Identification of 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside as a Glycine N-Methyltransferase Enhancer by High-Throughput Screening of Natural Products Inhibits Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results
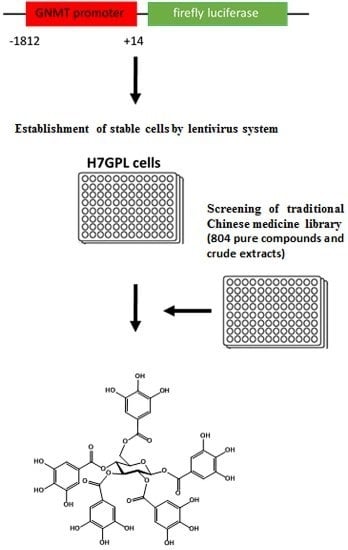
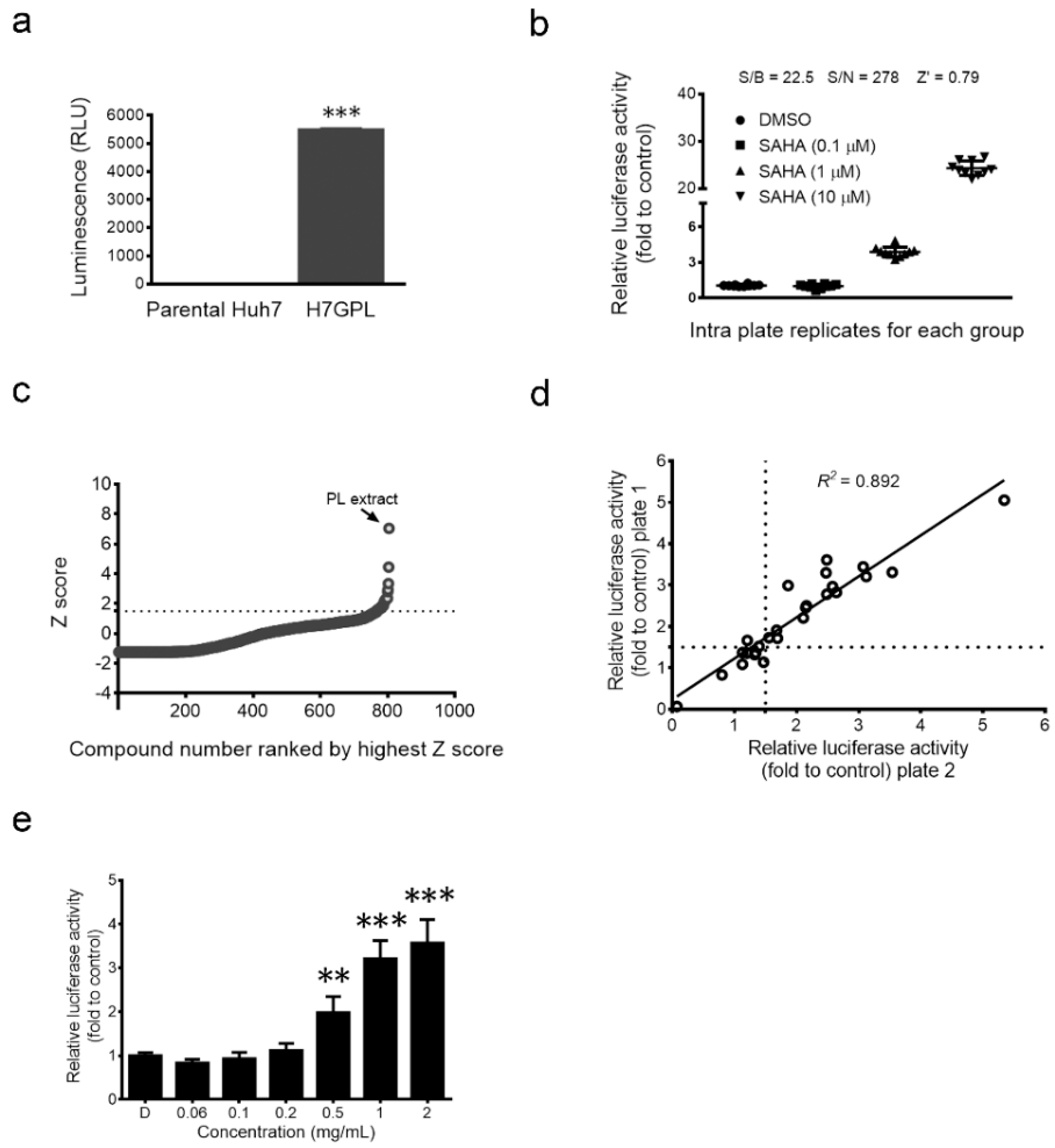
2.1. Establishment of Drug Screening Platform and Identification of Paeonia lactiflora Pall Extract as a Glycine N-Methyltransferase (GNMT) Inducer
2.2. The Anti-Hepatocellular Carcinomas (HCC) Effect of Paeonia lactiflora (PL) Extract-Derived Bioactive Fraction
2.3. 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside (PGG), the Active Component in F3-6, Inhibits Liver Cancer Cells’ Growth both in Vitro and in Vivo
2.4. PGG Treatment Induces Apoptosis in Huh7 Cells
2.5. PGG Sensitizes Huh7 Cells to Sorafenib Treatment
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Plasmids and Transfections
4.3. Development of GNMT Expression-Oriented Drug Screening Platform
4.4. Quantitative Real-Time PCR (qRT-PCR)
4.5. Immunoblotting
4.6. Cell Viability and Colony Formation Assay
4.7. Flow Cytometry
4.8. Caspase 3/7 Activity Assay
4.9. In Vivo Tumor Models
4.10. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GNMT | glycine N-methyltransferase |
| HCC | hepatocellular carcinomas |
| PL | Paeonia lactiflora Pall |
| PGG | 1,2,3,4,6-penta-O-galloyl-β-d-glucopyranoside |
| NFLD | nonalcoholic fatty liver |
| qPCR | quantitative real-time PCR |
| SAHA | suberoylanilide hydroxamic acid |
| TSA | trichostatin A |
| S/B | signal-to-background |
| S/N | signal-to-noise |
| mpk | mg per kg of body weight |
References
- Forner, A.; Llovet, J.M.; Bruix, J. Hepatocellular carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: Globocan 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Cervello, M.; McCubrey, J.A.; Cusimano, A.; Lampiasi, N.; Azzolina, A.; Montalto, G. Targeted therapy for hepatocellular carcinoma: Novel agents on the horizon. Oncotarget 2012, 3, 236–260. [Google Scholar] [CrossRef] [PubMed]
- Berasain, C. Hepatocellular carcinoma and sorafenib: Too many resistance mechanisms? Gut 2013, 62, 1674–1675. [Google Scholar] [CrossRef] [PubMed]
- Kalyan, A.; Nimeiri, H.; Kulik, L. Systemic therapy of hepatocellular carcinoma: Current and promising. Clin. Liver Dis. 2015, 19, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Scudellari, M. Drug development: Try and try again. Nature 2014, 516, S4–S6. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.J.; Liu, S.P.; Lee, C.M.; Yen, C.H.; Chuang, P.C.; Chen, C.Y.; Tsai, T.F.; Huang, S.F.; Lee, Y.H.; Chen, Y.M. Characterization of a glycine N-methyltransferase gene knockout mouse model for hepatocellular carcinoma: Implications of the gender disparity in liver cancer susceptibility. Int. J. Cancer 2009, 124, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.H.; Lin, Y.T.; Chen, H.L.; Chen, S.Y.; Chen, Y.M. The multi-functional roles of GNMT in toxicology and cancer. Toxicol. Appl. Pharmacol. 2013, 266, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.H.; Hung, J.H.; Ueng, Y.F.; Liu, S.P.; Chen, S.Y.; Liu, H.H.; Chou, T.Y.; Tsai, T.F.; Darbha, R.; Hsieh, L.L.; et al. Glycine N-methyltransferase affects the metabolism of aflatoxin B1 and blocks its carcinogenic effect. Toxicol. Appl. Pharmacol. 2009, 235, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Lin, J.R.; Darbha, R.; Lin, P.; Liu, T.Y.; Chen, Y.M. Glycine N-methyltransferase tumor susceptibility gene in the benzo(a)pyrene-detoxification pathway. Cancer Res. 2004, 64, 3617–3623. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Shiu, J.Y.; Tzeng, S.J.; Shih, L.S.; Chen, Y.J.; Lui, W.Y.; Chen, P.H. Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma. Int. J. Cancer 1998, 75, 787–793. [Google Scholar] [CrossRef]
- Avila, M.A.; Berasain, C.; Torres, L.; Martin-Duce, A.; Corrales, F.J.; Yang, H.; Prieto, J.; Lu, S.C.; Caballeria, J.; Rodes, J.; et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J. Hepatol. 2000, 33, 907–914. [Google Scholar] [CrossRef]
- Liao, Y.J.; Chen, T.L.; Lee, T.S.; Wang, H.A.; Wang, C.K.; Liao, L.Y.; Liu, R.S.; Huang, S.F.; Chen, Y.M. Glycine N-methyltransferase deficiency affects Niemann-Pick type C2 protein stability and regulates hepatic cholesterol homeostasis. Mol. Med. 2012, 18, 412–422. [Google Scholar] [PubMed]
- Martinez-Chantar, M.L.; Vazquez-Chantada, M.; Ariz, U.; Martinez, N.; Varela, M.; Luka, Z.; Capdevila, A.; Rodriguez, J.; Aransay, A.M.; Matthiesen, R.; et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology 2008, 47, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.H.; Lu, Y.C.; Li, C.H.; Lee, C.M.; Chen, C.Y.; Cheng, M.Y.; Huang, S.F.; Chen, K.F.; Cheng, A.L.; Liao, L.Y.; et al. Functional characterization of glycine N-methyltransferase and its interactive protein DEPDC6/DEPTOR in hepatocellular carcinoma. Mol. Med. 2012, 18, 286–296. [Google Scholar] [CrossRef] [PubMed]
- DebRoy, S.; Kramarenko, I.I.; Ghose, S.; Oleinik, N.V.; Krupenko, S.A.; Krupenko, N.I. A novel tumor suppressor function of glycine N-methyltransferase is independent of its catalytic activity but requires nuclear localization. PLoS ONE 2013, 8, e70062. [Google Scholar] [CrossRef] [PubMed]
- Kingston, D.G. Modern natural products drug discovery and its relevance to biodiversity conservation. J. Nat. Prod. 2011, 74, 496–511. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Graziose, R.; Lila, M.A.; Raskin, I. Merging traditional chinese medicine with modern drug discovery technologies to find novel drugs and functional foods. Curr. Drug Discov. Technol. 2010, 7, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Yang, W.L.; Lin, K.T.; Liu, C.H.; Liu, Y.W.; Huang, K.W.; Chang, P.M.; Lai, J.M.; Hsu, C.N.; Chao, K.M.; et al. Gene expression-based chemical genomics identifies potential therapeutic drugs in hepatocellular carcinoma. PLoS ONE 2011, 6, e27186. [Google Scholar] [CrossRef] [PubMed]
- Luka, Z.; Mudd, S.H.; Wagner, C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J. Biol. Chem. 2009, 284, 22507–22511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, X.; Wang, J.; Wen, R.; Jia, L.; Zhu, Y.; Li, R.; Wang, R.; Li, J.; Wang, L.; et al. Large dose means significant effect—Dose and effect relationship of Chi-Dan-Tui-Huang decoction on α-naphthylisothiocyanate-induced cholestatic hepatitis in rats. BMC Complement. Altern. Med. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Leung, P.S. Use of herbal medicines and natural products: An alternative approach to overcoming the apoptotic resistance of pancreatic cancer. Int. J. Biochem. Cell Biol. 2014, 53, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, L.; Kim, S.H.; Hagerman, A.E.; Lu, J. Anti-cancer, anti-diabetic and other pharmacologic and biological activities of penta-galloyl-glucose. Pharm. Res. 2009, 26, 2066–2080. [Google Scholar] [CrossRef] [PubMed]
- Oh, G.S.; Pae, H.O.; Oh, H.; Hong, S.G.; Kim, I.K.; Chai, K.Y.; Yun, Y.G.; Kwon, T.O.; Chung, H.T. In vitro anti-proliferative effect of 1,2,3,4,6-penta-O-galloyl-β-d-glucose on human hepatocellular carcinoma cell line, SK-HEP-1 cells. Cancer Lett. 2001, 174, 17–24. [Google Scholar] [CrossRef]
- Yin, S.; Dong, Y.; Li, J.; Lu, J.; Hu, H. Penta-1,2,3,4,6-O-galloyl-β-d-glucose induces senescence-like terminal S-phase arrest in human hepatoma and breast cancer cells. Mol. Carcinog. 2011, 50, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yin, S.; Jiang, C.; Luo, X.; Guo, X.; Zhao, C.; Fan, L.; Meng, Y.; Lu, J.; Song, X.; et al. Involvement of autophagy induction in penta-1,2,3,4,6-O-galloyl-β-d-glucose-induced senescence-like growth arrest in human cancer cells. Autophagy 2014, 10, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Huidobro, C.; Torano, E.G.; Fernandez, A.F.; Urdinguio, R.G.; Rodriguez, R.M.; Ferrero, C.; Martinez-Camblor, P.; Boix, L.; Bruix, J.; Garcia-Rodriguez, J.L.; et al. A DNA methylation signature associated with the epigenetic repression of glycine N-methyltransferase in human hepatocellular carcinoma. J. Mol. Med. 2013, 91, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACS) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Giannini, G.; Cabri, W.; Fattorusso, C.; Rodriquez, M. Histone deacetylase inhibitors in the treatment of cancer: Overview and perspectives. Future Med. Chem. 2012, 4, 1439–1460. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.M.; Shih, Y.P.; Wu, C.H.; Chen, Y.M. Characterization of the 5′ regulatory region of the human glycine N-methyltransferase gene. Gene 2009, 443, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.G.; Li, W.H.; He, W.G.; Jiang, N.; Zhang, G.X.; Chen, W.; Yang, H.F.; Liu, Q.L.; Huang, Y.N.; Zhang, L.; et al. MiR-184 post-transcriptionally regulates SOX7 expression and promotes cell proliferation in human hepatocellular carcinoma. PLoS ONE 2014, 9, e88796. [Google Scholar] [CrossRef] [PubMed]
- Malo, N.; Hanley, J.A.; Cerquozzi, S.; Pelletier, J.; Nadon, R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 2006, 24, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, M.; Yamagishi, T.; Nonaka, G.; Nishioka, I.; Nagasawa, T.; Oura, H. Tannins and related compounds. XII. Isolation and characterization of galloylglucoses from paeoniae radix and their effects on urea-nitrogen concentration in rat serum. Chem. Pharm. Bull. 1983, 31, 2593–2600. [Google Scholar] [CrossRef] [PubMed]
- Kiss, A.K.; Derwinska, M.; Dawidowska, A.; Naruszewicz, M. Novel biological properties of oenothera paradoxa defatted seed extracts: Effects on metallopeptidase activity. J. Agric. Food Chem. 2008, 56, 7845–7852. [Google Scholar] [CrossRef] [PubMed]
- Maron, D.M.; Ames, B.N. Revised methods for the salmonella mutagenicity test. Mutat. Res./Environ. Mutagen. Relat. Subj. 1983, 113, 173–215. [Google Scholar] [CrossRef]
- Test No. 471: Bacterial reverse mutation test. In OECD Guidelines for the Testing of Chemicals, Section 4: Health Effects; OECD Publishing: Paris, France, 2014.




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kant, R.; Yen, C.-H.; Lu, C.-K.; Lin, Y.-C.; Li, J.-H.; Chen, Y.-M.A. Identification of 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside as a Glycine N-Methyltransferase Enhancer by High-Throughput Screening of Natural Products Inhibits Hepatocellular Carcinoma. Int. J. Mol. Sci. 2016, 17, 669. https://doi.org/10.3390/ijms17050669
Kant R, Yen C-H, Lu C-K, Lin Y-C, Li J-H, Chen Y-MA. Identification of 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside as a Glycine N-Methyltransferase Enhancer by High-Throughput Screening of Natural Products Inhibits Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2016; 17(5):669. https://doi.org/10.3390/ijms17050669
Chicago/Turabian StyleKant, Rajni, Chia-Hung Yen, Chung-Kuang Lu, Ying-Chi Lin, Jih-Heng Li, and Yi-Ming Arthur Chen. 2016. "Identification of 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside as a Glycine N-Methyltransferase Enhancer by High-Throughput Screening of Natural Products Inhibits Hepatocellular Carcinoma" International Journal of Molecular Sciences 17, no. 5: 669. https://doi.org/10.3390/ijms17050669
APA StyleKant, R., Yen, C.-H., Lu, C.-K., Lin, Y.-C., Li, J.-H., & Chen, Y.-M. A. (2016). Identification of 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside as a Glycine N-Methyltransferase Enhancer by High-Throughput Screening of Natural Products Inhibits Hepatocellular Carcinoma. International Journal of Molecular Sciences, 17(5), 669. https://doi.org/10.3390/ijms17050669