Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors
Abstract
:1. Introduction
2. Results
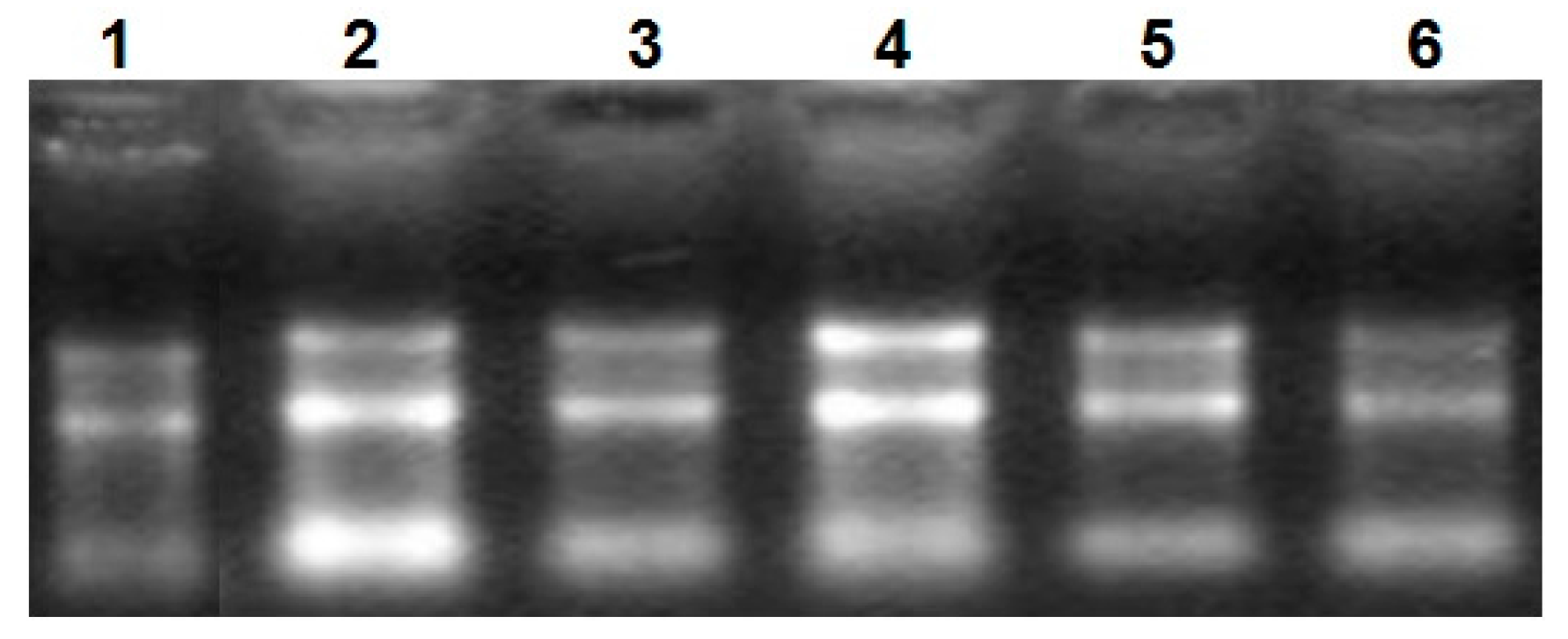
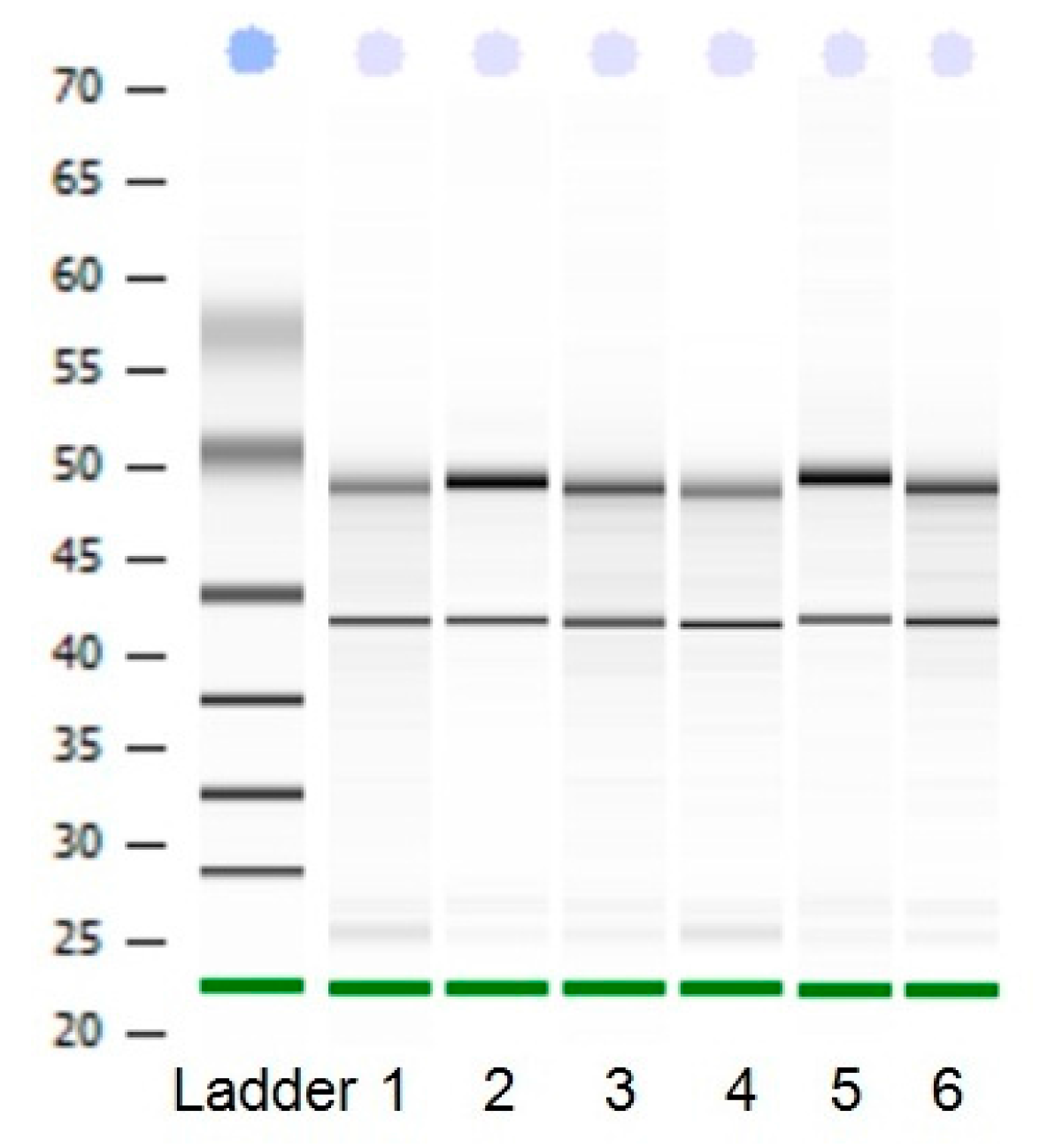
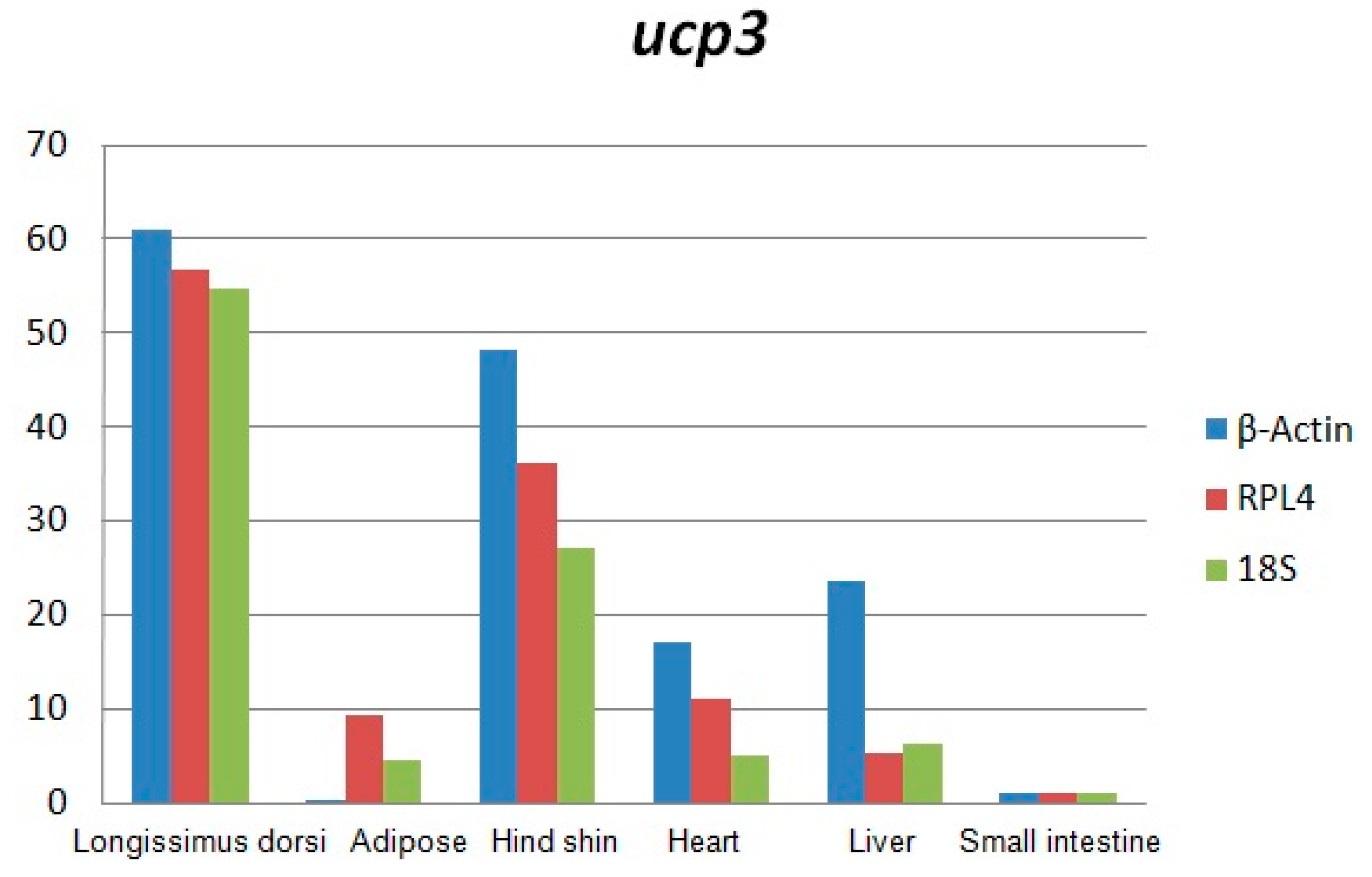
2.1. Analysis of Uncoupling Protein 3 (UCP3) Gene Using qRT-PCR
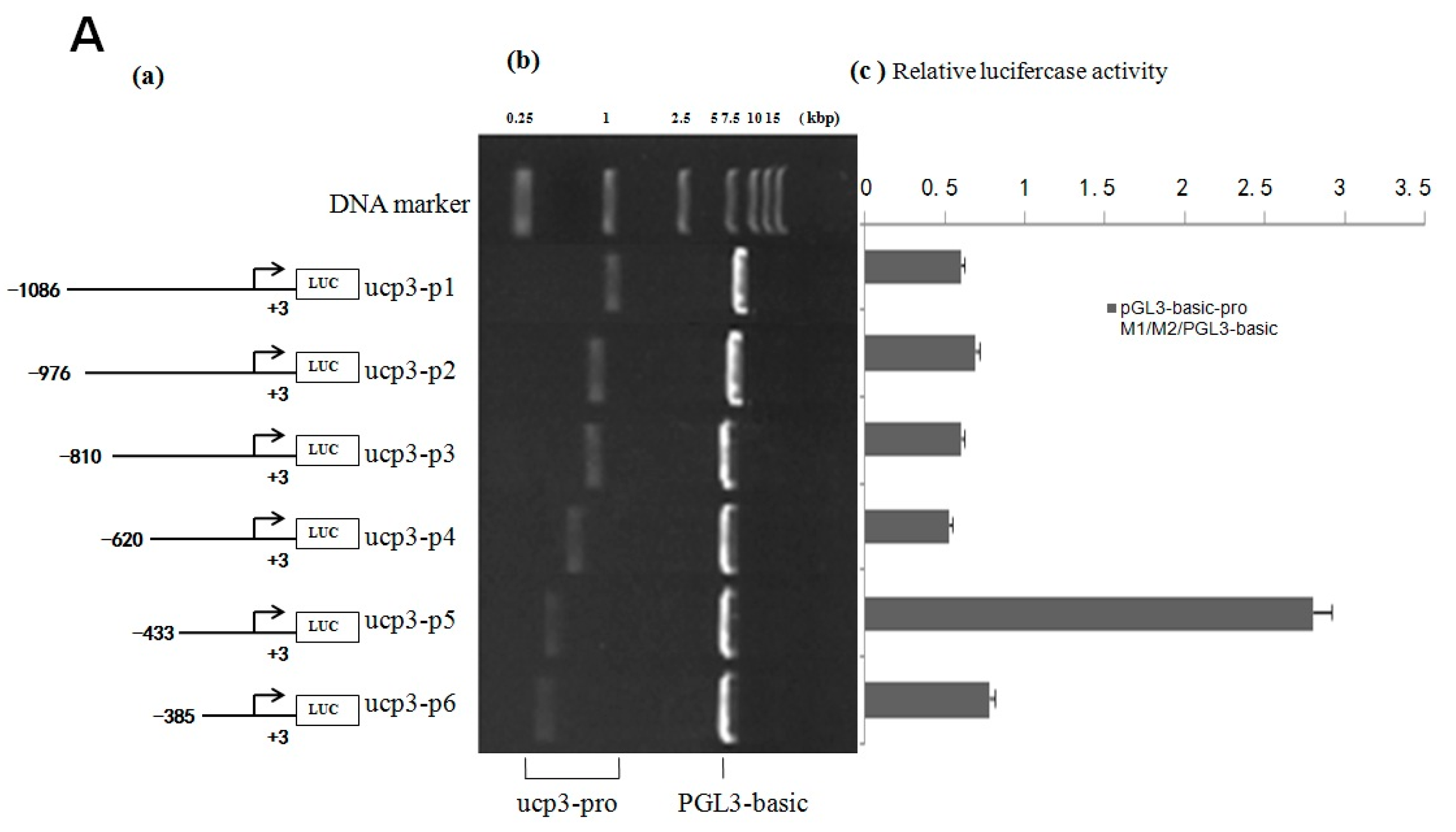
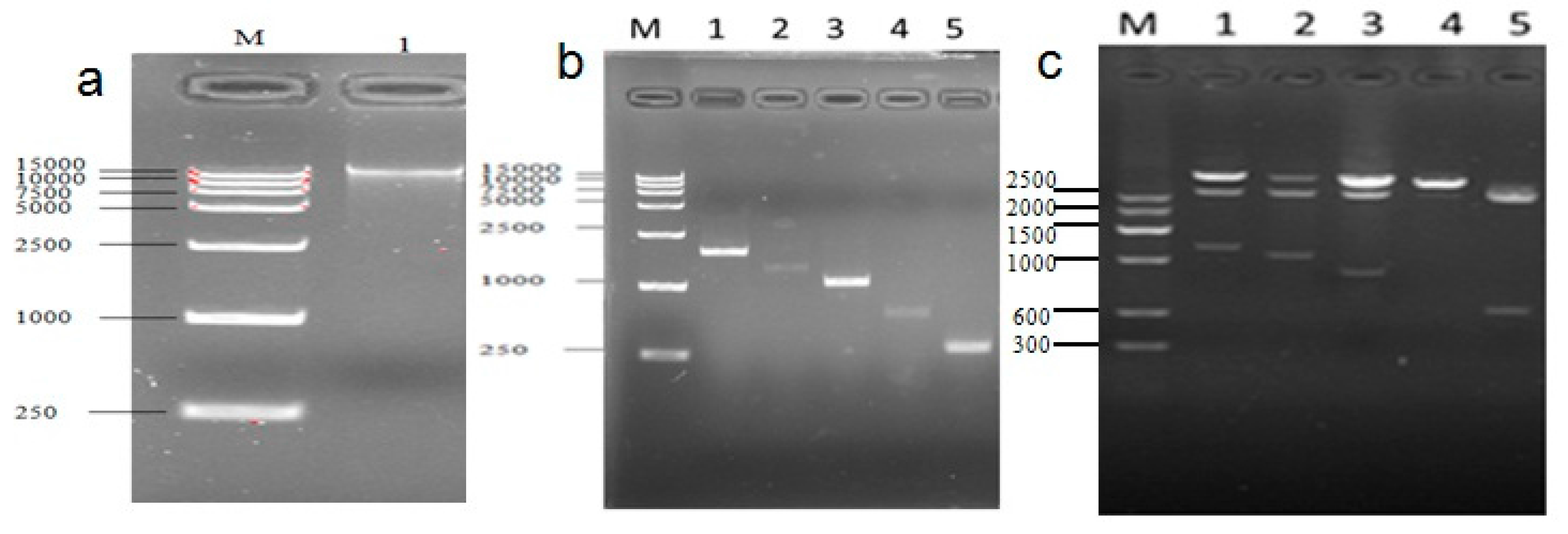
2.2. Cloning and Activity Analysis of the Cattle ucp3 Promoter
2.3. Expression of PEGFP-N3-ucp3-pro in C2C12 Cell Line
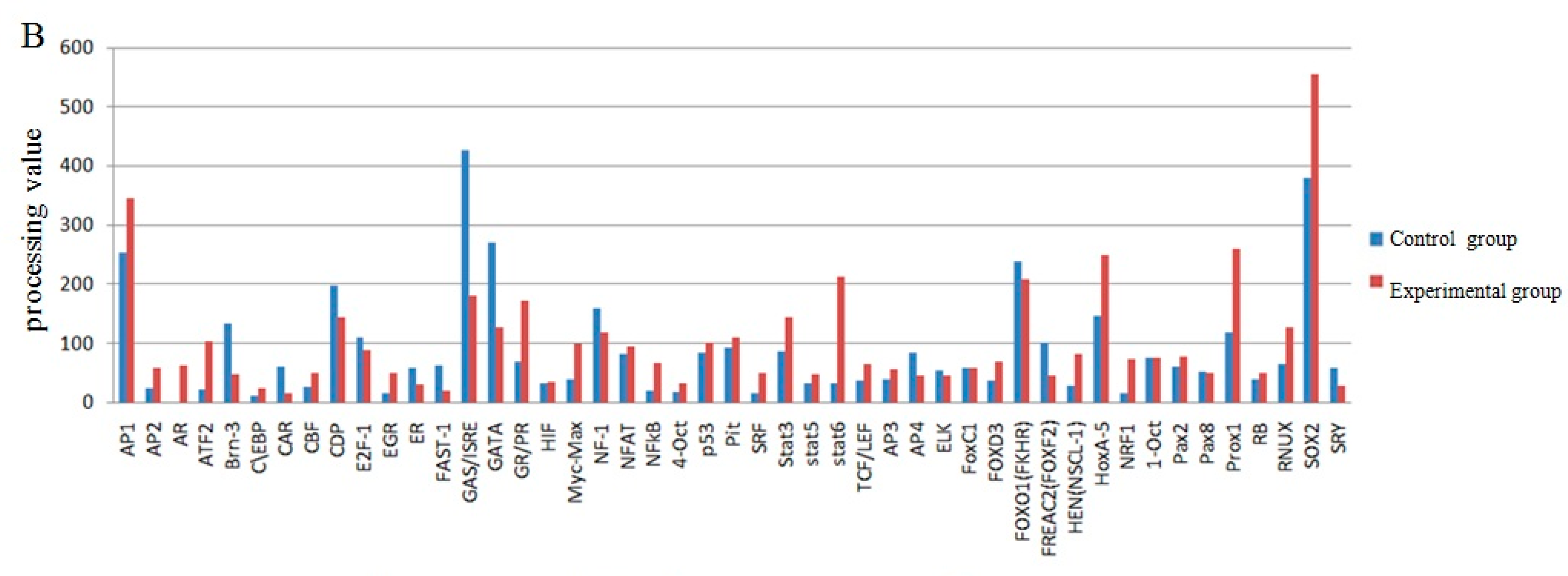
2.4. The Screening of Transcription Factor by Promoter-Binding Transcription Factors (TF) Profiling Assay II
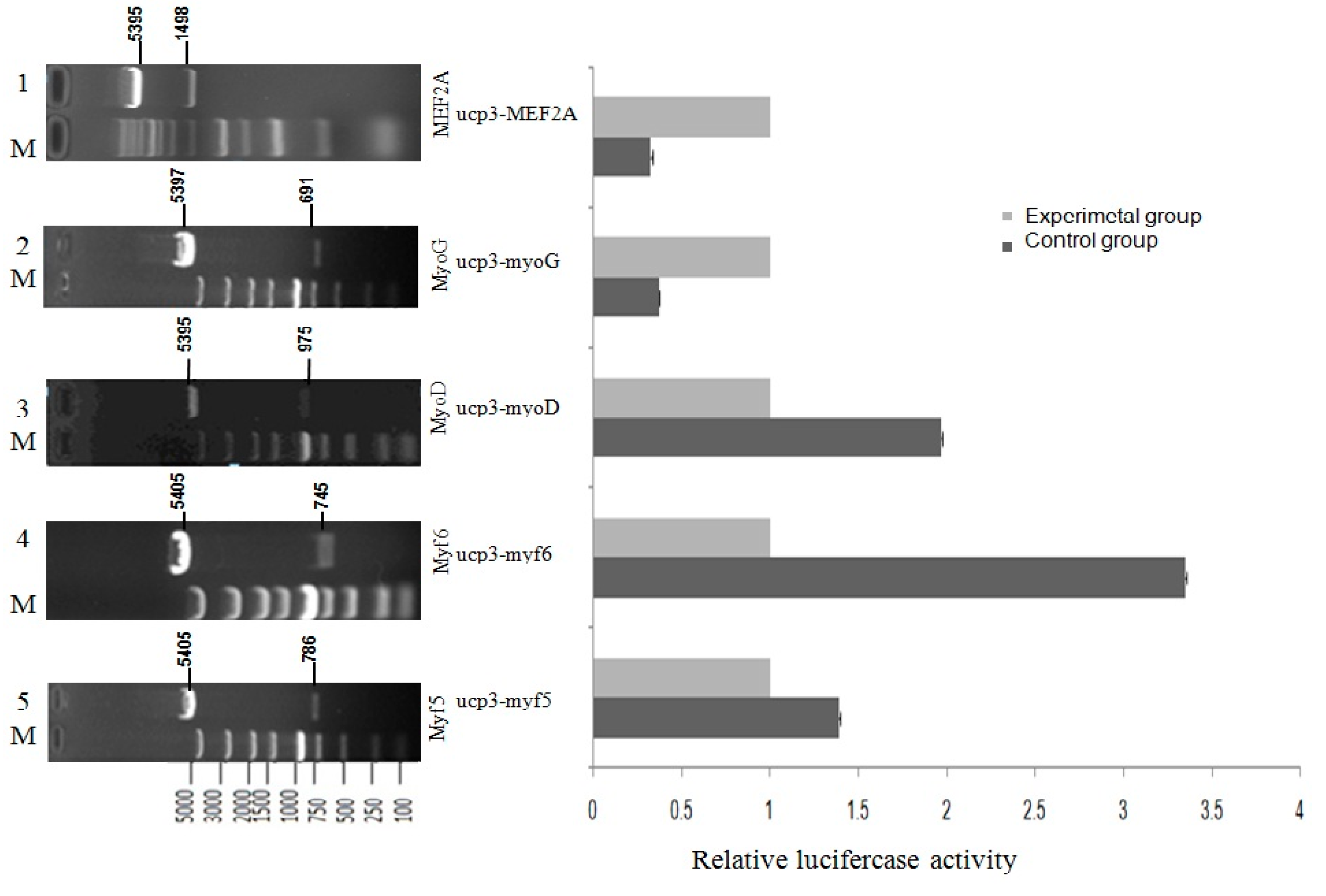
2.5. Effect of Transcription Factors on UCP3 Promoter Activity
3. Discussion
4. Experimental Section
4.1. Experimental Animals and Tissue Sampling
4.2. RNA Isolation and Synthesis of cDNA
4.3. Confirmation of Gene Expression Using qRT-PCR
4.4. Amplify UCP3 Promoter of Guanling Cattle
4.5. Transcription Factor Profiling of UCP3 Promoter by Filter Assay
4.6. Construction of Myogenic Regulatory Factors (MRFs) Family and MEF2A Transcription Factors Expression Vectors
4.7. Analyse Activity of UCP3 Promoter
4.8. The Data Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Daniel, R.; Frederic, B. The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem. J. 2000, 345, 161–179. [Google Scholar]
- Joseph, H.; Francois, C. Evolutionary history of the UCP gene family: Gene duplication and selection. BMC Evol. Biol. 2008, 8. [Google Scholar] [CrossRef]
- Boss, O.; Samec, S.; Dulloo, A.; Seydoux, J.; Muzzin, P.; Giacobino, J.P. Tissue-dependent upregulation of rat uncoupling protein-2 expression in response to fasting or cold. FEBS Lett. 1997, 412, 111–114. [Google Scholar] [CrossRef]
- Boss, O.; Samec, S.; Paoloni-Giacobino, A.; Rossier, C.; Dulloo, A.; Seydoux, J.; Muzzin, P.; Giacobino, J.P. Uncoupling protein-3, a new member of the mitochondrial carrier family with tissue specific expression. FEBS Lett. 1997, 408, 39–42. [Google Scholar] [CrossRef]
- Solanes, G.; Pedraza, N.; Iglesias, R.; Giralt, M.; Villarroya, F. Functional Relationship between MyoD and Peroxisome Proliferator-Activated Receptor Dependent Regulatory Pathways in the Control of the Human Uncoupling Protein-3 Gene Transcription. Mol. Endocrinol. 2003, 17, 1944–1958. [Google Scholar] [CrossRef] [PubMed]
- Nowacka-Woszuk, J.; Szczerbal, I.; Fijak-Nowak, H.; Switonski, M. Chromosomal localization of 13 candidate genes for human obesity in the pig genome. J. Appl. Gene 2008, 49, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, F.; Senese, R.; de Lange, P.; Goglia, F.; Lanni, A.; Lombardi, A. Uncoupling proteins: A complex journey to function discovery. Biofactors 2009, 35, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Nabben, M.; Hoeks, J.; Moonen-Kornips, E.; van Beurden, D.; Briedé, J.J.; Hesselink, M.K. Significance of uncoupling protein 3 in mitochondrial function upon mid- and long-term dietary high-fat exposure. FEBS Lett. 2011, 585, 4010–4017. [Google Scholar] [CrossRef] [PubMed]
- Senese, R.; Valli, V.; Moreno, M.; Lombardi, A.; Busiello, R.A.; Cioffi, F. Uncoupling protein 3 expression levels influence insulin sensitivity, fatty acid oxidation, and related signaling pathways. Pflugers Arch. 2011, 461, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Nabben, M.; van Bree, B.W.; Lenaers, E.; Hoeks, J.; Hesselink, M.K.; Schaart, G.; Gijbels, M.J.; Glatz, J.F.; da Silva, G.J.; de Windt, L.J.; et al. Lack of UCP3 does not affect skeletal muscle mitochondrial function under lipid-challenged conditions, but leads to sudden cardiac death. Basic Res. Cardiol. 2014, 109, 447. [Google Scholar] [CrossRef] [PubMed]
- Busiello, R.A.; Savarese, S.; Lombardi, A. Mitochondrial uncoupling proteins and energy metabolism. Front. Physiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.A.; Tjian, R. Unexpected roles for core promoter recog-nition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 2010, 11, 549–558. [Google Scholar] [PubMed]
- Colin, R.L.; Florian, M.; Sean, E.H.; James, G.M.; Jason, D.L. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 2012, 484, 251–255. [Google Scholar]
- De Lange, P.; Feola, A.; Ragni, M.; Senese, R.; Moreno, M.; Lombardi, A.; Silvestri, E.; Amat, R.; Villarroya, F.; Goglia, F.; et al. Differential 3,5,3-triiodothyronine-mediated regulation of uncoupling protein 3 transcription: Role of fatty acids. Endocrinology 2007, 148, 4064–4072. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liu, C.; Jia, M.; Yang, Y.; Ye, F.; Yan, Y. Progress of upstream transcriptional control elements in promoters of skeletal specific genes (in Chinese). Chin. J. Cell Biol. 2012, 34, 500–505. [Google Scholar]
- China National Commission of Animal Genetic Resources. Animal Genetic Resources in China Bovines; China Agriculture Press: Beijing, China, 2011; Volume 5, p. 151. [Google Scholar]
- Larkin, S.; Mull, E.; Miao, W.; Pittner, R.; Albrandt, K.; Moore, C.; Young, A.; Denaro, M.; Beaumont, K. Regulation of the third member of the uncoupling protein family, UCP3, by cold and thyroid hormone. Biochem. Biophys. Res. Commun. 1997, 240, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Samec, S.; Seydoux, J.; Dulloo, A.G. Role of UCP homologues in skeletal muscles and brown adipose tissue: Mediators of thermogenesis or regulators of lipids as fuel substrate? FASEB J. 1998, 12, 715–724. [Google Scholar] [PubMed]
- Weigle, D.S.; Selfridge, L.E.; Schwartz, M.W.; Seeley, R.J.; Cummings, D.E.; Havel, P.J.; Kuijper, J.L.; BeltrandelRio, H. Elevated free fatty acids induceuncoupling protein 3 expression in muscle: A potential explanation for the effect of fasting. Diabetes 1998, 47, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Kratky, D.; Strauss, J.G.; Zechner, R. Tissue-specific activity of lipoprotein lipase in skeletal muscle regulates the expression of uncoupling protein 3 in transgenic mouse models. Biochem. J. 2001, 355, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Jitrapakdee, S.; Thompson, M. Differential regulation of the promoter activity of the mouse UCP2 and UCP3 genes by MyoD and myogenin. J. Biochem. Mol. Biol. 2007, 40, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Girousse, A.; Tavernier, G.; Tiraby, C.; Lichtenstein, L.; Iacovoni, J.S.; Mairal, A.; Villarroya, F.; Langin, D. Transcription of the human uncoupling protein 3 gene is governed by a complex interplay between the promoter and intronic sequences. Diabetologia 2009, 52, 1638–1646. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.W.; Ho, J.W.; Liu, H.F.; So, D.H.; Tse, Z.H.; Chan, K.H.; Ramsden, D.B.; Ho, S.L. Mitochondrial neuronal uncoupling proteins: A target for potential disease-modification in Parkinson’s disease. Transl. Neurodegener. 2012, 1, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, F.; Iglesias, R.; Giralt, M. PPARs in the control of uncoupling proteins gene expression. PPAR Res. 2007, 12. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Bajard, L.; Chang, T.; Daubas, P.; Hadchouel, J.; Meilhac, S.; Montarras, D.; Rocancourt, D.; Relaix, F. The formation of skeletal muscle: From somite to limb. J. Anat. 2003, 202, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Boutet, S.C.; Biressi, S.; Iori, K.; Natu, V.; Rando, T.A. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol. Cell 2010, 40, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Gros, J.; Manceau, M.; Thomé, V.; Marcelle, C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 2005, 435, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005, 435, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Dodou, E.; Xu, S.M.; Black, B.L. MEF2c is activated directly by MyoGenic basic helix-loop-helixproteins during skeletal muscle development in vivo. Mech. Dev. 2003, 120, 1021–1032. [Google Scholar] [CrossRef]
- Hennebry, A.; Berry, C.; Siriett, V. Myostatin regμlates fiber-type composition of skeletal muscle by regμlating MEF2 and MyoD gene expression. Am. J. Physiol. Cell Physiol. 2009, 296, C525–C534. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.Y.; Ko, Y.G. TRIM72, a novel negative feedback regμlator of MyoGenesis, is transcriptionally activated by the synergism of MyoD (or MyoGenin) and MEF2. Biochem. Biophys. Res. Commun. 2010, 396, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Bryantsev, A.L.; Baker, P.W.; Lovato, T.L. Differential requirements for Myocyte Enhancer Factor-2 during adult MyoGenesis in Drosophila. Dev. Biol. 2012, 361, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cao, Y.; Li, S.; Tong, H.; Xing, X.; Li, G.; Yan, Y. Cloning and preliminary functional analysis of bovine MyoG promoter (in Chinese). Acta Vet. Zootech. Sin. 2012, 43, 37–43. [Google Scholar]
- Potthoff, M.J.; Olson, E.N. MEF2: A central regulator of diverse developmental programs. Development 2007, 134, 4131–4140. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Xia, X.; He, J.; Ren, Z.; Xu, D.; Xiong, Y.; Zuo, B. MyoD is a novel activator of porcine FIT1 gene by interacting with the canonical E-box element during myogenesis. Int. J. Mol. Sci. 2015, 16, 25014–25030. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Zimmermann, A.; Hinney, A.; Volckmar, A.L.; Jarrett, H.W.; Fromme, T.; Klingenspor, M. A novel SP1/SP3 dependent intronic enhancer governing transcription of the UCP3 gene in brown adipocytes. PLoS ONE 2013, 8, e83426. [Google Scholar] [CrossRef] [PubMed]
- Udagawa, C.; Tada, N.; Asano, J.; Ishioka, K.; Ochiai, K.; Bonkobara, M.; Tsuchida, S.; Omi, T. The genetic association study between polymorphisms in uncoupling protein 2 and uncoupling protein 3 and metabolic data in dogs. BMC Res. Notes 2014, 7, 904. [Google Scholar] [CrossRef] [PubMed][Green Version]
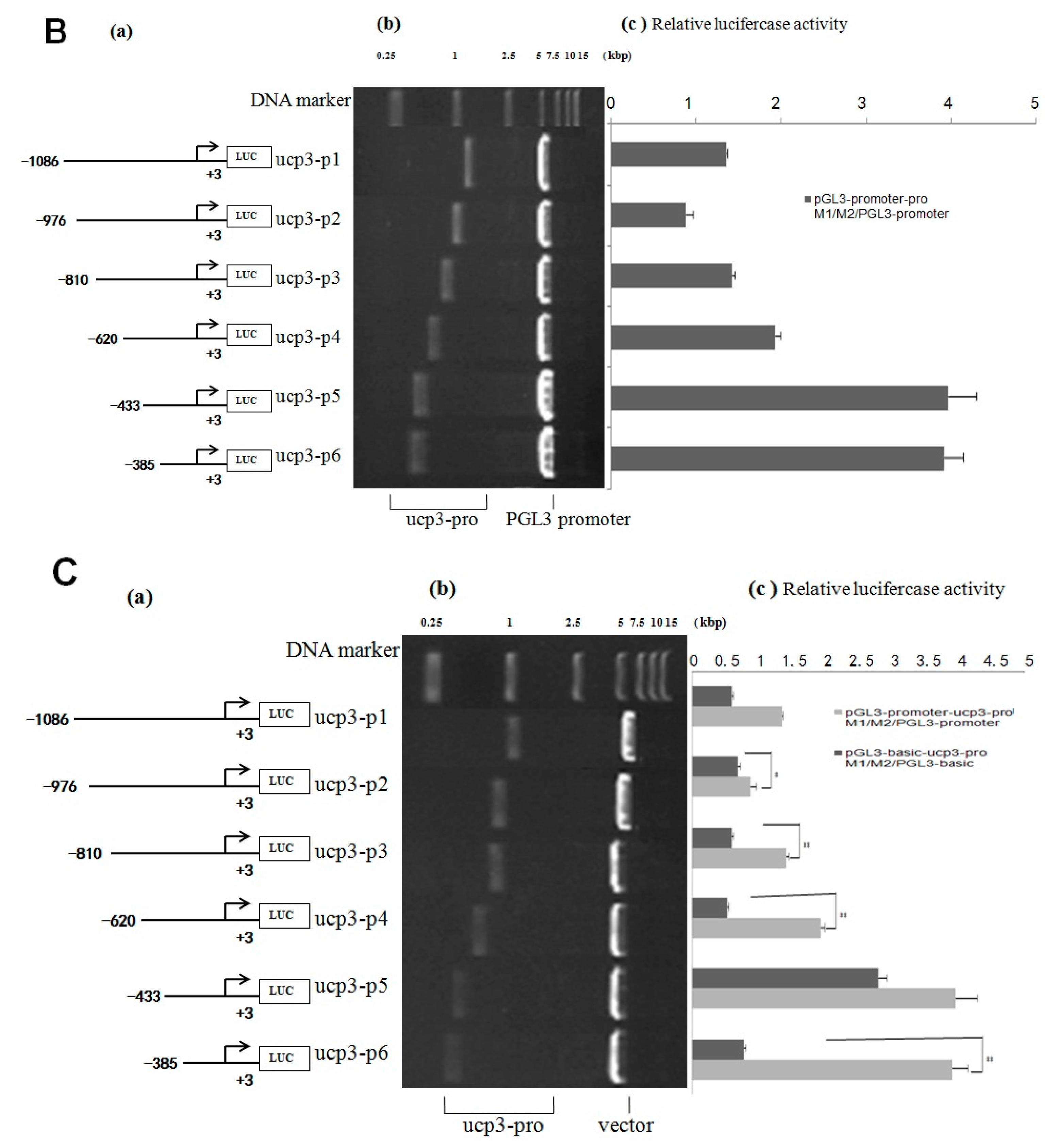
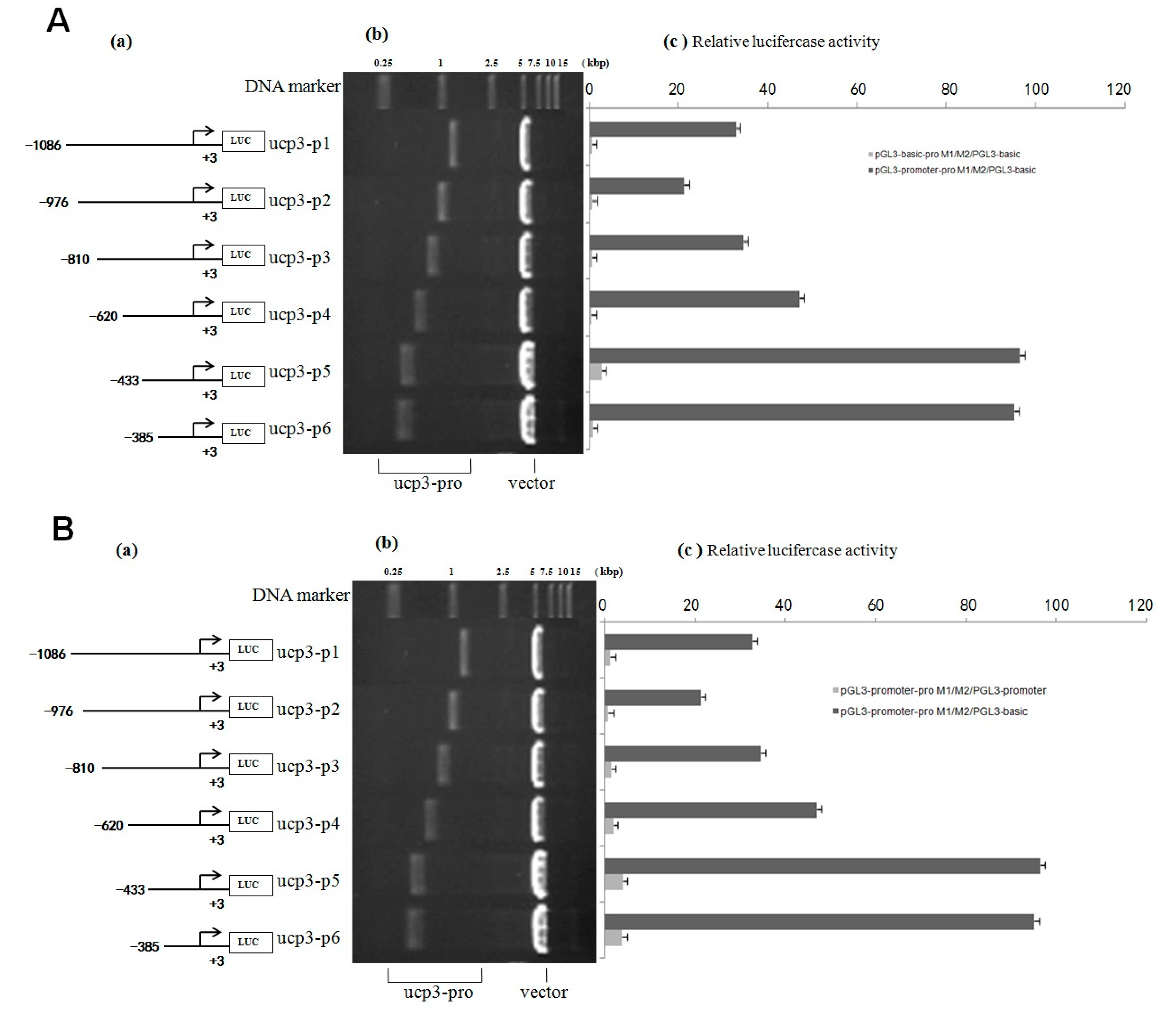
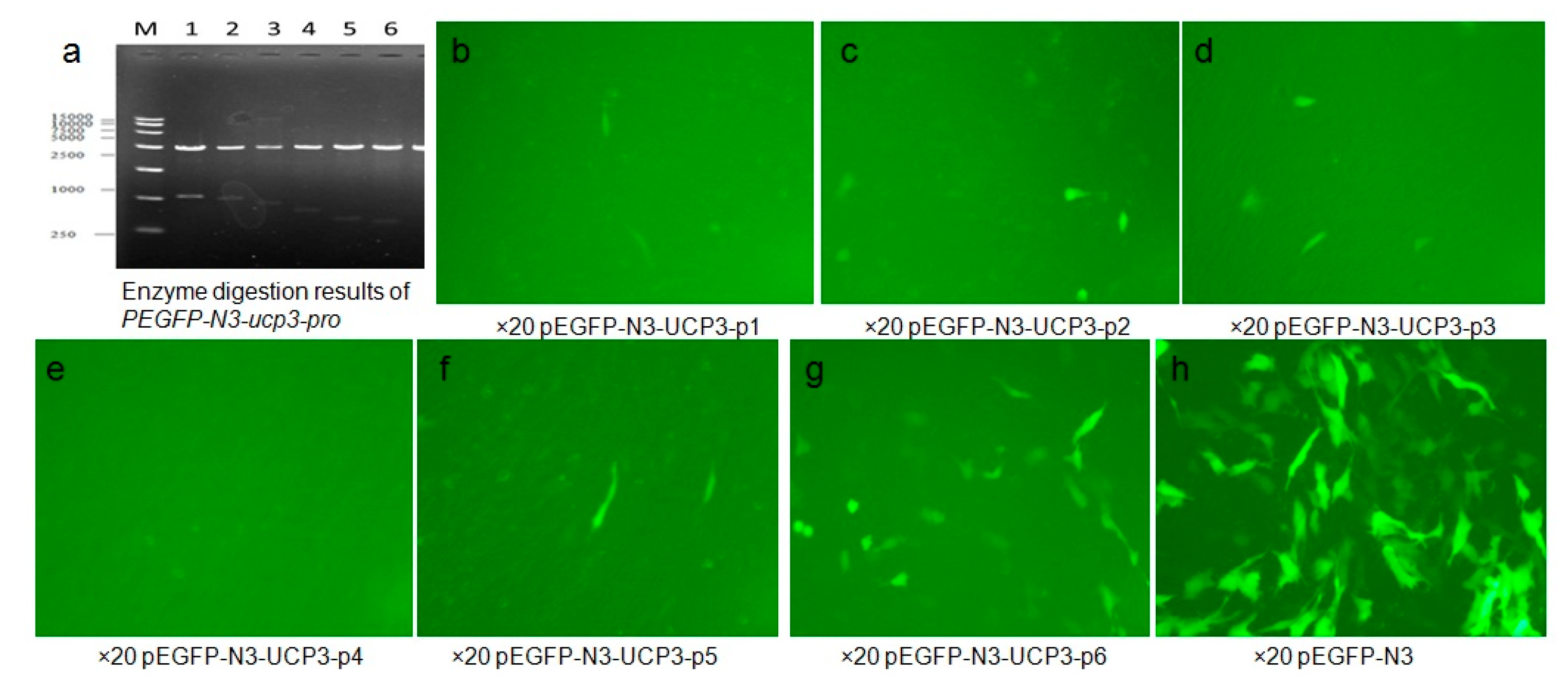
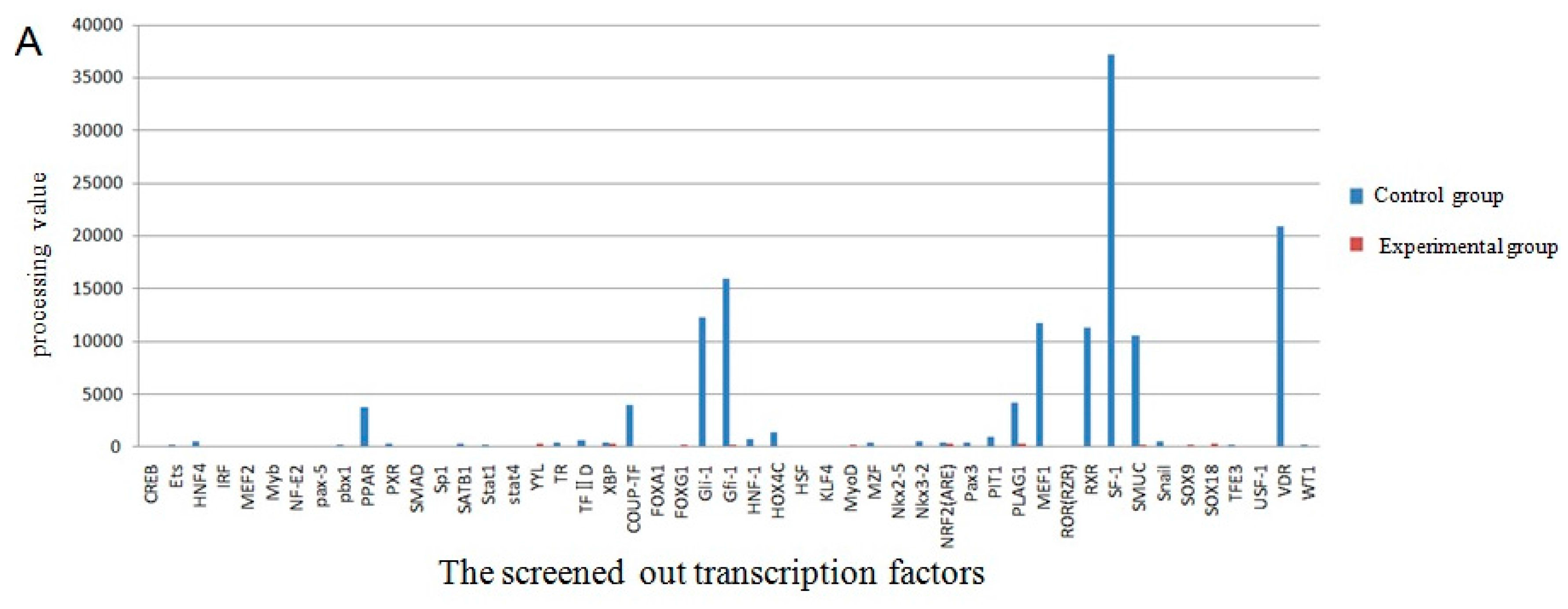
| Start | End | Score | Promoter Sequence (5′ to 3′) |
|---|---|---|---|
| −907 | −857 | 0.95 | ATTAAAATGCATGAATCCGCAGTCATAAAAAGGCACGAAGTGCTGACATA |
| −842 | −792 | 0.87 | AACTTGGAAAACACTTTGGTAAGTAAAAGAAGCCAGTCACAAAAGGTCA |
| −449 | −399 | 0.80 | GAATTAGCATCACCATTGTTACTATTAAGAGGAGGCCCAGGGGAACCCTG |
| −301 | −251 | 0.99 | TCATCACCTGCAATGGGGTGTTTTAAAGAAGCCCAAGTCAAGGGAACTGA |
| −154 | −104 | 1.00 | AAGGTTTCAGGTCAGCCAGAGTGTATAAAGACAGGTGCCAAGCCAGAGGC |
| −52 | −2 | 1.00 | CCTGGGATGGAGCCCTGAGGACCCTTAAAGAGGCCCCATGCTTCCGCCAC |
| Site | Transcription Factor Binding Sites |
|---|---|
| −1300~−1200 bp | GATA-1, p300, MZF1 |
| −1200~−1100 bp | GATA-1, GATA-2, GATA-3, GATA-X, C/EBPb, C/EBP, MZF1 |
| −1100~−1000 bp | CdxA, Nkx-2, AML-1a, GATA-1, GATA-2, GATA-3, GATA-X, Cdx, COUP-T |
| −1000~−900 bp | NF-kap, Lyf-1, CdxA, NF-kap, Sox-5, GATA-1, Pbx-1 |
| −900~−800 bp | CdxA, C/EBP |
| −800~−700 bp | GATA-X, GATA-2, CdxA, SRY, Egr-1, Egr-2, Egr-3, NGFI-C |
| −700~−600 bp | MZF1, Oct-1, GATA-1, CdxA, AML-1a, Nkx-2 |
| −600~−500 bp | CdxA, C/EBP, USF, Nkx-2, SRY, C/EBPb, HNF-3b, S8, HFH-2 |
| −500~−400 bp | Sox-5, GATA-1, S8, Oct-1, Nkx-2, CdxA |
| −400~−300 bp | CdxA, GATA-X, AML-1a, HNF-4, GATA-1, deltaE, C/EBPb, USF, Lyf-1 |
| −300~−200 bp | TATA, SRY , AML-1a, Nkx-2, USF, Sp1 |
| −200~−100 bp | Nkx-2, USF, deltaE, CdxA, TATA, MyoD |
| −100~+1 bp | Nkx-2, USF, deltaE, CdxA, TATA, MyoD |
| +1~+100 bp | GATA-1, GATA-2, CdxA |
| Gene Symbol | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Length (bp) |
|---|---|---|---|
| β-actin | GCAGGTCATCACCATCGG | GCTGTCACCTTCACCGTTC | 564 bp |
| RPL4 | AGGAGGCTGTGTTGCTTCTG | CGACGGTTTCTCATCTTGC | 106 bp |
| 18SRNA | GACTCAACACGGGAAACCTC | AGACAAATCGCTCCACCAAC | 120 bp |
| UCP3 | TCTACGACTCCGTCAAGC | CTGGCGATGGTCCTGT | 469 bp |
| Gene Symbol | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Length (bp) | Tm (°C) |
|---|---|---|---|---|
| UCP3-P1 | CTAGCTAGCTACCCTCAGTGCCTATCA | CCGCTCGAGGCCATTTGCTGGTGTCT | 1080 | 56 |
| UCP3-P2 | CTAGCTAGCGCCTCCAAGTGTCATGT | CCGCTCGAGGCCATTTGCTGGTGTCT | 980 | 60 |
| UCP3-P3 | CTAGCTAGCTGAATCCGCAGTCATAAA | CCGCTCGAGGCCATTTGCTGGTGTCT | 814 | 60 |
| UCP3-P4 | CTAGCTAGCTGAAGGGAAGAGGAAACA | CCGCTCGAGGCCATTTGCTGGTGTCT | 624 | 59.4 |
| UCP3-P5 | CTAGCTAGCCCTACCTCGTAAGAGTTGTG | CCGCTCGAGGCCATTTGCTGGTGTCT | 437 | 60 |
| UCP3-P6 | CTAGCTAGCCACATAGTAGGCACCAAGA | CCGCTCGAGGCCATTTGCTGGTGTCT | 389 | 59.3 |
| Reagent | Experimental Group (μL) | Compare Group (μL) |
|---|---|---|
| TF Binding buffer mix | 15 | 15 |
| TF Probe mix | 3 | 3 |
| UCP3 promoter PCR fragment | 5 | 0 |
| Nuclear extract | 6 | 6 |
| ddH2O | 1 | 6 |
| Total Volume | 30 | 30 |
| Gene Symbol | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Length (bp) |
|---|---|---|---|
| MyoD | EcoRI: CCGGAATTCATGGAGCTGCTGTCGC | XhoI: CCGCTCGAGTCAGAGCACCTGGTAAAT | 975 |
| Myf5 | EcoRI: CCGGAATTCATGGACATGATGGACGGCTG | XhoI: CCGCTCGAGTCATAGCACATGATAGATG | 786 |
| Myf6 | KpnI: CGGGATCCATGATGATGGACCTTTTTGAAACTGGC | EcoRI: CGGAATTCTTACTTCTCCACCACCTCCTCCACGCAG | 745 |
| MyoG | BamHI: GGGGTACCATGGAGCTGTATGAGACCTCT | EcoRI: CGGAATTCTCAGTTTGGTATGGTTTCATCTGG | 691 |
| MEF2A | EcoRI: CCGGAATTCATGGGGCGGAAGAAAATACAA | XhoI: CCGCTCGAGGTTAGGTCACCCACGCATC | 1498 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Xu, H.; Chen, X.; Liu, Z.; Zhang, W.; Xia, D. Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors. Int. J. Mol. Sci. 2016, 17, 682. https://doi.org/10.3390/ijms17050682
Chen W, Xu H, Chen X, Liu Z, Zhang W, Xia D. Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors. International Journal of Molecular Sciences. 2016; 17(5):682. https://doi.org/10.3390/ijms17050682
Chicago/Turabian StyleChen, Wei, Houqiang Xu, Xiang Chen, Zhongwei Liu, Wen Zhang, and Dan Xia. 2016. "Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors" International Journal of Molecular Sciences 17, no. 5: 682. https://doi.org/10.3390/ijms17050682
APA StyleChen, W., Xu, H., Chen, X., Liu, Z., Zhang, W., & Xia, D. (2016). Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors. International Journal of Molecular Sciences, 17(5), 682. https://doi.org/10.3390/ijms17050682





