J. Cancer 1990, 45, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Davidson, B.R.; Goldin, R.; Pereira, S.P.; Rosenberg, W.M.; Taylor-Robinson, S.D.; Thillainayagam, A.V.; Thomas, H.C.; Thursz, M.R.; Wasan, H. Guidelines for the diagnosis and treatment of cholangiocarcinoma: Consensus document. Gut 2002, 51 (Suppl. 6), VI1–VI9. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. Microrna expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [PubMed]
- Cheng, A.M.; Byrom, M.W.; Shelton, J.; Ford, L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005, 33, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. Microrna-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef] [PubMed]
- Sassen, S.; Miska, E.A.; Caldas, C. Microrna: Implications for cancer. Virchows Arch. 2008, 452, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lynam-Lennon, N.; Maher, S.G.; Reynolds, J.V. The roles of microRNA in cancer and apoptosis. Biol Rev. Camb. Philos. Soc. 2009, 84, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.G.; Urazova, L.N.; Stegny, V.N. MicroRNAs and human cancer. Exp. Oncol. 2012, 34, 2–8. [Google Scholar] [PubMed]
- Jeffrey, S.S. Cancer biomarker profiling with microRNAs. Nat. Biotechnol 2008, 26, 400–401. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Motawi, T.K.; Rizk, S.M.; Ibrahim, T.M.; Ibrahim, I.A. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell. Biochem. Funct. 2016. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. Micrornas: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Cojocneanu-Petric, R.; Chira, S.; Truta, A.; Floares, A.; Petrut, B.; Achimas-Cadariu, P.; Berindan-Neagoe, I. Clinical and pathological implications of miRNA in bladder cancer. Int J. Nanomed. 2015, 10, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Kuwano, K.; Ochiya, T.; Takeshita, F. The impact of extracellular vesicle-encapsulated circulating microRNAs in lung cancer research. BioMed Res. Int. 2014, 2014, 486413. [Google Scholar] [CrossRef] [PubMed]
- Letelier, P.; Garcia, P.; Leal, P.; Alvarez, H.; Ili, C.; Lopez, J.; Castillo, J.; Brebi, P.; Roa, J.C. miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int. J. Clin Exp. Pathol. 2014, 7, 1849–1867. [Google Scholar] [PubMed]
- Selaru, F.M.; Olaru, A.V.; Kan, T.; David, S.; Cheng, Y.; Mori, Y.; Yang, J.; Paun, B.; Jin, Z.; Agarwal, R.; et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology 2009, 49, 1595–1601. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.A.; Werner, J.; Willenbrock, H.; Roslind, A.; Giese, N.; Horn, T.; Wojdemann, M.; Johansen, J.S. Microrna expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod. Pathol. 2012, 25, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Masica, D.; Ishida, M.; Tomuleasa, C.; Umegaki, S.; Kalloo, A.N.; Georgiades, C.; Singh, V.K.; Khashab, M.; Amateau, S.; et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology 2014, 60, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Hsing, A.W.; Rashid, A.; Devesa, S.S.; Fraumeni, J.F. Biliary tract cancer. In Cancer Epidemiology and Prevention, 3rd ed.; Schottenfeld, D., Fraumeni, J.F., Jr., Eds.; Oxford University Press: New York, NY, USA, 2006; pp. 878–900. [Google Scholar]
- De Groen, P.C.; Gores, G.J.; LaRusso, N.F.; Gunderson, L.L.; Nagorney, D.M. Biliary tract cancers. N. Engl. J. Med. 1999, 341, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Shaib, Y.; El-Serag, H.B. The epidemiology of cholangiocarcinoma. Semin. Liver Dis. 2004, 24, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.L.; El-Serag, H.B. Risk factors for cholangiocarcinoma. Hepatology 2011, 54, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Stinton, L.M.; Shaffer, E.A. Epidemiology of gallbladder disease: Cholelithiasis and cancer. Gut Liver 2012, 6, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Eguchi, H.; Nagano, H.; Kobayashi, S.; Akita, H.; Hama, N.; Wada, H.; Kawamoto, K.; Tomokuni, A.; Tomimaru, Y.; et al. Plasma miR-21 is a novel diagnostic biomarker for biliary tract cancer. Cancer Sci. 2013, 104, 1626–1631. [Google Scholar] [CrossRef] [PubMed]
- Rakic, M.; Patrlj, L.; Kopljar, M.; Klicek, R.; Kolovrat, M.; Loncar, B.; Busic, Z. Gallbladder cancer. Hepatobiliary Surg. Nutr. 2014, 3, 221–226. [Google Scholar] [PubMed]
- Misra, S.; Chaturvedi, A.; Misra, N.C.; Sharma, I.D. Carcinoma of the gallbladder. Lancet Oncol. 2003, 4, 167–176. [Google Scholar] [CrossRef]
- Soto, S.; Herrera, H.; Rosas, C.; Lilayú, D. Cáncer de vesícula biliar durante el trienio 2001–2003 en el hospital base de osorno. Cuadernos de cirugia UACH 2004, 18, 21–26. [Google Scholar] [CrossRef]
- Lazcano-Ponce, E.C.; Miquel, J.F.; Munoz, N.; Herrero, R.; Ferrecio, C.; Wistuba, I.I.; Alonso de Ruiz, P.; Aristi Urista, G.; Nervi, F. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J. Clin. 2001, 51, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Jarnagin, W.R.; Shoup, M. Surgical management of cholangiocarcinoma. Semin. Liver Dis. 2004, 24, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Malhi, H.; Gores, G.J. Cholangiocarcinoma: Modern advances in understanding a deadly old disease. J. Hepatol. 2006, 45, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Thomas, H.C.; Davidson, B.R.; Taylor-Robinson, S.D. Cholangiocarcinoma. Lancet 2005, 366, 1303–1314. [Google Scholar] [CrossRef]
- Blechacz, B.; Komuta, M.; Roskams, T.; Gores, G.J. Clinical diagnosis and staging of cholangiocarcinoma. Nat. Rev. Gastroenterol Hepatol. 2011, 8, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, V.; Semeraro, R.; Torrice, A.; Gatto, M.; Napoli, C.; Bragazzi, M.C.; Gentile, R.; Alvaro, D. Intra-hepatic and extra-hepatic cholangiocarcinoma: New insight into epidemiology and risk factors. World J. Gastrointest. Oncol. 2010, 2, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Plieskatt, J.; Rinaldi, G.; Feng, Y.; Peng, J.; Easley, S.; Jia, X.; Potriquet, J.; Pairojkul, C.; Bhudhisawasdi, V.; Sripa, B.; et al. A microRNA profile associated with opisthorchis viverrini-induced cholangiocarcinoma in tissue and plasma. BMC Cancer 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Perysinakis, I.; Margaris, I.; Kouraklis, G. Ampullary cancer—a separate clinical entity? Histopathology 2014, 64, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.H.; Bekaii-Saab, T. Ampullary cancer: An overview. Am. Soc. Clin. Oncol. Educ. Book 2014, 34, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Kala, Z.; Weber, P.; Hemmelova, B.; Marek, F.; Hlavsa, J.; Sobotka, M. Ampullary tumours (ampullomas) in the elderly—an interdisciplinary problem. Indian J. Med. Res. 2010, 131, 418–421. [Google Scholar] [PubMed]
- Ito, F.; Agni, R.; Rettammel, R.J.; Been, M.J.; Cho, C.S.; Mahvi, D.M.; Rikkers, L.F.; Weber, S.M. Resection of hilar cholangiocarcinoma: Concomitant liver resection decreases hepatic recurrence. Ann. Surg. 2008, 248, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Kai, Z.S.; Pasquinelli, A.E. Microrna assassins: Factors that regulate the disappearance of miRNAs. Nat. Struct. Mol. Biol. 2010, 17, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Jarnagin, W.R. Gallbladder carcinoma. Eur. J. Surg. Oncol. 2008, 34, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Mayo, S.C.; Shore, A.D.; Nathan, H.; Edil, B.; Wolfgang, C.L.; Hirose, K.; Herman, J.; Schulick, R.D.; Choti, M.A.; Pawlik, T.M. National trends in the management and survival of surgically managed gallbladder adenocarcinoma over 15 years: A population-based analysis. J.Gastrointest. Surg. 2010, 14, 1578–1591. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, S.D.; Johnson, P.T.; Sheth, S.; Hruban, R.; Fishman, E.K. Ct of gallbladder cancer and its mimics: A pattern-based approach. Abdom. Imaging 2013, 38, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Lee, J.M.; Lee, E.S.; Han, J.K.; Choi, B.I. Preoperative staging of gallbladder carcinoma using biliary mr imaging. J. Magn. Reson. Imaging JMRI 2015, 41, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. Micrornas modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M.; Calin, G.A. MiRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Micrornas as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C. Oncomirs: The discovery and progress of microRNAs in cancers. Mol. Cancer 2007, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zarate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzo, M.; et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Voortman, J.; Giovannetti, E.; Steinberg, S.M.; Leon, L.G.; Kim, Y.T.; Funel, N.; Park, J.K.; Kim, M.A.; Kang, G.H.; et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE 2010, 5, e10630. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, A.; Sakatani, T.; Ushiku, T.; Hino, R.; Isogai, M.; Ishikawa, S.; Uozaki, H.; Takada, K.; Fukayama, M. Downregulation of microRNA-200 in ebv-associated gastric carcinoma. Cancer Res. 2010, 70, 4719–4727. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, Z.; Zhao, Y.; Ding, Y.; Liu, H.; Xi, Y.; Xiong, W.; Li, G.; Lu, J.; Fodstad, O.; et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1). J. Biol. Chem. 2010, 285, 21496–21507. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear rnase iii drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase iii transcribes human microRNAs. Nat. Struct Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer. Adv. Exp. Med. Biol 2013, 774, 1–20. [Google Scholar] [PubMed]
- Filippov, V.; Solovyev, V.; Filippova, M.; Gill, S.S. A novel type of RNAse iii family proteins in eukaryotes. Gene 2000, 245, 213–221. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human dicer. Nat. Struct. Mol. Biol. 2013, 19, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.W.; MacRae, I.J. The molecular machines that mediate microRNA maturation. J. Cell. Mol. Med. 2009, 13, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for risc loading and slicer-independent unwinding. Nat. Struct. Mol. Biol 2009, 16, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.O.; Lai, E.C. Mirtrons: MicroRNA biogenesis via splicing. Biochimie 2011, 93, 1897–1904. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate miR-451 provides a platform for dicer-independent, ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 15163–15168. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Chung, W.J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Mol. Cell 2007, 28, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Steitz, J.A. Versatile microRNA biogenesis in animals and their viruses. RNA Biol. 2014, 11, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. MicroRNA-dependent localization of targeted mrnas to mammalian p-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ Utr as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef] [PubMed]
- Duursma, A.M.; Kedde, M.; Schrier, M.; le Sage, C.; Agami, R. miR-148 targets human dnmt3b protein coding region. RNA 2008, 14, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Jin, L.; Zhang, F.; Sarnow, P.; Kay, M.A. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mrnas. Nat. Struct. Mol. Biol. 2009, 16, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Voinnet, O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009, 10, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Long, D.; Lee, R.; Williams, P.; Chan, C.Y.; Ambros, V.; Ding, Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007, 14, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ding, X.C.; Weiler, J.; Grosshans, H. Regulating the regulators: Mechanisms controlling the maturation of microRNAs. Trends Biotechnol. 2009, 27, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Jay, C.; Nemunaitis, J.; Chen, P.; Fulgham, P.; Tong, A.W. MiRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007, 26, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Esteller, M. CPG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle 2007, 6, 1455–1459. [Google Scholar] [CrossRef] [PubMed]
- Davis-Dusenbery, B.N.; Hata, A. Mechanisms of control of microRNA biogenesis. J. Biochem. 2010, 148, 381–392. [Google Scholar] [PubMed]
- Han, L.; Witmer, P.D.; Casey, E.; Valle, D.; Sukumar, S. DNA methylation regulates microRNA expression. Cancer Biol. Ther. 2007, 6, 1284–1288. [Google Scholar] [CrossRef] [PubMed]
- Hudson, R.S.; Yi, M.; Esposito, D.; Watkins, S.K.; Hurwitz, A.A.; Yfantis, H.G.; Lee, D.H.; Borin, J.F.; Naslund, M.J.; Alexander, R.B.; et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2011, 40, 3689–3703. [Google Scholar] [CrossRef] [PubMed]
- Datta, J.; Kutay, H.; Nasser, M.W.; Nuovo, G.J.; Wang, B.; Majumder, S.; Liu, C.G.; Volinia, S.; Croce, C.M.; Schmittgen, T.D.; et al. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008, 68, 5049–5058. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Takatsuka, S.; Akashi, H.; Yamamoto, E.; Nojima, M.; Maruyama, R.; Kai, M.; Yamano, H.O.; Sasaki, Y.; Tokino, T.; et al. Genome-wide profiling of chromatin signatures reveals epigenetic regulation of microRNA genes in colorectal cancer. Cancer Res. 2011, 71, 5646–5658. [Google Scholar] [CrossRef] [PubMed]
- Krzeminski, P.; Sarasquete, M.E.; Misiewicz-Krzeminska, I.; Corral, R.; Corchete, L.A.; Martin, A.A.; Garcia-Sanz, R.; San Miguel, J.F.; Gutierrez, N.C. Insights into epigenetic regulation of microRNA-155 expression in multiple myeloma. Biochim. Biophys. Acta 2014, 1849, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Dakhlallah, D.; Batte, K.; Wang, Y.; Cantemir-Stone, C.Z.; Yan, P.; Nuovo, G.; Mikhail, A.; Hitchcock, C.L.; Wright, V.P.; Nana-Sinkam, S.P.; et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2013, 187, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006, 20, 2202–2207. [Google Scholar] [CrossRef] [PubMed]
- Karube, Y.; Tanaka, H.; Osada, H.; Tomida, S.; Tatematsu, Y.; Yanagisawa, K.; Yatabe, Y.; Takamizawa, J.; Miyoshi, S.; Mitsudomi, T.; et al. Reduced expression of dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005, 96, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.X.; Fan, L.; Lu, R.N.; Fang, C.; Shen, W.Y.; Zou, Z.J.; Wang, Y.H.; Zhu, H.Y.; Miao, K.R.; Liu, P.; et al. Downregulated dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci. 2012, 103, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Gillies, J.K.; Lorimer, I.A. Regulation of p27kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 2007, 6, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. Dicer1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.D.; Ross, S.A. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr. Rev. 2008, 66, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, S.D.; Carletti, M.Z.; Hong, X.; Christenson, L.K. Hormonal regulation of microRNA expression in periovulatory mouse mural granulosa cells. Biol. Reprod. 2008, 79, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, R.; Ferracin, M.; Negrini, M.; Calin, G.A.; Davuluri, R.V.; Ivan, M. Regulation of microRNA expression: The hypoxic component. Cell Cycle 2007, 6, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell. Mol. Med. 2014, 18, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Bu, X.; Dai, C.; Shang, C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour Biol. 2015, 36, 4773–4776. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [PubMed]
- Sun, T.; Kalionis, B.; Lv, G.; Xia, S.; Gao, W. Role of exosomal noncoding rnas in lung carcinogenesis. BioMed Res. Int. 2015, 2015, 125807. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mrnas and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Farber, E.L.; Rapoport, A.L.; Tejada, D.; Deniskin, R.; Akhmedov, N.B.; Farber, D.B. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS ONE 2009, 4, e4722. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mrna and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Khoury, S.; Tran, N. Circulating microRNAs: Potential biomarkers for common malignancies. Biomark. Med. 2009, 9, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Shigehara, K.; Yokomuro, S.; Ishibashi, O.; Mizuguchi, Y.; Arima, Y.; Kawahigashi, Y.; Kanda, T.; Akagi, I.; Tajiri, T.; Yoshida, H.; et al. Real-time pcr-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS ONE 2011, 6, e23584. [Google Scholar] [CrossRef] [PubMed]
- Caby, M.P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yang, J.; Xie, R.; Gao, L.; Yang, Y.; Fan, H.; Qian, K. Exosomal-like vesicles with immune-modulatory features are present in human plasma and can induce CD4+ T-cell apoptosis in vitro. Transfusion 2011, 51, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma extracellular rna profiles in healthy and cancer patients. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small rna composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Messina, L.; Gutierrez-Vazquez, C.; Rivas-Garcia, E.; Sanchez-Madrid, F.; de la Fuente, H. Immunomodulatory role of microRNAs transferred by extracellular vesicles. Biol. Cell 2015, 107, 61–77. [Google Scholar] [CrossRef] [PubMed]
- de Candia, P.; Torri, A.; Gorletta, T.; Fedeli, M.; Bulgheroni, E.; Cheroni, C.; Marabita, F.; Crosti, M.; Moro, M.; Pariani, E.; et al. Intracellular modulation, extracellular disposal and serum increase of miR-150 mark lymphocyte activation. PLoS ONE 2013, 8, e75348. [Google Scholar] [CrossRef] [PubMed]
- Ostenfeld, M.S.; Jeppesen, D.K.; Laurberg, J.R.; Boysen, A.T.; Bramsen, J.B.; Primdal-Bengtson, B.; Hendrix, A.; Lamy, P.; Dagnaes-Hansen, F.; Rasmussen, M.H.; et al. Cellular disposal of mir23b by rab27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014, 74, 5758–5771. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.L.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Zernecke, A.; Bidzhekov, K.; Noels, H.; Shagdarsuren, E.; Gan, L.; Denecke, B.; Hristov, M.; Koppel, T.; Jahantigh, M.N.; Lutgens, E.; et al. Delivery of microRNA-126 by apoptotic bodies induces cxcl12-dependent vascular protection. Sci. Signal. 2009, 2, ra81. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Riwanto, M.; Besler, C.; Knau, A.; Fichtlscherer, S.; Roxe, T.; Zeiher, A.M.; Landmesser, U.; Dimmeler, S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Henson, R.; Lang, M.; Wehbe, H.; Maheshwari, S.; Mendell, J.T.; Jiang, J.; Schmittgen, T.D.; Patel, T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006, 130, 2113–2129. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Pu, Y. MicroRNA signatures in total peripheral blood of gallbladder cancer patients. Tumour Biol. 2015, 36, 6985–6990. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yan, H.X.; Yang, W.; Hu, L.; Yu, L.X.; Liu, Q.; Li, L.; Huang, D.D.; Ding, J.; Shen, F.; et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J. Hepatol. 2009, 50, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yin, J.; Li, T.; Yuan, L.; Wang, D.; He, J.; Du, X.; Lu, J. Upregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinoma. Oncol. Rep. 2015, 33, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Silakit, R.; Loilome, W.; Yongvanit, P.; Chusorn, P.; Techasen, A.; Boonmars, T.; Khuntikeo, N.; Chamadol, N.; Pairojkul, C.; Namwat, N. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: A prospective prognostic indicator. J. Hepatobiliary Pancreat. Sci. 2014, 21, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Voigtlander, T.; Gupta, S.K.; Thum, S.; Fendrich, J.; Manns, M.P.; Lankisch, T.O.; Thum, T. Micrornas in serum and bile of patients with primary sclerosing cholangitis and/or cholangiocarcinoma. PLoS ONE 2015, 10, e0139305. [Google Scholar] [CrossRef] [PubMed]
- Bernuzzi, F.; Marabita, F.; Lleo, A.; Carbone, M.; Mirolo, M.; Marzioni, M.; Alpini, G.; Alvaro, D.; Muri Boberg, K.; Locati, M.; et al. Serum microRNAs as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Clin. Exp. Immunol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Sudo, H.; Kawauchi, J.; Takizawa, S.; Kondou, S.; Nobumasa, H.; Ochiai, A. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS ONE 2015, 10, e0118220. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Feng, F.; Zhu, L.; Zheng, Y.; Luo, X.; Liu, C.; Yi, B.; Jiang, X. Circulating miR-106a is a novel prognostic and lymph node metastasis indicator for cholangiocarcinoma. Sci. Rep. 2015, 5, 16103. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chen, Y.; Wu, W.; Ouyang, N.; Chen, J.; Li, H.; Liu, X.; Su, F.; Lin, L.; Yao, Y. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS ONE 2013, 8, e80707. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Jin, H.; Yang, Z.; Luo, G.; Lu, Y.; Li, K.; Ren, G.; Su, T.; Pan, Y.; Feng, B.; et al. miR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene egr2. Biochem. Biophys. Res. Commun. 2010, 392, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yang, Y.; Zhang, P.; Wang, F.; Ma, Y.; Qin, H.; Wang, Y. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-myb. J. Cell. Mol. Med. 2014, 18, 2125–2134. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Pan, S.; Kang, M.; Dong, R.; Zhao, J. MicroRNA-150 functions as a tumor suppressor in osteosarcoma by targeting IGF2BP1. Tumour Biol. 2016, 37, 5275–5284. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wu, X.; Cao, H.; Zhan, Q.; Xia, M.; Zhou, Q.; Cai, X.; An, F. Regulatory role of serum miR-224 in invasiveness and metastasis of cholangiocarcinoma. Zhonghua Gan Zang Bing Za Zhi 2015, 23, 748–753. (In Chinese) [Google Scholar] [PubMed]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, A.; Szura, M.; Gil, K.; Brzozowska, I.; Maduzia, D.; Mizia, E.; Walocha, K.; Matyja, A. Metabolism of bile with respect to etiology of gallstone disease—Systematic review. Folia Med. Cracov. 2014, 54, 5–16. [Google Scholar] [PubMed]
- Munoz-Garrido, P.; Garcia-Fernandez de Barrena, M.; Hijona, E.; Carracedo, M.; Marin, J.J.; Bujanda, L.; Banales, J.M. Micrornas in biliary diseases. World J. Gastroenterol. 2012, 18, 6189–6196. [Google Scholar] [CrossRef] [PubMed]
- Haga, H.; Yan, I.; Takahashi, K.; Wood, J.; Patel, T. Emerging insights into the role of microRNAs in the pathogenesis of cholangiocarcinoma. Gene Expr. 2014, 16, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Seo, N.; Kim, S.Y.; Lee, S.S.; Byun, J.H.; Kim, J.H.; Kim, H.J.; Lee, M.G. Sclerosing cholangitis: Clinicopathologic features, imaging spectrum, and systemic approach to differential diagnosis. Korean J. Radiol. 2016, 17, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Hundal, R.; Shaffer, E.A. Gallbladder cancer: Epidemiology and outcome. Clin. Epidemiol. 2014, 6, 99–109. [Google Scholar] [PubMed]
- Lamont, J.T.; Carey, M.C. Cholesterol gallstone formation. 2. Pathobiology and pathomechanics. Prog. Liver Dis. 1992, 10, 165–191. [Google Scholar] [PubMed]
- Yang, B.; Liu, B.; Bi, P.; Wu, T.; Wang, Q.; Zhang, J. An integrated analysis of differential miRNA and mRNA expressions in human gallstones. Mol. Biosyst. 2015, 11, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Fanyin Meng, H.F.; Han, Y.; Glaser, S.; Alpini, G. Functional role of microRNA-200 family in human gall bladder cancer stem cells. FASEB J. 2015, 29 (Suppl. 45), 45–47. [Google Scholar]
- Xiao, Y.; Wang, J.; Yan, W.; Zhou, Y.; Chen, Y.; Zhou, K.; Wen, J.; Wang, Y.; Cai, W. Dysregulated miR-124 and miR-200 expression contribute to cholangiocyte proliferation in the cholestatic liver by targeting il-6/stat3 signalling. J. Hepatol. 2014, 62, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Zahm, A.M.; Hand, N.J.; Boateng, L.A.; Friedman, J.R. Circulating microRNA is a biomarker of biliary atresia. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kemper, J.K. Controlling sirt1 expression by microRNAs in health and metabolic disease. Aging (Albany NY) 2010, 2, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, R.M.; van Mil, S.W.; Oldenburg, B.; Siersema, P.D.; Klomp, L.W.; van Erpecum, K.J. Bile acids and their nuclear receptor FXR: Relevance for hepatobiliary and gastrointestinal disease. Biochim. Biophys Acta 2010, 1801, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Tarling, E.J.; Ahn, H.; de Aguiar Vallim, T.Q. The nuclear receptor FXR uncouples the actions of miR-33 from srebp-2. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.M.; Marquart, T.J.; Albert, C.J.; Suchy, F.J.; Wang, D.Q.; Ananthanarayanan, M.; Ford, D.A.; Baldan, A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol. Med. 2012, 4, 882–895. [Google Scholar] [CrossRef] [PubMed]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Kim, T.; Civelek, M.; Baldan, A.; Esau, C.; Edwards, P.A. Microrna-144 regulates hepatic atp binding cassette transporter a1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid x receptor. Circ. Res. 2013, 112, 1602–1612. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Li, T.; Owsley, E.; Chiang, J.Y. A putative role of microRNA in regulation of cholesterol 7α-hydroxylase expression in human hepatocytes. J. Lipid Res. 2010, 51, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. MicroRNA-370 controls the expression of microRNA-122 and cpt1alpha and affects lipid metabolism. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Gerin, I.; Bommer, G.T.; McCoin, C.S.; Sousa, K.M.; Krishnan, V.; MacDougald, O.A. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E198–E206. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Gao, Z.; Alarcon, R.M.; Ye, J.; Yun, Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009, 276, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, N.; Nakagawa, Y.; Tokushige, N.; Aoki, N.; Matsuzaka, T.; Ishii, K.; Yahagi, N.; Kobayashi, K.; Yatoh, S.; Takahashi, A.; et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem. Biophys. Res. Commun. 2009, 385, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. miR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Horie, T.; Nishino, T.; Baba, O.; Kuwabara, Y.; Yokode, M.; Kita, T.; Kimura, T. Microrna-33a/b in lipid metabolism—Novel “Thrifty” Models. Circ. J. 2015, 79, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.L.; Khosroheidari, M.; Eddy, E.; Done, S.C. MicroRNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis 2015, 242, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Nishi, H.; Fisher, E.A. Cholesterol homeostasis regulation by miR-223: Basic science mechanisms and translational implications. Circ. Res. 2015, 116, 1112–1114. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.; Wu, T.; Miao, L.; Mei, Y.; Wu, M. miR-181a regulates lipid metabolism via IDH1. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Zhan, X.R.; Li, X.Y.; Yu, J.J.; Liu, X.M. MicroRNA-185 regulates expression of lipid metabolism genes and improves insulin sensitivity in mice with non-alcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 17914–17923. [Google Scholar] [PubMed]
- Vickers, K.C.; Landstreet, S.R.; Levin, M.G.; Shoucri, B.M.; Toth, C.L.; Taylor, R.C.; Palmisano, B.T.; Tabet, F.; Cui, H.L.; Rye, K.A.; et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef] [PubMed]
- Stokes, C.S.; Krawczyk, M.; Lammert, F. Gallstones: Environment, lifestyle and genes. Dig. Dis. 2011, 29, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Di Ciaula, A.; Wang, D.Q.; Bonfrate, L.; Portincasa, P. Current views on genetics and epigenetics of cholesterol gallstone disease. Cholesterol 2013, 2013, 298421. [Google Scholar] [CrossRef] [PubMed]
- Eiholm, S.; Thielsen, P.; Kromann-Andersen, H. Endoscopic brush cytology from the biliary duct system is still valuable. Dan Med. J. 2013, 60, A4656. [Google Scholar] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef]
- Shen, J.; Stass, S.A.; Jiang, F. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013, 329, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large b-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Guo, W.; Zhao, Y.; Wang, Y.; Zha, R.; Ding, J.; Liang, L.; Hu, J.; Shen, H.; Chen, Z.; et al. MicroRNA-26a acts as a tumor suppressor inhibiting gallbladder cancer cell proliferation by directly targeting HMGA2. Int. J. Oncol. 2014, 44, 2050–2058. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Guo, W.; Zhao, Y.; Wang, Y.; Zha, R.; Ding, J.; Liang, L.; Yang, G.; Chen, Z.; Ma, B.; et al. MicroRNA-135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci. 2014, 105, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Xiang, Y.; Tang, J.; Wu, G.; Li, J.; Xiao, H.; Li, C.; Chen, Y.; Zhao, J. miR-34 is associated with poor prognosis of patients with gallbladder cancer through regulating telomere length in tumor stem cells. Tumour Biol. 2013, 35, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.H.; Zhang, Y.D.; Gong, L.S.; Liu, W.D.; Zhang, Y. Increased expression of microRNA-335 predicts a favorable prognosis in primary gallbladder carcinoma. Onco. Targets Ther. 2013, 6, 1625–1630. [Google Scholar] [PubMed]
- Ma, M.Z.; Li, C.X.; Zhang, Y.; Weng, M.Z.; Zhang, M.D.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA hotair, a c-myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer 2014, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.Z.; Chu, B.F.; Zhang, Y.; Weng, M.Z.; Qin, Y.Y.; Gong, W.; Quan, Z.W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218–5p. Cell. Death Dis. 2015, 6, e1583. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Nakamura, M.; Ohtsuka, T.; Nagayoshi, Y.; Mori, Y.; Takahata, S.; Aishima, S.; Tanaka, M. High expression of microRNA-155 is associated with the aggressive malignant behavior of gallbladder carcinoma. Oncol. Rep. 2013, 30, 17–24. [Google Scholar] [PubMed]
- Chang, Y.; Liu, C.; Yang, J.; Liu, G.; Feng, F.; Tang, J.; Hu, L.; Li, L.; Jiang, F.; Chen, C.; et al. miR-20a triggers metastasis of gallbladder carcinoma. J. Hepatol. 2013, 59, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Luo, X.; Kan, T.; Zhang, Y.; Yu, W.; Wei, Y.; Shen, N.; Yi, B.; Jiang, X. TGF-β upregulates miR-182 expression to promote gallbladder cancer metastasis by targeting cadm1. Mol. Biosyst. 2014, 10, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Han, C.; Wu, T. MicroRNA-26a promotes cholangiocarcinoma growth by activating β-catenin. Gastroenterology 2012, 143, 246–256.e8. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, J.; Liu, X.; Guo, Z.; Sun, X.; Zhang, J. The real-time dynamic monitoring of microRNA function in cholangiocarcinoma. PLoS ONE 2014, 9, e99431. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.H.; Park, S.J.; Kim, H.K. miR-215 overexpression distinguishes ampullary carcinomas from pancreatic carcinomas. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 325–329. [Google Scholar] [CrossRef]
- Witek, R.P.; Yang, L.; Liu, R.; Jung, Y.; Omenetti, A.; Syn, W.K.; Choi, S.S.; Cheong, Y.; Fearing, C.M.; Agboola, K.M.; et al. Liver cell-derived microparticles activate hedgehog signaling and alter gene expression in hepatic endothelial cells. Gastroenterology 2009, 136, 320–330.e2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Pigati, L.; Yaddanapudi, S.C.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.L.; Wojcik, S.; Liu, J.; Frankel, W.L.; Alder, H.; Yu, L.; Schmittgen, T.D.; Croce, C.M.; Bloomston, M. A differential microRNA profile distinguishes cholangiocarcinoma from pancreatic adenocarcinoma. Ann. Surg. Oncol. 2014, 21, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Shimada, Y.; Nagata, T.; Sawada, S.; Yoshioka, I.; Matsui, K.; Moriyama, M.; Omura, T.; Osawa, S.; Shibuya, K.; et al. Role of aquaporin-5 in gallbladder carcinoma. Eur. Surg. Res. 2013, 51, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Paraskevi, A.; Theodoropoulos, G.; Papaconstantinou, I.; Mantzaris, G.; Nikiteas, N.; Gazouli, M. Circulating microRNA in inflammatory bowel disease. J. Crohns Colitis 2012, 6, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Sandanayake, N.S.; Sinclair, J.; Andreola, F.; Chapman, M.H.; Xue, A.; Webster, G.J.; Clarkson, A.; Gill, A.; Norton, I.D.; Smith, R.C.; et al. A combination of serum leucine-rich α-2-glycoprotein 1, CA19–9 and interleukin-6 differentiate biliary tract cancer from benign biliary strictures. Br. J. Cancer 2011, 105, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Morimura, R.; Nagata, H.; Kosuga, T.; Iitaka, D.; Konishi, H.; Shiozaki, A.; et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer 2011, 105, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Yang, L.F.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhu, Y.P.; Shen, Y.J.; Shi, G.H.; Ye, D.W. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011, 71, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.M.; Lin, P.M.; Wang, Y.M.; Chen, Z.J.; Lin, S.F.; Yang, M.Y. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol. 2012, 33, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Tsujiura, M.; Ichikawa, D.; Komatsu, S.; Shiozaki, A.; Takeshita, H.; Kosuga, T.; Konishi, H.; Morimura, R.; Deguchi, K.; Fujiwara, H.; et al. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer 2010, 102, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; Bian, H.B.; Wang, J.R.; Cheng, Z.X.; Wang, K.M.; De, W. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J. Surg. Oncol. 2011, 104, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Kanaan, Z.; Rai, S.N.; Eichenberger, M.R.; Roberts, H.; Keskey, B.; Pan, J.; Galandiuk, S. Plasma miR-21: A potential diagnostic marker of colorectal cancer. Ann. Surg. 2012, 256, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, V.J.; Maxey, R.W.; Levy, R.A. Racial and ethnic differences in response to medicines: Towards individualized pharmaceutical treatment. J. Natl. Med. Assoc. 2002, 94, 1–26. [Google Scholar] [PubMed]
- Chapman, M.H.; Tidswell, R.; Dooley, J.S.; Sandanayake, N.S.; Cerec, V.; Deheragoda, M.; Lee, A.J.; Swanton, C.; Andreola, F.; Pereira, S.P. Whole genome rna expression profiling of endoscopic biliary brushings provides data suitable for biomarker discovery in cholangiocarcinoma. J. Hepatol. 2012, 56, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wu, Y.; Wang, D.; Huang, S.; Wen, Y.; Cao, J.; Zhang, L. Establishment of an experimental method for detecting circulating miRNAs in BDL mice. Clin Exp. Med. 2011, 12, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
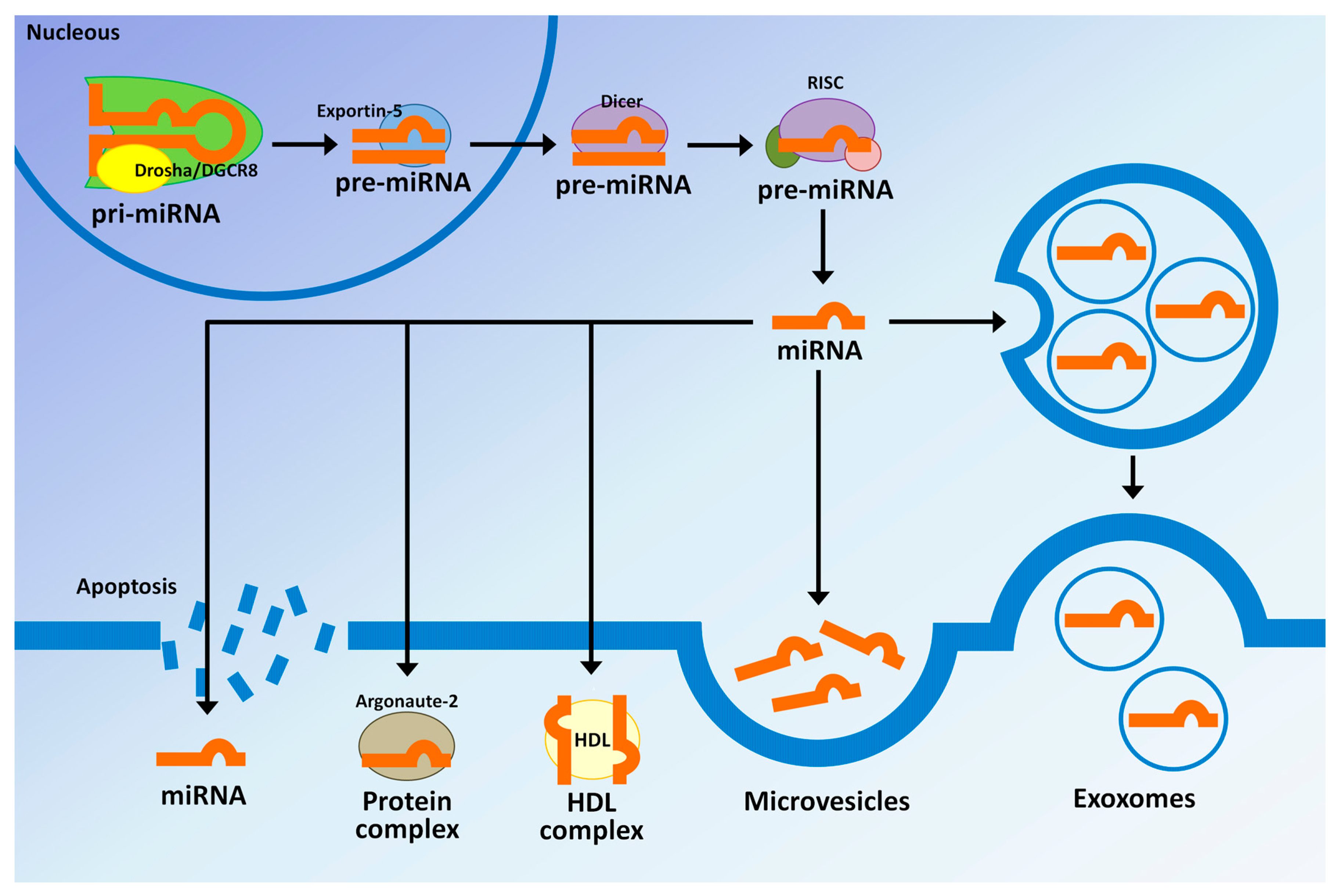
| miRNAs | AUC | Sensitivity | Specificity | Reference |
|---|---|---|---|---|
| BTCs vs. HVs | Kishimoto et al. [30] | |||
| miR-21 | 0.93 | 85.1% | 100% | |
| BTCs vs. BBD | ||||
| miR-21 | 0.83 | 72.3% | 91.3% | |
| iCCA vs. controls | Wang et al. [134] | |||
| miR-150 | 0.764 | 80.6% | 58.1% | |
| iCCA vs. HVs | Silakit et al. [135] | |||
| miR-192 | 0.803 | 74% | 72% | |
| PSC vs. CCA (serum) | Voigtlander et al. [136] | |||
| miR-1281 | 0.83 | 55% | 90% | |
| miR-126 | 0.87 | 68% | 93% | |
| miR-26a | 0.78 | 52% | 93% | |
| miR-30b | 0.78 | 52% | 88% | |
| miR-122 | 0.65 | 32% | 90% | |
| PSC vs. CCA (bile) | ||||
| miR-412 | 0.81 | 50% | 89% | |
| miR-640 | 0.81 | 50% | 92% | |
| miR-1537 | 0.78 | 67% | 90% | |
| miR-3189 | 0.80 | 67% | 89% | |
| PSC vs. HVs | Bernuzzi et al. [137] | |||
| miR-200c | 0.74 | -- | -- | |
| CCA vs. HVs | ||||
| miR-483-5p | 0.77 | -- | -- | |
| miR-194 | 0.74 | -- | -- | |
| miR-483-5p and miR-194 | 0.81 | -- | -- | |
| CCA vs. PSC | ||||
| miR-222 | 0.71 | -- | -- | |
| miR-483-5p | 0.70 | -- | -- | |
| miR-222 and miR-483-5p | 0.77 | -- | -- | |
| PBC vs. HVs | Kojima et al. [138] | |||
| Combination of eight | 0.953 | 80.3% | 97.6% | |
| miRNAs (miR-6075, miR-4294, miR-6880-5p, miR-6799-5p, miR-125a-3p, miR-4530, miR-6836-3p, and miR-4476) | ||||
| BTCs vs. choledocholithiasis | Shigehara et al. [114] | |||
| miR-9 | 0.975 | 88.9% | 100% | |
| miR-145* | 0.975 | 77.8% | 100% | |
| miR-944 | 0.765 | 77.8% | 100% | |
| CCA vs. HVs | Cheng et al. [139] | |||
| miR-106a | 0.89 | 81.6% | 85% |
| miRNAs: Up-/Down-Regulation | Samples Number | Type of Sample | Diagnosis/Prognosis Potential | Origin of Specimen | Relevance | Reference |
|---|---|---|---|---|---|---|
| Up: miR-21, miR-187, and miR-202 Down: let-7a, miR-143 and miR-335 | GBC (40); HVs (40) | Plasma | All diagnosis/only miR-187, miR-202 and miR-143 prognosis | China | GBC versus HVs | Li et al. [132] |
| Up: miR-21 | BTCs (94) including CCA, GBC and AC; HVs (50); BBD (23) | Plasma | Diagnosis and prognosis | Japan | BTCs versus HVs and BTCs versus BBD | Kishimoto et al. [30] |
| Up: miR-483-5p, miR505-3p, miR874, miR885-5p, miR-320b, miR-92b-3p, miR1275 and miR1307-3p | iCCA (13); HVs (5) | Plasma | Diagnosis | Thailand | iCCA versus HVs | Plieskatt et al. [40] |
| Up: miR-150 | iCCA (15) | Plasma | Diagnosis | China | iCCA versus controls | Wang et al. [134] |
| Up: miR-192 | iCCA (51); HVs (32) | Serum | Diagnosis and prognosis | Thailand | iCCA versus HVs | Silakit et al. [135] |
| Up (serum samples): miR-1281, miR-126, miR-26a, miR-30b and miR-122 Down (bile samples): miR-412, miR-640, miR-1537 Up (bile samples): miR-3189 | PSC (40 serum,52bile); CCA (31 serum, 19 bile); PSC/CCA (12 bile); HVs (12 serum) | Serum/bile | Diagnosis | Germany | PSC versus CCA | Voigtlander et al. [136] |
| Down: miR-200c (PSC vs. HVs) Up: miR-483-5p and miR-194 (CCA vs. HVs) Up: miR-222 and miR-483-5p (CCA vs. PSC) | CCA (70); PSC (70); HVs (70) | Serum | Diagnosis | Italy | PSC versus HVs; CCA versus HVs; CCA versus PSC | Bernuzzi et al. [137] |
| Down: miR-125a-3p and miR-6893-5p | BTCs (98) including iCCA, eCCA, GBC, HBD and AC; HVs (150) | Serum | Diagnosis | Japan | BTCs versus HVs | Kojima et al. [138] |
| Up: miR-191, miR-486-3p, miR-1274b, miR-16 and miR-484 | CCA (46); Control group (50) including BBO, PSC, SOD, CP and cholangitis | Bile in EVs | Diagnosis | USA (Including Caucasian;African American; Asian and Hispanic) | CCA versus PSC and BBO | Li et al. [24] |
| Up: miR-9, miR-145 *, miR-105, miR-147b, let-7f-2*, let-7i*, miR-302c*, miR-199a-3p, miR-222* and miR-942 | BTCs (9) including CCA and GBC; Choledocholithiasis (9) | Bile | Diagnosis | Japan | BTCs versus choledocholithiasis | Shigehara et al. [114] |
| Down:miR-106a Up: miR-21 (no significant difference) | CCA (103); BBD (34); HVs (20) | Serum | Diagnosis and Prognosis | China | CCA versus BBD and HVs | Cheng et al. [139] |
| Down: miR-21 and miR-17-5p (no significant difference) | PC (22); AC (6); HVs (8) | Serum in EVs | -- | China | AC versus PC | Que et al. [145] |
| Up: miR-210, miR-200c, miR-194 and miR-192 Down: miR-133a and miR-891a | Choledocholithiasis and HVs | Gallstones | -- | China | Choledocholithiasis versus HVs | Yang et al. [152] |
| Up: miR-224 | CCA (30); HVs (50) | Serum | Diagnosis | China | CCA versus HVs | Huang et al. [144] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Letelier, P.; Riquelme, I.; Hernández, A.H.; Guzmán, N.; Farías, J.G.; Roa, J.C. Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers. Int. J. Mol. Sci. 2016, 17, 791. https://doi.org/10.3390/ijms17050791
Letelier P, Riquelme I, Hernández AH, Guzmán N, Farías JG, Roa JC. Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers. International Journal of Molecular Sciences. 2016; 17(5):791. https://doi.org/10.3390/ijms17050791
Chicago/Turabian StyleLetelier, Pablo, Ismael Riquelme, Alfonso H. Hernández, Neftalí Guzmán, Jorge G. Farías, and Juan Carlos Roa. 2016. "Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers" International Journal of Molecular Sciences 17, no. 5: 791. https://doi.org/10.3390/ijms17050791
APA StyleLetelier, P., Riquelme, I., Hernández, A. H., Guzmán, N., Farías, J. G., & Roa, J. C. (2016). Circulating MicroRNAs as Biomarkers in Biliary Tract Cancers. International Journal of Molecular Sciences, 17(5), 791. https://doi.org/10.3390/ijms17050791