Guanabenz Downregulates Inflammatory Responses via eIF2α Dependent and Independent Signaling
Abstract
:1. Introduction
2. Results
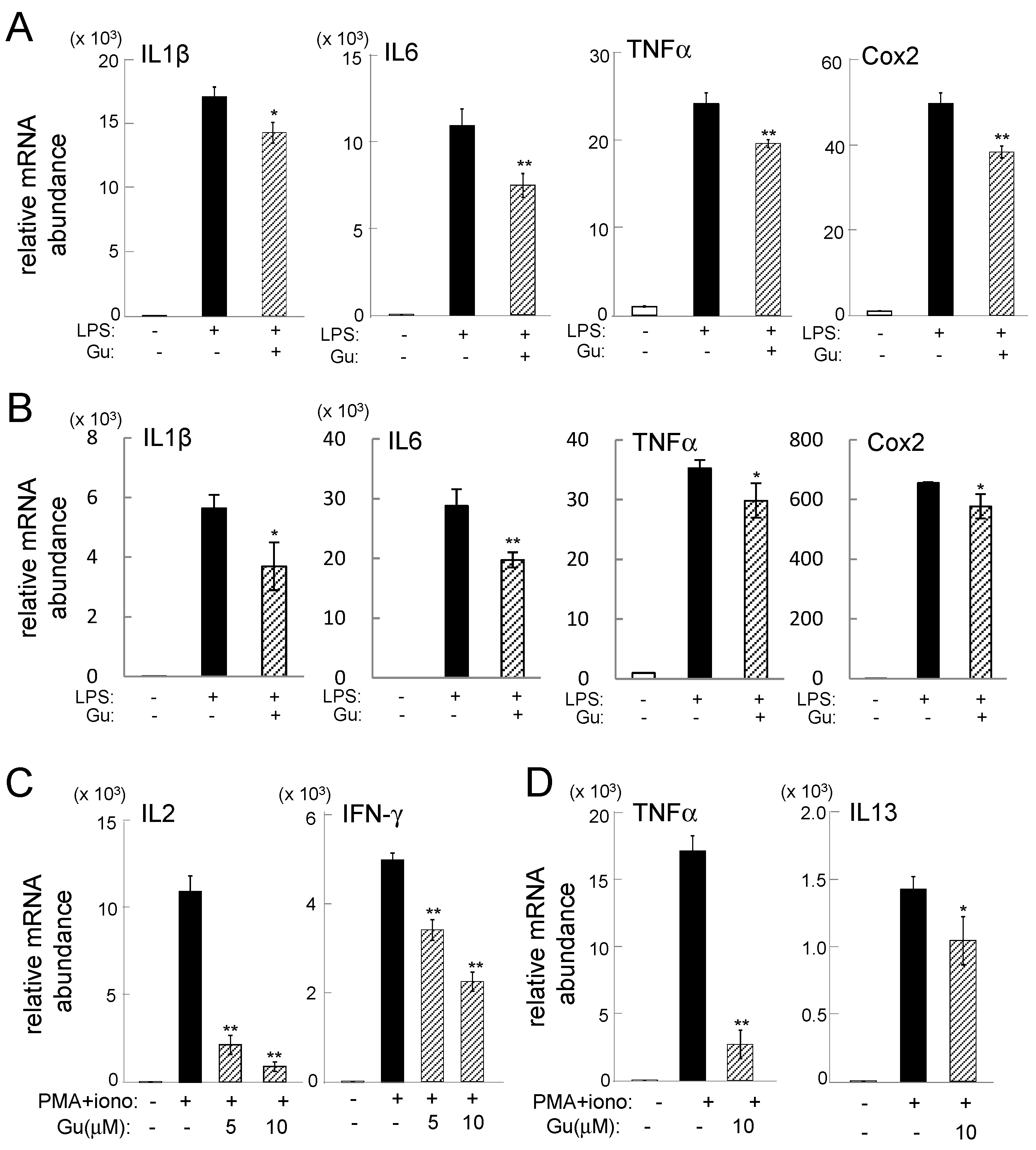
2.1. Guanabenz-Driven Suppression of Inflammatory Gene Expression
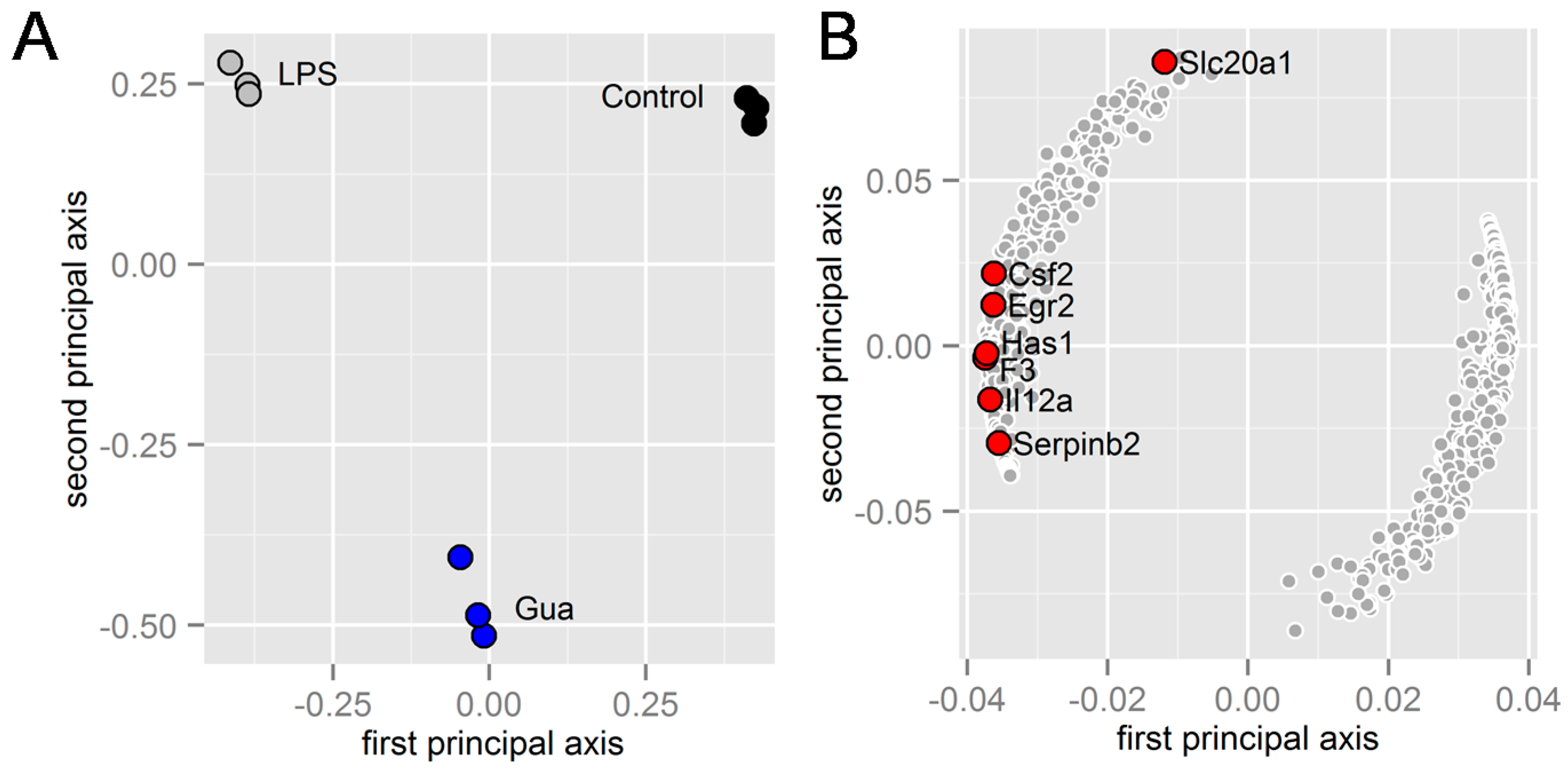
2.2. Prediction of the Pathways and Genes Involved in the Responses to Guanabenz
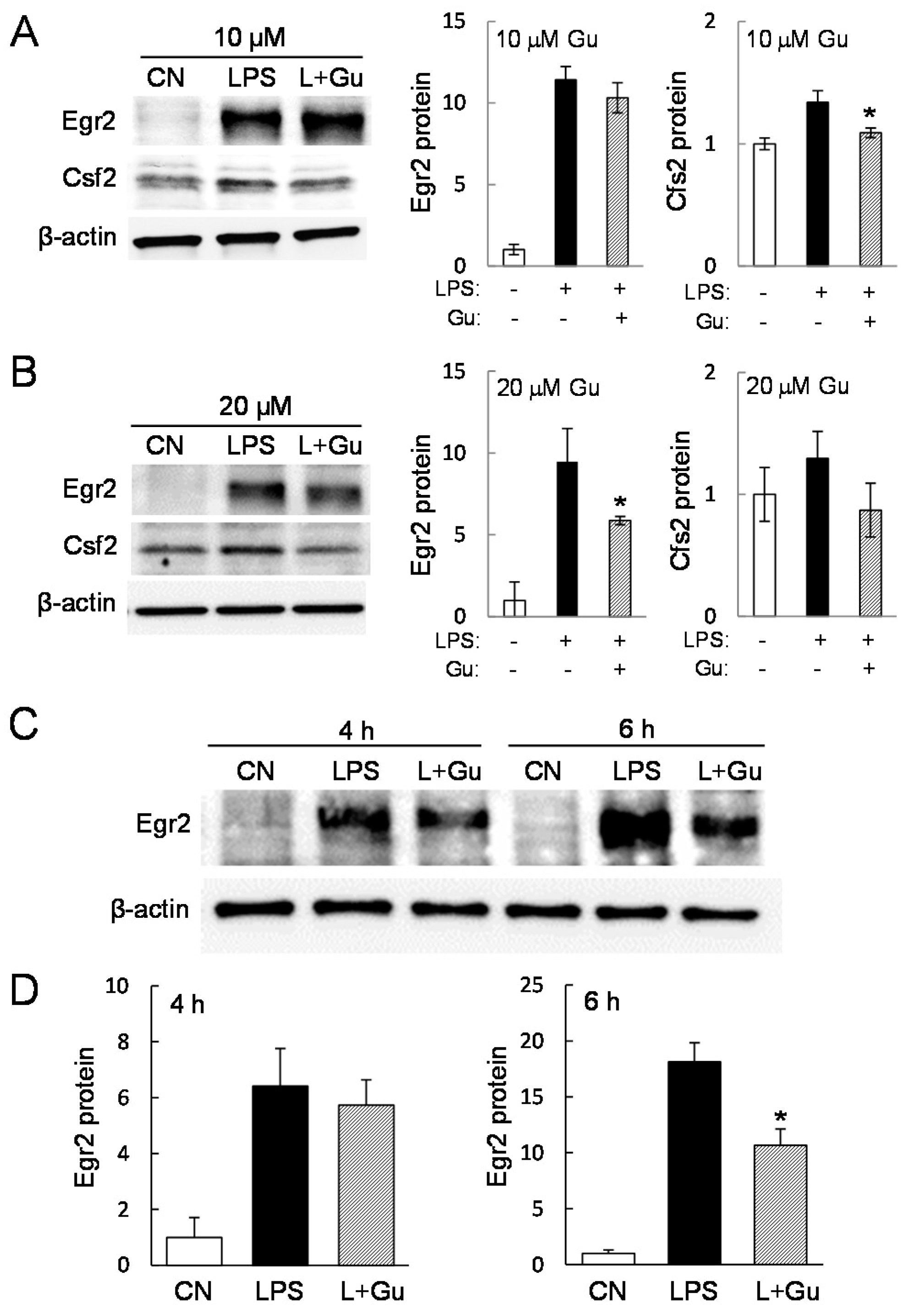
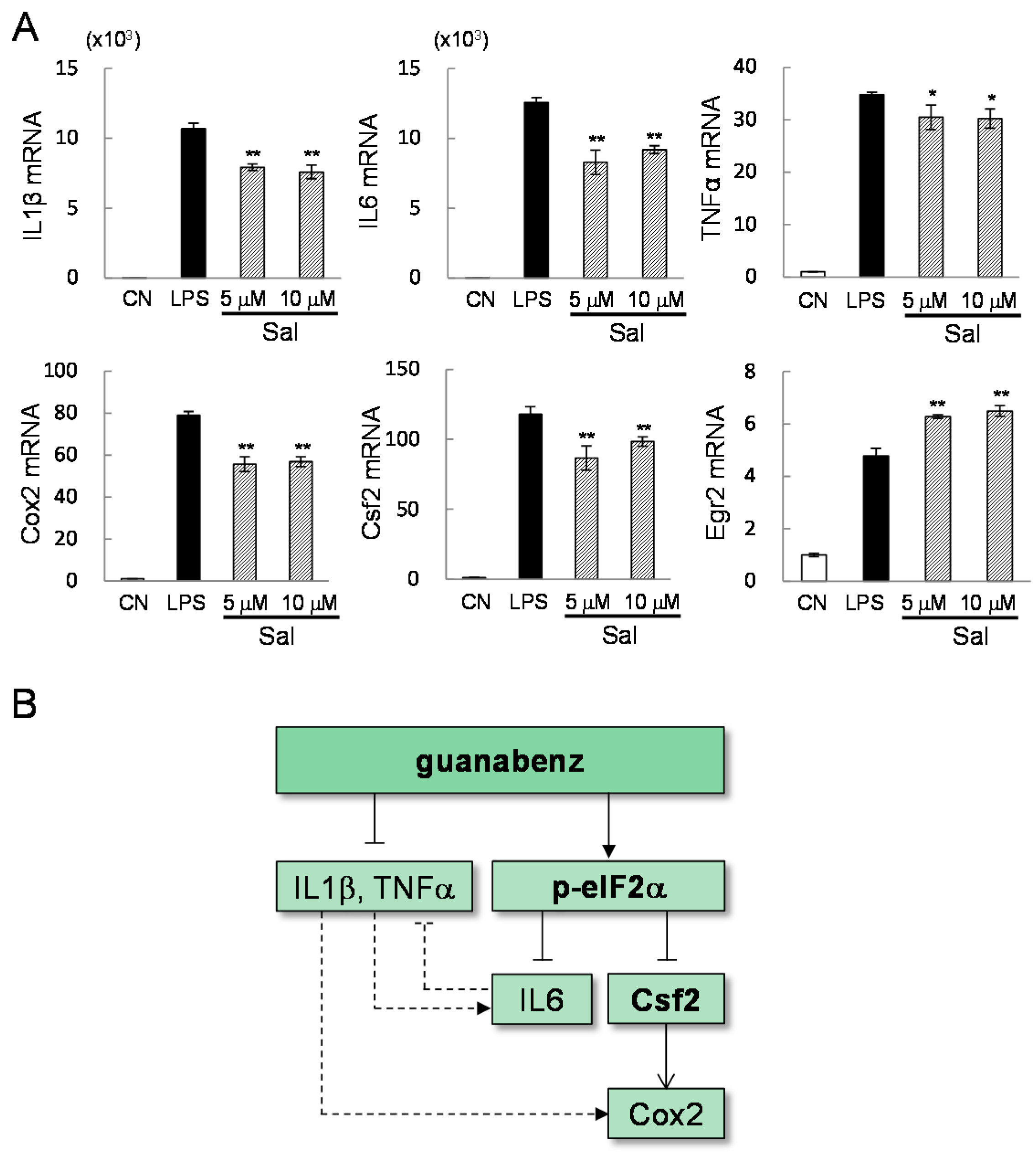
2.3. Guanabenz-Driven Downregulation of Egr2 and Csf2 in Lipopolysaccharide (LPS)-Stimulated Cells
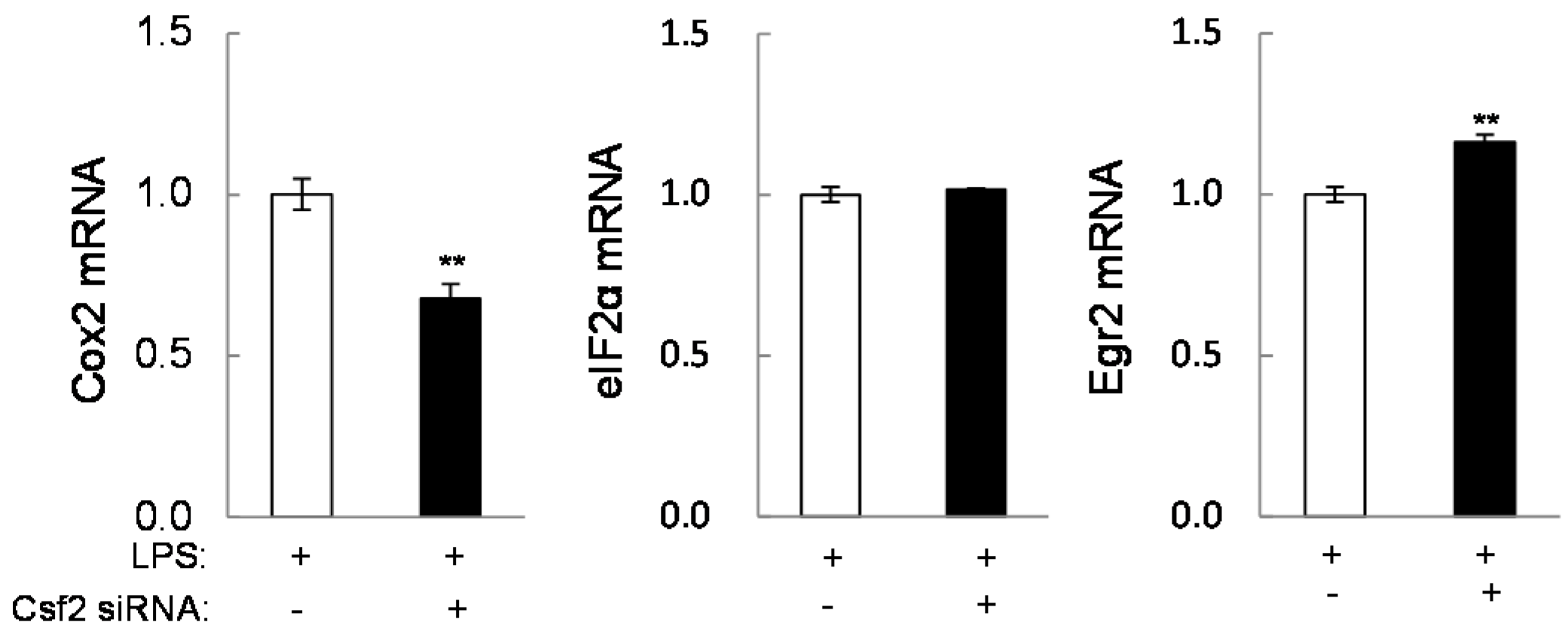
2.4. Potential Linkage of Egr2 and Csf2 to IL1β, TNFα, and Cox2
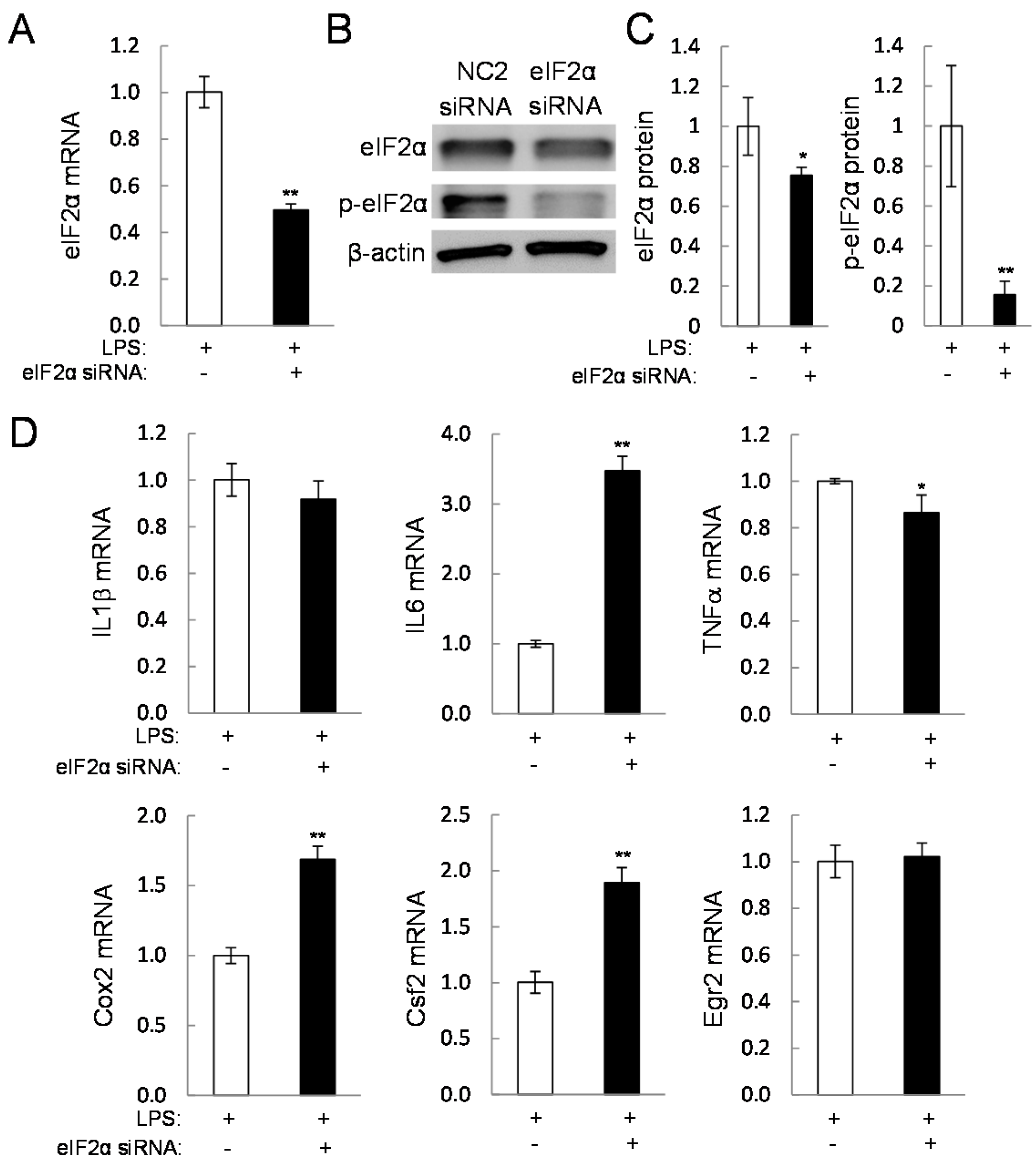
2.5. Direct Linkage of eif2α Signaling to IL6, Cox2, and Csf2
3. Discussion
4. Experimental Section
4.1. Cell Culture
4.2. Microarray
4.3. qPCR and Western Blot Analysis
4.4. RNA Silencing Using siRNAs
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wek, R.C.; Jiang, H.Y.; Anthony, T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006, 34, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Tabas, I.; Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011, 13, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Boyce, M.; Bryant, K.F.; Jousse, C.; Long, K.; Harding, H.P.; Scheuner, D.; Kaufman, R.J.; Ma, D.; Coen, D.M.; Ron, D.; et al. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 2005, 307, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Tsaytler, P.; Harding, H.P.; Ron, D.; Bertolotti, A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 2011, 332, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, K.; Tanjung, N.; Yokota, H. Suppression of osteoclastogenesis through phosphorylation of eukaryotic translation initiation factor 2 α. J. Bone Miner. Metab. 2013, 31, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Yokota, H.; Hamamura, K.; Chen, A.; Dodge, T.R.; Tanjung, N.; Abedinpoor, A.; Zhang, P. Effects of salubrinal on development of osteoclasts and osteoblasts from bone marrow derived cells. BMC Musculoskelet. Disord. 2013, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, K.; Chen, A.; Tanjung, N.; Takigawa, S.; Sudo, A.; Yokota, H. In vitro and in silico analysis of an inhibitory mechanism of osteoclastogenesis by salubrinal and guanabenz. Cell Signal. 2015, 27, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, K.; Chen, A.; Uto, Y.; Yokota, H. Potential therapeutic applications of salubrinal for skeletal diseases and beyond. J. Nat. Sci. 2015, 1, e151. [Google Scholar] [CrossRef]
- Hamamura, K.; Lin, C.C.; Yokota, H. Salubrinal reduces expression and activity of MMP13 in chondrocytes. Osteoarthr. Cartil. 2013, 21, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, K.; Nishimura, A.; Iino, T.; Takigawa, S.; Sudo, A.; Yokota, H. Chondroprotective effects of salubrinal in a mouse model of osteoarthritis. Bone Jt. Res. 2105, 4, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.S.; Walker, B.R.; Vanov, S.K.; Helfant, R.H. Guanabenz effects on blood pressure and noninvasive parameters of cardiac performance in patients with hypertension. Clin. Pharmacol. Ther. 1976, 19, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Way, S.W.; Podojil, J.R.; Clayton, B.L.; Zaremba, A.; Collins, T.L.; Kunjamma, R.B.; Robinson, A.P.; Brugarolas, P.; Miller, R.H.; Miller, S.D.; et al. Pharmaceutical integrated stress response enhancement protects oligodendrocytes and provides a potential multiple sclerosis therapeutic. Nat. Commun. 2015, 6, 6532. [Google Scholar] [CrossRef] [PubMed]
- Van Neeuwenhuijze, A.; Koenders, M.; Roeleveld, D.; Sleeman, M.A.; van den Berg, W.; Wicks, I.P. GM-CSF as a therapeutic target in inflammatory diseases. Mol. Immunol. 2013, 56, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Vincenti, M.P.; Yokota, H. Principal component analysis for predicting transcription-factor binding motifs from array-derived data. BMC Bioinform. 2005, 6, 276. [Google Scholar] [CrossRef] [PubMed]
- Joseph, L.J.; Le Beau, M.M.; Jamieson, G.A.; Acharya, S.; Shows, T.B.; Rowley, J.D.; Sukhatme, V.P. Molecular cloning, sequencing, and mapping of EGR2, a human early growth response gene encoding a protein with “zinc-binding finger” structure. Proc. Natl. Acad. Sci. USA 1988, 85, 7164–7168. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Rezk, A.; Miyazaki, Y.; Hilgenberg, E.; Touil, H.; Shen, P.; Moore, C.S.; Michel, L.; Althekair, F.; Rejasekharan, S.; et al. Proinflammatory GM-CSF-producing B cells in multiple sclerosis and B cell depletion therapy. Sci. Transl. Med. 2015, 7, 310. [Google Scholar] [CrossRef] [PubMed]
- Tanikawa, M.; Lee, H.Y.; Watanabe, K.; Majewska, M.; Skarzynski, D.J.; Park, S.B.; Lee, D.S.; Park, C.K.; Acosta, T.J.; Okuda, K. Regulation of prostaglandin biosynthesis by interleukin-1 in cultured bovine endometrial cells. J. Endocrinol. 2008, 199, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Rossi, J.F.; Lu, Z.Y.; Jourdan, M.; Klein, B. Interleukin-6 as a therapeutic target. Clin. Cancer Res. 2015, 21, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Blaser, H.; Mak, T.W. Regulation of tumour necrosis factor signalling: Live or let die. Nat. Rev. Immunol. 2015, 15, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Lan, S.; Zhu, J.; Siclari, V.A.; Qin, L. Epidermal growth factor receptor signaling promotes proliferation and survival in osteoprogenitors by increasing early growth response 2 expression. J. Biol. Chem. 2013, 288, 20488–20498. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, N.; Luppen, C.A.; Ho, V.V.; Nagpal, S.; Hacia, J.G.; Smith, E.; Frenkel, B. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J. Mol. Endocrinol. 2004, 33, 175–193. [Google Scholar] [CrossRef] [PubMed]
- Stenson, W.F.; Nickells, M.W.; Atkinson, J.P. Metabolism of exogenous arachidonic acid by murine macrophage-like tumor cell lines. Prostaglandins 1981, 21, 675–689. [Google Scholar] [CrossRef]
- Cassel, D.L.; Hoxie, J.A.; Cooper, R.A. Phorbol ester modulation of T-cell antigens in the Jurkat lymphoblastic leukemia cell line. Cancer Res. 1983, 43, 4582–4586. [Google Scholar] [PubMed]
- Butterfield, J.H.; Weiler, D.A.; Hunt, L.W.; Wynn, S.R.; Roche, P.C. Purification of tryptase from a human mast cell line. J. Leukoc. Biol. 1990, 47, 409–419. [Google Scholar] [PubMed]



| Pathway | Ratio | p |
|---|---|---|
| Lysosome | 3.27 | 0.0007 |
| T cell receptor signaling pathway | 3.29 | 0.0011 |
| Hematopoietic cell lineage | 3.70 | 0.0011 |
| Cytokine-cytokine receptor interaction | 2.38 | 0.0011 |
| Fc gamma R-mediated phagocytosis | 3.39 | 0.0017 |
| Rheumatoid arthritis | 3.39 | 0.0026 |
| Chemokine signaling pathway | 2.42 | 0.0044 |
| MAPK signaling pathway | 2.06 | 0.0077 |
| ErbB signaling pathway | 2.84 | 0.0154 |
| NOD-like receptor signaling pathway | 3.45 | 0.0154 |
| Gene | Description | p1 | C1 | C2 | C3 | L1 | L2 | L3 | G1 | G2 | G3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F3 | coagulation factor III | −0.997 | |||||||||
| Has1 | hyaluronan synthase1 | −0.992 | |||||||||
| Il12a | interleukin 12a | −0.979 | |||||||||
| Egr2 | early growth response 2 | −0.967 | |||||||||
| Csf2 | colony stimulating factor 2 | −0.964 | |||||||||
| Serpinb2 | serine/cysteine peptidase inhibitor, B2 | −0.946 | |||||||||
| Slc20a1 | solute carrier family 20, member 1 | −0.317 |
| Gene | Accession Number | Forward Primer | Backward Primer |
|---|---|---|---|
| Mouse Primers | |||
| IL1β | NM_008361 | 5′-GCCCATCCTCTGTGACTCAT-3′ | 5′-AGGCCACAGGTATTTTGTCG-3′ |
| IL6 | NM_031168 | 5′-TTCCATCCAGTTGCCTTCTT-3′ | 5′-TCCACGATTTCCCAGAGAAC-3′ |
| TNFα | NM_013693 | 5′-GAACTGGCAGAAGAGGCACT-3′ | 5′-AGGGTCTGGGCCATAGAACT-3′ |
| Cox2 | AF378830 | 5′-CCCCCACAGTCAAAGACACT-3′ | 5′-CTCATCACCCCACTCAGGAT-3′ |
| Csf2 | NM_009969 | 5′-GAGGCCATCAAAGAAGCCCT-3′ | 5′-AAATTGCCCCGTAGACCCTG-3′ |
| eIF2α | NM_026114 | 5′-GAATGTACTCCAGATTGGCTGACTAC-3′ | 5′-CCTCAATGTGAAGACCTGTATCGA-3′ |
| Egr2 | NM_010118 | 5′-GATCTGCATGCGAAACTTCA-3′ | 5′-CACTGCTCTTCCGTTCCTTC-3′ |
| GAPDH | NM_008084 | 5′-TGCACCACCAACTGCTTAG-3′ | 5′-GGATGCAGGGATGATGTTC-3′ |
| Human Primers | |||
| IFNγ | NM_000619 | 5′-TTCAGCTCTGCATCGTTTTG-3′ | 5′-TCTTTTGGATGCTCTGGTCA-3′ |
| IL2 | NM_000586 | 5′-GCAACTCCTGTCTTGCATTG-3′ | 5′-GCCTTCTTGGGCATGTAAAA-3′ |
| IL13 | NM_002188 | 5′-GTACTGTGCAGCCCTGGAAT-3′ | 5′-TTTACAAACTGGGCCACCTC-3′ |
| TNFα | NM_000594 | 5′-CAGAGGGCCTGTACCTCATC-3′ | 5′-GGAAGACCCCTCCCAGATAG-3′ |
| GAPDH | NM_001289745 | 5′-GCACCGTCAAGGCTGAGAAC-3′ | 5′-ATGGTGGTGAAGACGCCAGT-3′ |
| Mouse siRNAs | |||
| Csf2 (GM-CSF) | 5′-GCGGAAGACAAACGAGAGA-3′ | 5′-GCCUGAAGAUAUUCGAGCA-3′ | |
| 5′-AUGAAGAGGUAGAAGUCGU-3′ | 5′-CCAGCUACUACCAGACAUA-3′ | ||
| eIF2α (eIF2s1) | 5′-UCGAGCAGAUAUUGAAGUA-3′ | 5′-CAUGAUUCUUCUUAGUGAA-3′ | |
| 5′-UGUCACAAGUUAAAGCCAA-3′ | 5′-GAACUCAAUGGGCAAGUAA-3′ | ||
| Egr2 | 5′-CGACCUCGAAAGUACCCUA-3′ | ||
| Control #1 (NC1) for Egr2 | 5′-UGUACUGCUUACGAUUCGG-3′ | ||
| Control #2 (NC2) for Csf2 & eIF2α | 5′-UGGUUUACAUGUCGACUAA-3′ | 5′-UGGUUUACAUGUUGUGUGA-3′ | |
| 5′-UGGUUUACAUGUUUUCUGA-3′ | 5′-UGGUUUACAUGUUUUCCUA-3′ | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takigawa, S.; Chen, A.; Nishimura, A.; Liu, S.; Li, B.-Y.; Sudo, A.; Yokota, H.; Hamamura, K. Guanabenz Downregulates Inflammatory Responses via eIF2α Dependent and Independent Signaling. Int. J. Mol. Sci. 2016, 17, 674. https://doi.org/10.3390/ijms17050674
Takigawa S, Chen A, Nishimura A, Liu S, Li B-Y, Sudo A, Yokota H, Hamamura K. Guanabenz Downregulates Inflammatory Responses via eIF2α Dependent and Independent Signaling. International Journal of Molecular Sciences. 2016; 17(5):674. https://doi.org/10.3390/ijms17050674
Chicago/Turabian StyleTakigawa, Shinya, Andy Chen, Akinobu Nishimura, Shengzhi Liu, Bai-Yan Li, Akihiro Sudo, Hiroki Yokota, and Kazunori Hamamura. 2016. "Guanabenz Downregulates Inflammatory Responses via eIF2α Dependent and Independent Signaling" International Journal of Molecular Sciences 17, no. 5: 674. https://doi.org/10.3390/ijms17050674
APA StyleTakigawa, S., Chen, A., Nishimura, A., Liu, S., Li, B.-Y., Sudo, A., Yokota, H., & Hamamura, K. (2016). Guanabenz Downregulates Inflammatory Responses via eIF2α Dependent and Independent Signaling. International Journal of Molecular Sciences, 17(5), 674. https://doi.org/10.3390/ijms17050674






