Abstract
The zebrafish (Danio rerio) is an ideal vertebrate model to investigate the developmental molecular mechanism of organogenesis and regeneration. Recent innovation in genome editing technologies, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) system, have allowed researchers to generate diverse genomic modifications in whole animals and in cultured cells. The CRISPR/Cas9 and TALEN techniques frequently induce DNA double-strand breaks (DSBs) at the targeted gene, resulting in frameshift-mediated gene disruption. As a useful application of genome editing technology, several groups have recently reported efficient site-specific integration of exogenous genes into targeted genomic loci. In this review, we provide an overview of TALEN- and CRISPR/Cas9-mediated site-specific integration of exogenous genes in zebrafish.
1. Introduction
The zebrafish is a useful model organism for basic biology and applied research, including in vivo drug screening, owing to its remarkable characteristics, such as its small size, rapid generation time and optical transparency during early embryogenesis [1,2]. Both knockdown and knockout analyses are useful for examining the in vivo function of uncharacterized genes. The injection of antisense morpholino oligonucleotides (MO) in zebrafish embryos, which causes suppression of translation of the target gene, is generally used to knock down the endogenous target gene [3]. However, undesirable off-target effects of MO injection have been reported and MO injection in zebrafish embryos frequently gives rise to the ectopic induction of p53 [4]. In the case of knockout analysis, genome editing technologies efficiently induce DNA double-strand breaks (DSBs) in the targeted gene, leading to frameshift-mediated gene disruption [5,6]. Two groups firstly reported the zinc finger nuclease (ZFN)-mediated gene disruptions in zebrafish [7,8]. Recent accumulating evidence shows that genetically gene-disrupted mutants and morphants (MO-injected embryos) often exhibit distinct phenotypes [4,9,10,11]. The complete suppression of maternal factors by MO is difficult, limiting the use of MO-based knockdown analysis. The functional analysis of zebrafish maternal-zygotic mutants established through genome editing technologies revealed novel developmental functions of maternal factors [12,13]. Therefore, these technologies are indispensable for the loss-of-function analysis of maternal and/or zygotic factors in zebrafish.
Genome editing technologies (ZFN, transcription activator-like effector nuclease (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) enable us to manipulate various genomic modifications in model organisms and in cultured cells [5,14,15,16]. Both ZFNs and TALENs are chimeric proteins fusing the DNA-binding domains required for the protein-DNA interaction and the FokI nuclease catalytic domain [17,18,19]. Because FokI nuclease functions as a dimer, DSBs are produced in the spacer region located between the DNA recognition sites of a pair of ZFNs or TALENs. A ZFN possesses four to six Cys2-His2 zinc finger motifs, one of which recognizes three nucleotides, and each zinc finger domain often interferes with nucleotide recognition specificity [20]. Each TALEN possesses 14.5–19.5 TALE effector repeats, one of which independently recognizes a single nucleotide. Each TALE repeat comprises 34 highly conserved residues except the two repeat-variable di-residues (RVDs) at amino acid positions 12 and 13. Therefore, the RVDs are essential for the nucleotide recognition specificity known as the TALE code: NG = T, HD = C, NI = A, NN = G or A. Efficient assembly methods for ordering TALE repeats have been developed [18,19,21,22]; therefore, the construction of functional TALENs is much easier than the construction of functional ZFNs. Recently, the CRISPR/Cas9 system has emerged as a more convenient technique for genome editing [15,23,24]. The type 2 CRISPR/Cas9 system originally consisted of three components: a Cas9 nuclease and two short RNAs, the CRISPR RNA (crRNA) and the trans-activating crRNA (tracrRNA) [15]. The crRNA is essential for the RNA–DNA interaction and possesses a complementary stretch of RNA (20 bases) to generate specificity to the targeted genome sequences followed by the protospacer-adjacent motif (PAM) sequence NGG (N: any nucleotide). The tracrRNA is required to interact with the crRNA and Cas9 nuclease. We and another group have reported a ready-to-use method consisting of synthetic crRNA, synthetic tracrRNA and recombinant Cas9; this complex cleaves the targeted genomic locus with high frequency [25,26]. Importantly, a single guide RNA (gRNA), combining the crRNA and tracrRNA, was elegantly developed and has been widely used as a simplified two-component system [23,24]. More recently, Zetsche et al. reported a novel class 2 CRISPR effector, Cpf1, which is a single RNA-guided endonuclease [27].
Genome editing technologies efficiently produce site-specific DSBs that are usually repaired by non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ) and homologous recombination (HR) (Figure 1) [28,29]. In the absence of donor DNA templates, the error-prone NHEJ pathway directly connects the ends of broken strands, leading to the production of insertion and/or deletion (indel) mutations. DSBs can be repaired by HR in the presence of donor DNA templates with large homology to the target site. Recently, an alternative end-joining pathway was characterized; MMEJ joins the exposed ends at microhomology regions (five to 25 bases) on the target locus, which are annealed and filled in by DNA polymerases. In mouse or rat, the HR-mediated knock-in of homologous fragments derived from a donor vector functions well. However, HR-dependent knock-in events are restricted in zebrafish as described below. One possible explanation of such a difference is that initial mitoses of zebrafish embryonic cleavages occur faster than those of mouse. Here, we review two HR-independent knock-in technologies, NHEJ- and MMEJ-mediated targeted integrations of exogenous genes, in zebrafish (Figure 2).
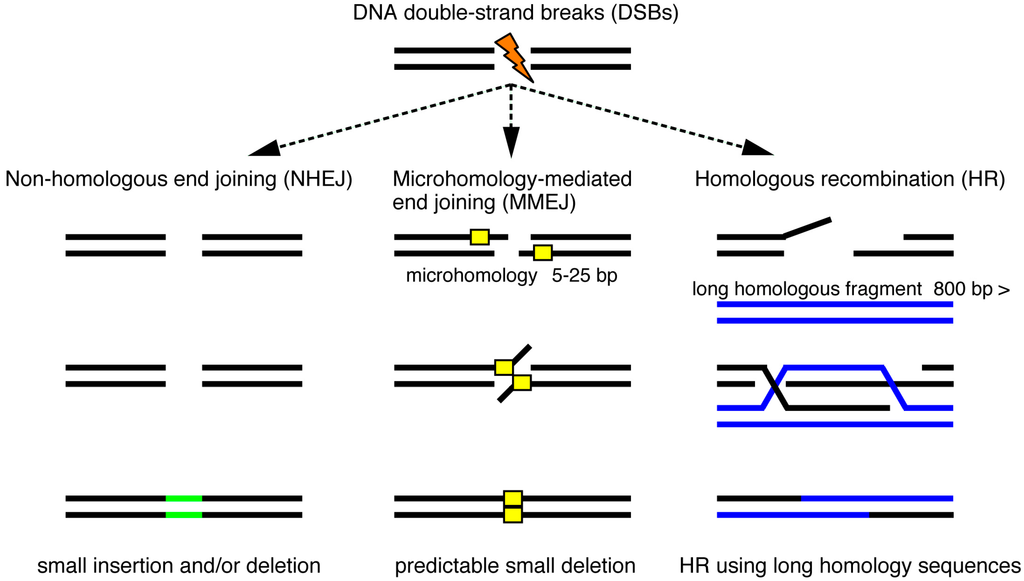
Figure 1.
Targeted genomic modifications using genome editing technologies. DNA double-strand breaks (DSBs) induced by genome editing technologies are repaired by non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ) and homologous recombination (HR). NHEJ repair, which connects the ends of the broken strands, leads to unpredictable insertion and/or deletion mutations (green bar), while MMEJ repair uses microhomology sequences (yellow box) and often causes a predictable small deletion. HR repair requires long double-strand DNA fragments (blue bar) that possess homology to the targeted genomic locus. Site-specific integrations of donor DNA are mediated by these DNA repair mechanisms.
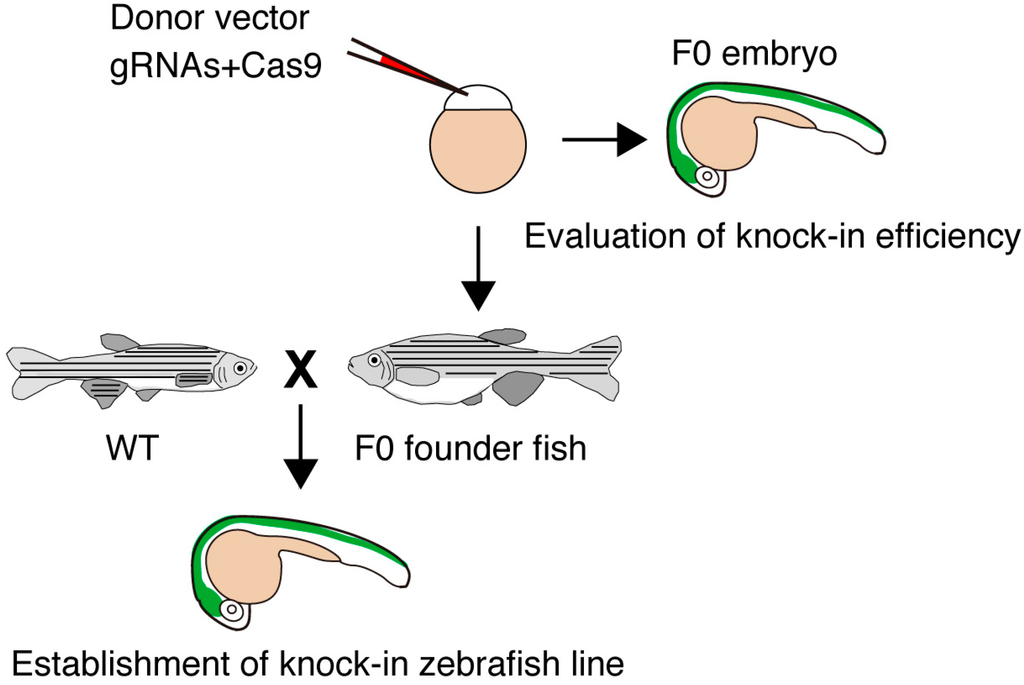
Figure 2.
Strategy for establishing knock-in fish. Donor vector, gRNAs and Cas9 mRNA are injected into zebrafish embryos. The knock-in event is estimated by examining the expression of the fluorescent gene (green area). Potential F0 founders are mated with wild-type (WT) fish, and the knock-in lines expressing the fluorescent gene are selected. The targeted knock-in at the targeted locus is determined by genomic PCR and sequencing analysis.
2. Genomic Insertion of Single-Stranded Oligodeoxynucleotides (ssODNs)
It has been shown that short oligonucleotide templates, called single-stranded oligodeoxynucleotides (ssODNs), can be used to introduce genomic alterations, including single nucleotide substitutions, to model organisms and cultured cells [30,31]. Using TALENs and the CRISPR/Cas9 system, ssODNs can be integrated at a targeted genomic locus in zebrafish [30,32,33,34]. Donor ssODNs possess approximately 20 to 50 base homologous sequences to the target site and the designed DNA fragments, such as loxP, restriction enzyme sites and hemagglutinin (HA) tags. Precise genomic alteration with ssODNs can be introduced at the targeted loci, whereas undesired mutations, including indel mutations and tandem integration of ssODNs, are simultaneously observed [30,32,33]. Interestingly, Yoshimi et al. demonstrated that the CAG-GFP donor vector was efficiently integrated into the rat Rosa26 locus using two ssODNs (80 bases) that bridged the linearized donor vector and the targeted genomic locus [31]. It is noteworthy that site-directed genomic modifications and donor DNA insertion using ssODNs are useful to establish model organisms for human genetic diseases, which includes the various mutations responsible for genetic disorders.
3. Precise Site-Specific Integration of Donor DNA by Homologous Recombination (HR)
Genome editing technologies (TALEN and CRISPR/Cas9) were applied to perform HR-mediated integration of donor DNA templates because the HR of the donor vector at the target site is largely enhanced by the production of site-specific DSBs. The eGFP reporter donor containing approximately 1 kb of homology arms was integrated at the target site using HR in zebrafish [35]; however, its efficacy for knock-in use was relatively low to put into practical use. In this report, the authors confirmed the germline transmission of the knock-in allele at the tyrosine hydroxylase locus in four of 275 potential F0 zebrafish founders. Subsequently, Shin et al. reported the efficient TALEN-mediated knock-in of donor DNA templates by optimizing the length of the homology arms and the configuration of the donor DNA construct [36]. They used various sizes of homology arms (ranging from 0.3 to 3.7 kb) in the donor vectors and found that the long homology arms (more than 2 kb) may be sufficient to achieve the HR-mediated knock-in. Additionally, superfolder GFP (sfGFP) was integrated by TALENs into the sox2 locus, and 29 of 363 F0 founders produced sfGFP-positive F1 embryos. Furthermore, tandem dimeric Tomato (tdTomato) was integrated into the gfap locus, and five of 44 F0 founders produced tdTomato-positive F1 embryos [36]. More recently, Irion et al. reported the CRISPR/Cas9-mediated knock-in of a donor vector [37]. To examine the knock-in efficacy, they chose the albino mutant (albb4), which carries a nonsense mutation in the slc45a2 gene essential for melanin production. Because the alb mutant embryos exhibit a pale phenotype with pigment defects, the phenotypic rescue by the HR-mediated knock-in of a donor vector containing the wild-type slc45a2 gene is easily determined by the appearance of the pigmented cells. They demonstrated that the frequency of phenotypic rescue was increased using circular donor DNA containing CRISPR target sites. In the rescue experiment, three of 28 F0 founders produced pigmented embryos with the wild-type phenotype [37]. Although the construction of donor vectors containing long homology arms is very complicated and time-consuming, HR-mediated genome editing in zebrafish is a powerful tool for precise genomic modifications.
4. Genomic Insertion of Donor DNA by Non-Homologous End Joining (NHEJ)
Recently, site-specific insertion via homology-independent repair mechanisms, presumably mediated by NHEJ, was developed. When the targeted genomic locus and the donor vector containing the ZFN or TALEN target sequences were simultaneously cleaved by ZFN or TALEN, the NHEJ-mediated insertion of donor DNA was observed in cultured cells at the target site [38,39]. One advantage of this method is the simple construction of the donor vector because there is no requirement for long homology arms. Auer et al. reported the efficient CRISPR/Cas9-mediated knock-in of donor vectors at targeted sites in zebrafish [40]. In this case, the donor vector contained a bait sequence efficiently cleaved by CRISPR/Cas9. When the donor vector containing a CRISPR target site was co-injected with gRNAs and Cas9 mRNA, concurrent cleavage of the donor vector and the targeted genomic locus caused the efficient genomic insertion of the donor vector at the target site. The authors targeted the eGFP locus in an eGFP transgenic line, and the donor vector containing the bait sequence and the transcriptional activator Gal4 gene was co-injected with gRNAs and Cas9 mRNA into the embryos containing the eGFP and UAS promoter-RFP transgenes. Successful in-frame Gal4 integration, but not out-of-frame and reverse integration, was easily visualized by a switch from eGFP to RFP expression. The authors demonstrated that the germline transmission rate of forward donor DNA integration was 31% (9 of 29 F0 founders) [40]. When they selected the embryos expressing RFP and allowed them to grow into adult fish, the germline transmission rate of in-frame integration increased to 40% (2 of 5 F0 founders). It should be noted that the ratio of in-frame integration of the donor vector is theoretically one-sixth of all integration events because out-of-frame and reverse integrations are not functional.
More recently, Kimura et al. improved the NHEJ-mediated knock-in of donor vectors for establishing transgenic zebrafish [41]. They prepared the donor vector containing a heat shock protein 70 (hsp) promoter and reporter/driver genes (fluorescent protein genes or the Gal4 gene) (Figure 3) and designed gRNA targeting 200–600 bp upstream of the target gene transcription start site. Because the hsp promoter in the donor DNA can receive the enhancer activity of the endogenous target gene, both forward and reverse genomic integration of the donor DNA should be functional. In fact, they established the Tg[evx2-hs:Gal4]; Tg[UAS:eGFP] knock-in line in the reverse direction and the eGFP expression was comparable to the endogenous expression of the evx2 gene. Furthermore, the introduction of the hsp promoter usually enhances the basal expression of the integrated reporter/driver gene. Immunostaining using an anti-Evx2 antibody revealed that the eGFP expression generally overlapped with Evx2 protein expression. However, the expression of the Evx2 protein was not detected in some of the eGFP-positive cells, presumably owing to the enhanced leaky expression of the hsp-dependent promoter activity. Using this strategy, the authors consistently established stable knock-in lines in four endogenous loci (evx2, eng1b, glyt2 and vglut2a) [41]. Importantly, the donor vector containing the reporter/driver gene is acceptable for genomic integration of any target genes. This makes this technique very convenient compared with the conventional BAC (bacterial artificial chromosome) method that requires complicated and tailored BAC constructs to individual target genes [42]. The CRISPR/Cas9-mediated knock-in of donor DNA becomes a powerful genetic tool for the generation of transgenic animals with cell-type-specific gene expression.
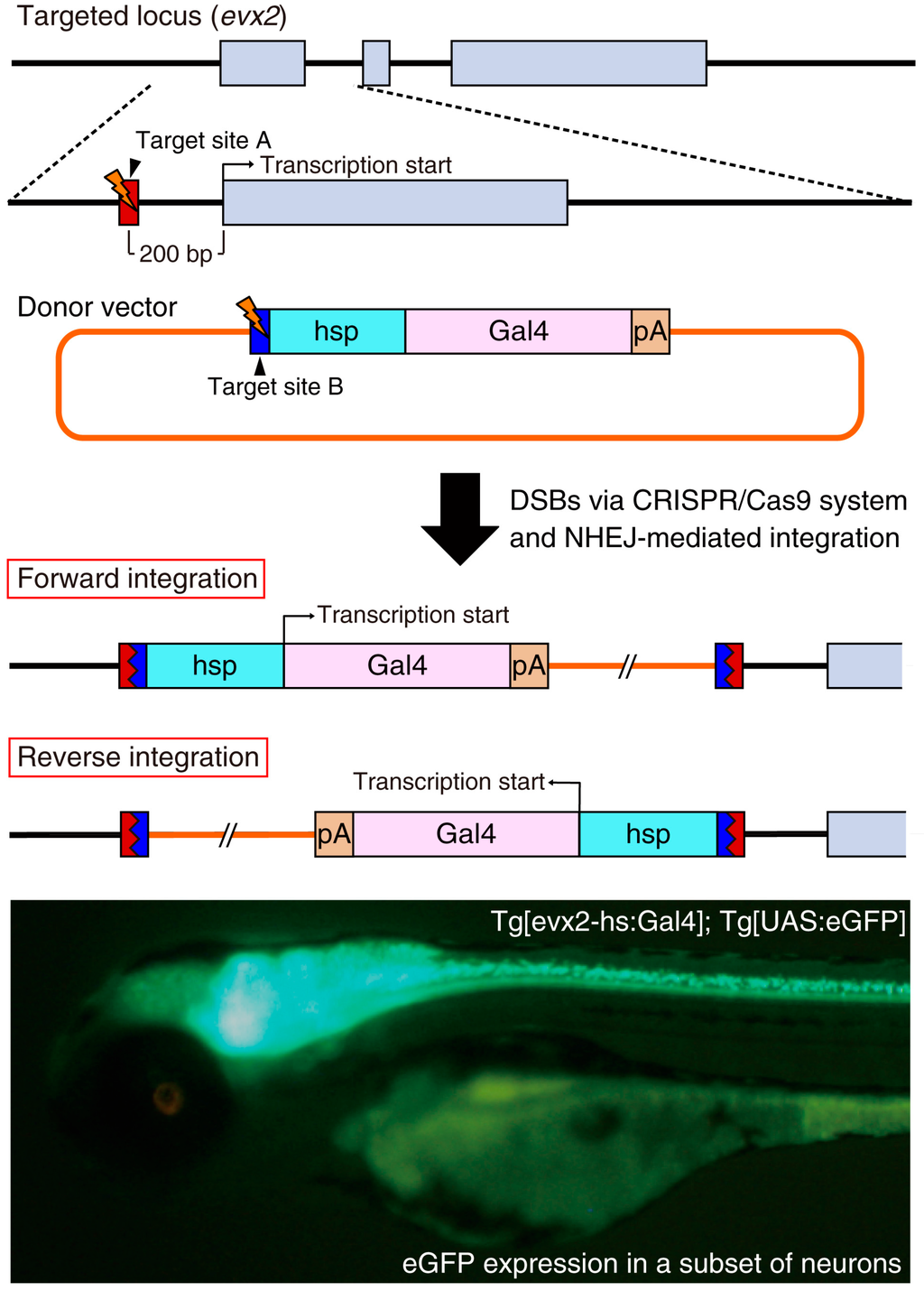
Figure 3.
Site-specific insertion of the hsp promoter-Gal4 donor into the evx2 locus. The donor vector consists of a bait sequence (blue box; target site B for Gbait-gRNA), the hsp promoter (light blue box), a Gal4 driver (pink box) and a polyA signal (pA) (orange box). The target site A for evx2-gRNA (red box) is located in the promoter region of the evx2 gene. When the CRISPR/Cas9 system simultaneously cleaves the target site A and B, the Gal4 driver is unpredictably integrated into the evx2 locus. Not only forward and reverse integrations but also tandem donor vector integrations can occur. We observed eGFP expression in a subset of neurons of the Tg[evx2-hs:Gal4]; Tg[UAS:eGFP] transgenic line because Gal4 driven by the endogenous evx2 enhancer induced eGFP expression as described previously [41]. Gray boxes; exons in the evx2 gene.
5. Precise Targeted Integration of Donor DNA by Microhomology-Mediated End Joining (MMEJ)
It has been recently shown that DSBs can be repaired by MMEJ that uses the existing microhomology sequences (five to 25 bases) around the broken DNA ends [29]. Both Dr. Yamamoto’s group and our group have developed novel knock-in methods acceptable for the precise targeted integration of exogenous genes mediated by the MMEJ pathway [43,44,45]. Using TALEN or the CRISPR/Cas9 system, they have succeeded in the MMEJ-mediated knock-in of donor vectors in cultured cells and in animals, including silkworms and frogs [43,45]. We designed a more versatile donor vector, which possesses a bait sequence (a functional gRNA target sequence) and short homology arms (10–40 bp) flanking the targeted genomic locus. We prepared a control vector without homology arms to examine the NHEJ-mediated knock-in of the donor vector. In the case of the tyrosinase locus, the NHEJ-mediated knock-in of the donor vector in the forward direction was detected in 53% of the injected F0 embryos using PCR, while the addition of 10–40 bp homology arms in the donor vector increased its efficiency to 85%. Genomic sequence analysis revealed that the MMEJ-mediated knock-in with precise targeted integration was 60%–77% of analyzed knock-in events. Therefore, the introduction of short homology arms promotes precise targeted integration of exogenous genes. In order to eliminate the undesired integration of the backbone vector sequences, we designed an improved donor vector that configures the eGFP and polyA signal between two bait sequences in the case of the keratin type 1 c19e/krtt1c19e locus (Figure 4). We observed eGFP expression in the epidermis at a high frequency (201/529 injected F0 embryos). We established stable zebrafish lines expressing eGFP in the epidermis, indicating that the MMEJ-mediated targeted integration of the exogenous gene is heritable. We found that the pre-screening of F0 embryos exhibiting appropriate eGFP expression is beneficial for the efficient identification of the knock-in allele. We propose that the MMEJ-mediated targeted integration of exogenous genes is a powerful genetic tool for precise genomic modifications in various model organisms.
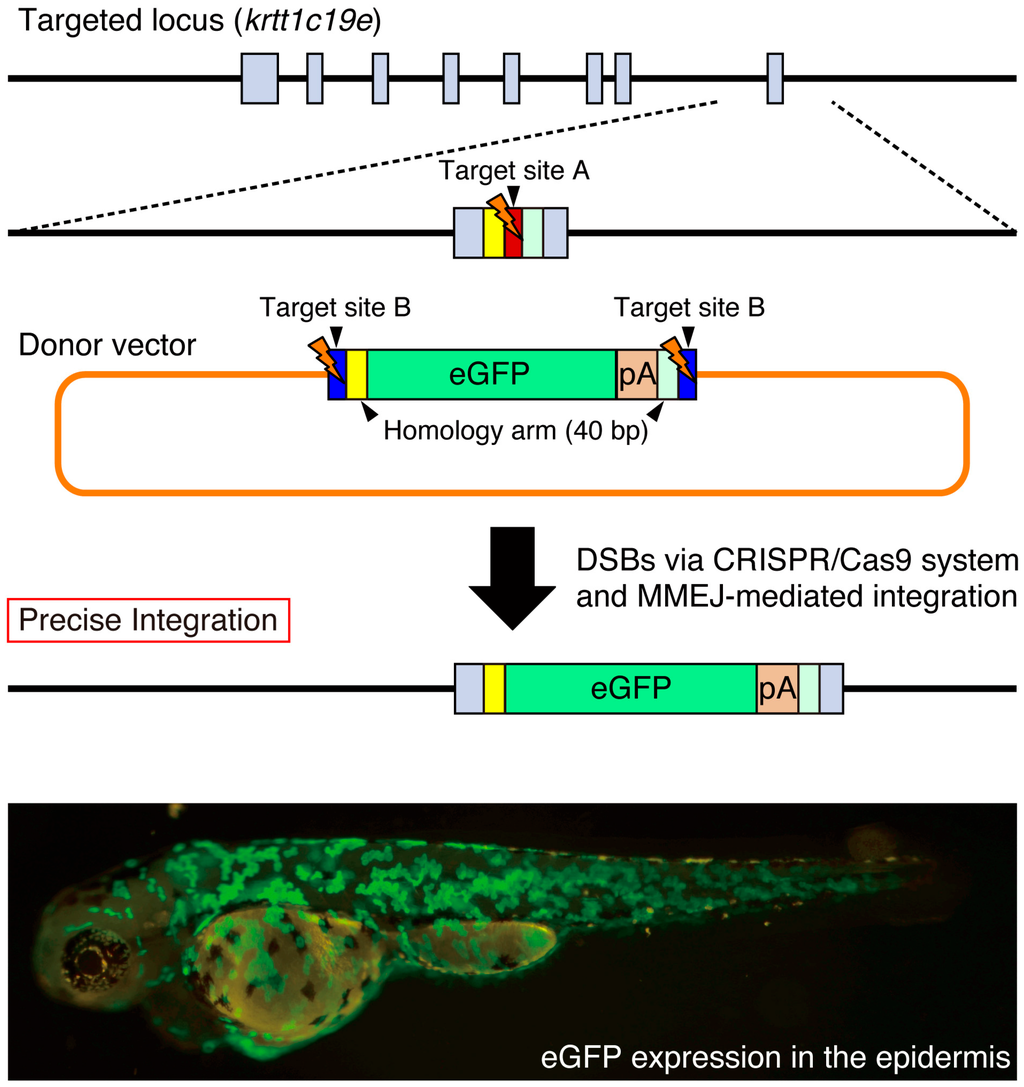
Figure 4.
Precise site-specific integration of eGFP into the krtt1c19e locus. The donor vector consists of two bait sequences (blue box; target site B for Gbait-gRNA), a left homology arm (yellow box; 40 bp), and an eGFP (green box) and polyA signal (pA) (orange box) and right homology arm (light green box). The target site A for krtt1c19e-gRNA (red box) is located near the stop codon of the krtt1c19e gene. When the CRISPR/Cas9 system simultaneously cleaves the target sites A and B, the eGFP reporter between the microhomology arms is precisely integrated into the krtt1c19e locus. Broad eGFP expression was detected in the epidermis of the injected F0 embryo as described previously [44]. Gray boxes; exons in the krtt1c19e gene.
6. Concluding Remarks and Future Perspective
Genetic manipulation in whole organisms plays an important role in basic, applied and medical sciences. Recent advances in genome editing technologies (ZFN, TALEN and CRISPR/Cas9) have enabled researchers to make precise genomic modifications that were previously difficult or impossible in various organisms including humans. In this review, we summarized the targeted integration of exogenous genes using genome editing technologies mediated by DNA repair mechanisms, such as NHEJ, MMEJ and HR. Because integration directions become random and various indel mutations occur at the junction in the process of NHEJ-mediated knock-in, we recommend that MMEJ-mediated knock-in technology is suitable for the precise targeted integration of donor DNA templates. Furthermore, the construction of a donor vector for MMEJ-mediated knock-in is much easier than that of HR-mediated knock-in. Therefore, we predict that the MMEJ-mediated targeted integration could be widely applied in various organisms. Importantly, genome editing technologies can be used to precisely model human genetic disorders in animals including zebrafish, which could lead to novel findings with respect to the molecular mechanisms of human diseases. Furthermore, these technologies make it easier to correct serious genetic disorders in affected patients and in induced pluripotent stem (iPS) cells derived from patients. Further innovation of genome editing technologies will accelerate various applications in basic biology, biotechnology and medicine, including potential therapeutic strategies for human genetic disorders.
Acknowledgments
We thank Higashijima for his kind permission to share the evx2 results. This work was supported in part by the Grants-in-Aid for Scientific Research of the Japan Society for the Promotion of Science, SENSHIN Medical Research Foundation and the Special Postdoctoral Researcher Program.
Author Contributions
Atsuo Kawahara, Yu Hisano, Satoshi Ota and Kiyohito Taimatsu wrote the manuscript. Atsuo Kawahara and Kiyohito Taimatsu created the figures.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ZFN | zinc finger nuclease |
| TALEN | transcription activator-like effector nuclease |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| MO | morpholino oligonucleotide |
| RVD | repeat-variable di-residue |
| crRNA | CRISPR RNA |
| tracrRNA | trans-activating crRNA |
| PAM | protospacer-adjacent motif |
| gRNA | guide RNA |
| DSB | double-strand break |
| NHEJ | non-homologous end joining |
| MMEJ | microhomology-mediated end joining |
| HR | homologous recombination |
| indel | insertion and/or deletion |
| ssODN | single-stranded oligodeoxynucleotide |
| HA | hemagglutinin |
| BAC | bacterial artificial chromosome |
| iPS | induced pluripotent stem |
References
- Ota, S.; Kawahara, A. Zebrafish: A model vertebrate suitable for the analysis of human genetic disorders. Congenit. Anom. (Kyoto) 2014, 54, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Chiang, C.Y.; Tsai, H.-J. Zebrafish and Medaka: New model organisms for modern biomedical research. J. Biomed. Sci. 2016, 23, 19. [Google Scholar] [CrossRef] [PubMed]
- Nasevicius, A.; Ekker, S.C. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 2000, 26, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Robu, M.E.; Larson, J.D.; Nasevicius, A.; Beiraghi, S.; Brenner, C.; Farber, S.A.; Ekker, S.C. p53 activation by knockdown technologies. PLoS Genet. 2007, 3, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Hisano, Y.; Ota, S.; Kawahara, A. Genome editing using artificial site-specific nucleases in zebrafish. Dev. Growth Differ. 2014, 56, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.H.; Kim, J.M.; Kim, H.T.; Lee, J.; Jeon, J.; Jin, Y.; Choi, J.H.; Ban, Y.H.; Ha, S.J.; Kim, C.H.; et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014, 24, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Noyes, M.B.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Mccammon, J.M.; Miller, J.C.; Faraji, F.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; Gregory, P.D.; et al. Heritable Targeted Gene Disruption in Zebrafish Using Designed Zinc Finger Nucleases. Nat. Biotechnol. 2009, 26, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Van Impel, A.; Zhao, Z.; Hermkens, D.M.; Roukens, M.G.; Fischer, J.C.; Peterson-Maduro, J.; Duckers, H.; Ober, E.A.; Ingham, P.W.; Schulte-Merker, S. Divergence of zebrafish and mouse lymphatic cell fate specification pathways. Development 2014, 141, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Kok, F.O.; Shin, M.; Ni, C.W.; Gupta, A.; Grosse, A.S.; VanImpel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I.; et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 2015, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Hisano, Y.; Inoue, A.; Taimatsu, K.; Ota, S.; Ohga, R.; Kotani, H.; Muraki, M.; Aoki, J.; Kawahara, A. Comprehensive analysis of sphingosine-1-phosphate receptor mutants during zebrafish embryogenesis. Genes Cells 2015, 20, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Hisano, Y.; Inoue, A.; Okudaira, M.; Taimatsu, K.; Matsumoto, H.; Kotani, H.; Ohga, R.; Aoki, J.; Kawahara, A. Maternal and Zygotic Sphingosine Kinase 2 are Indispensable for Cardiac Development in Zebrafish. J. Biol. Chem. 2015, 290, 14841–14851. [Google Scholar] [CrossRef] [PubMed]
- Mendelson, K.; Lan, Y.; Hla, T.; Evans, T. Maternal or zygotic sphingosine kinase is required to regulate zebrafish cardiogenesis. Dev. Dyn. 2015, 244, 948–954. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Ansai, S.; Kinoshita, M. Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 2014, 3, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Bibikova, M.; Carroll, D.; Segal, D.J.; Trautman, J.K.; Smith, J.; Kim, Y.G.; Chandrasegaran, S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001, 21, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Hosoi, S.; Woltjen, K.; Suzuki, K.I.; Kashiwagi, K.; Wada, H.; Ochiai, H.; Miyamoto, T.; Kawai, N.; Sasakura, Y.; et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 2013, 18, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Gonçalves, M.A.F.V. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015, 33, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Streubel, J.; Blücher, C.; Landgraf, A.; Boch, J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 2012, 30, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Hisano, Y.; Ota, S.; Arakawa, K.; Muraki, M.; Kono, N.; Oshita, K.; Sakuma, T.; Tomita, M.; Yamamoto, T.; Okada, Y.; et al. Quantitative assay for TALEN activity at endogenous genomic loci. Biol. Open 2013, 2, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ann Ran, F.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; Zhang, F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Aida, T.; Chiyo, K.; Usami, T.; Ishikubo, H.; Imahashi, R.; Wada, Y.; Tanaka, K.F.; Sakuma, T.; Yamamoto, T.; Tanaka, K. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 2015, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kotani, H.; Taimatsu, K.; Ohga, R.; Ota, S.; Kawahara, A. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 Protein complex in zebrafish. PLoS ONE 2015, 10, e0128319. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Bedell, V.M.; Wang, Y.; Campbell, J.M.; Poshusta, T.L.; Starker, C.G.; Krug, R.G.; Tan, W.; Penheiter, S.G.; Ma, A.C.; Leung, A.Y.H.; et al. In vivo genome editing using a high-efficiency TALEN system. Nature 2012, 491, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, K.; Kunihiro, Y.; Kaneko, T.; Nagahora, H.; Voigt, B.; Mashimo, T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun. 2016, 7, 10431. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Kaini, P.; Sander, J.D.; Joung, J.K.; Peterson, R.T.; Yeh, J.R.J. Heritable and Precise Zebrafish Genome Editing Using a CRISPR-Cas System. PLoS ONE 2013, 8, e68708. [Google Scholar] [CrossRef] [PubMed]
- Hruscha, A.; Krawitz, P.; Rechenberg, A.; Heinrich, V.; Hecht, J.; Haass, C.; Schmid, B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 2013, 140, 4982–4987. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013, 23, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Tong, X.; Wang, Z.; Liu, D.; Pan, R.; Li, Z.; Hu, Y.; Luo, Z.; Huang, P.; Wu, Q.; et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 2013, 10, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Chen, J.; Solnica-Krezel, L. Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development 2014, 141, 3807–3818. [Google Scholar] [CrossRef] [PubMed]
- Irion, U.; Krauss, J.; Nusslein-Volhard, C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 2014, 141, 4827–4830. [Google Scholar] [CrossRef] [PubMed]
- Cristea, S.; Freyvert, Y.; Santiago, Y.; Holmes, M.C.; Urnov, F.D.; Gregory, P.D.; Cost, G.J. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol. Bioeng. 2013, 110, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M.; Lin, V.G.; Guo, N.; Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013, 23, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Auer, T.O.; Duroure, K.; De Cian, A.; Concordet, J.P.; Del Bene, F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014, 24, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Hisano, Y.; Kawahara, A.; Higashijima, S.-I. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014, 4, 6545. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Okamura, Y.; Higashijima, S.I. alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J. Neurosci. 2006, 26, 5684–5697. [Google Scholar] [CrossRef] [PubMed]
- Nakade, S.; Tsubota, T.; Sakane, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T.; et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 5560. [Google Scholar] [CrossRef] [PubMed]
- Hisano, Y.; Sakuma, T.; Nakade, S.; Ohga, R.; Ota, S.; Okamoto, H.; Yamamoto, T.; Kawahara, A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 2015, 5, 8841. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Takenaga, M.; Kawabe, Y.; Nakamura, T.; Kamihira, M.; Yamamoto, T. Homologous recombination-independent large gene cassette knock-in in CHO cells using TALEN and MMEJ-directed donor plasmids. Int. J. Mol. Sci. 2015, 16, 23849–23866. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).